Abstract
Pateamine A inhibits translation by preventing proper translational initiation complex formation. In the December issue of Chemistry & Biology, Bordeleau et al. demonstrated that the effects of Pate-mine A on translation are mediated through the interaction between the RNA helicase eIF4A and mRNA [1].
Many aspects of malignantly transformed phenotypes are due to the dys-regulation of translation. The switch from normal growth to either malignant transformation or apoptosis is determined by the spectrum of mRNAs that are translated. Many growth-promoting and apoptotic mRNAs are poorly translated under normal physiological conditions, due to factors such as a highly structured 5′ -untranslated region (UTR) or an internal ribosomal entry site (IRES). Translation of most cellular mRNA, and especially those which contain a highly structured 5′ -UTR or an IRES, depends on the integrity and activity of the eIF4F complex, consisting of eIF4A, eIF4E, and eIF4G. eIF4G functions as the pivotal factor, acting through direct interactions with mRNA, eIF4E, eIF4A and another initiation factor, eIF3, and providing a link between the mRNA and the 40S ribosomal subunit. The recruitment of mRNA to the 40S ribosomal subunit to form the 48S initiation complex is catalyzed by the eIF4 group of factors: eIF4B, eIF4F, and eIF4H. eIF4F recognizes the cap structure at the 5′ -end of mRNA through eIF4E, unwinds the secondary structure of the 5′ -UTR region through the helicase activity of eIF4A, and binds the 43S complex through interactions between eIF4G and eIF3 (Figure 1A). eIF4H and eIF4B increase the processivity of eIF4A (reviewed in [2]).
Figure 1. Proposed Models of Translational Dysregulation by Pateamine A.
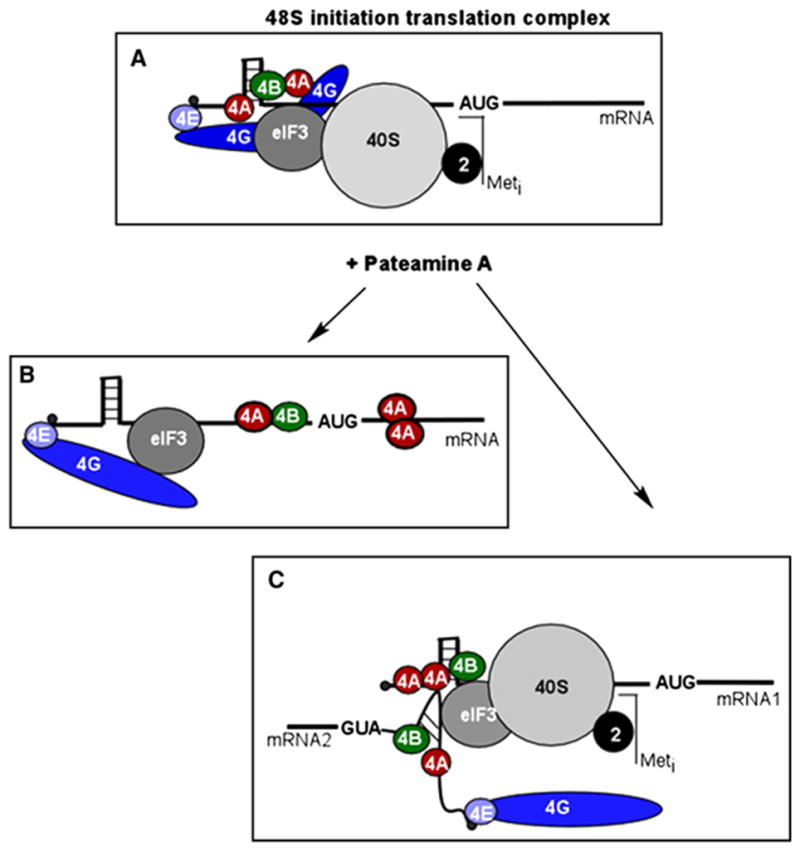
(A) 48S complex formation under normal conditions.
(B) Pateamine A sequesters eIF4A from eIF4F and stabilizes eIF4A on mRNA preventing 48S complex formation.
(C) Pateamine A induces aberrant, eIF4F-independent 48S initiation complexes that load at random locations on mRNA and prevent translation.
Pateamine A (PatA) is an immunosuppressive and antiproliferative agent that was isolated from the marine sponge Mycale sp. PatA was found to induce apoptosis in certain mammalian cell lines, especially those that were transformed by either ras or bcr-abl oncogenes [3]. PatA affects both transcriptional and translational processes with the greater inhibitory effect being on translation [4, 5]. At concentrations less than 0.1 μ M, PatA inhibits proliferation of cancer cells, suppresses cap-dependent and eIF4G-dependent IRES-driven translation, disrupts polysomes, and induces stress granule formation [4, 6, 7]. The ability of PatA to induce apoptosis in transformed cells but not in normal cells suggests that is has therapeutic potential in cancer treatment.
MALDI-TOF mass spectrometry and immunological analysis have identified eIF4A as the biological target of PatA [4, 8]. eIF4A is a bidirectional RNA helicase. It binds to and unwinds RNA in an ATP-dependent manner and hydrolyzes ATP in an RNA-dependent manner. It is the prototype for the DEAD/H-box protein family and contains at least nine motifs conserved in nucleic acid helicases. In contrast to other helicases, eIF4A does not have extra domains that regulate substrate specificity or stimulate strand separation. eIF4A alone is a weak ATPase and helicase, but these activities are stimulated by eIF4G, eIF4B, and eIF4H, suggesting that they supply the missing functions (reviewed in [9]). There are three eIF4A family members in mammals. eIF4AI and eIF4AII are both involved in RNA unwinding during the initiation of translation as a part of the eIF4F complex. eIF4AIII is a nucleocytoplasmic shuttling protein associated with the exon junction complex and is essential for nonsense-mediated mRNA decay [10]. Pelletier and colleagues have previously shown that all three eIF4A family members are captured from HL-60 cell extracts by a PatA-affinity resin and that PatA affects translation by specifically targeting free eIF4A [8]. During mRNA unwinding, eIF4A cycles between the eIF4F complex and the free eIF4A pool [11, 12], a process that is critical for the unwinding of mRNA. Interestingly, PatA stimulates the intrinsic ATPase and helicase activities of eIF4A rather than inhibiting them, by stabilizing the eIF4A:mRNA complex [8].
A new observation made by Pelletier and co-workers [1] is that eIF4A-mRNA stabilization allows attachment of additional factors to mRNA. Western blot analysis of eIF4A distribution throughout a sucrose gradient revealed the sedimentation of eIF4A in complexes larger than 48S in the presence of PatA [4]. The observation that such eIF4A sedimentation was sensitive to nuclease treatment [1] further supports the idea that, in the presence of PatA, eIF4A is part of complexes containing mRNA as well as other mRNA binding proteins. The authors suggest that the sequestration of eIF4A in new mRNA-protein complexes limits the availability of eIF4A for incorporation into eIF4F complex (Figure 1B).
One protein that interacts with the eIF4A:mRNA complex through the mRNA component is eIF4B [1]. eIF4B has been shown to interact with two different mRNA molecules simultaneously and to anneal complementary RNA strands. It may also facilitate binding of the 40S ribosomal subunit to mRNA [13]. The central part of mammalian eIF4B contains a DRYG motif, which facilitates its homodimerization and binding to eIF3. It has been suggested that in addition to binding mRNA and stimulating the helicase activity of eIF4A, eIF4B serves as a bridge between eIF3 and the 40S subunit [13]. Since eIF4A is one of the most abundant initiation factors in the cell (3 to 50 μ M, depending on the cell type) [14, 15], it is likely that the 10–20 nM PatA that is capable of disrupting polysomes in vivo does so not only by limiting the pool of free eIF4A that is recycled through eIF4F, but also by producing aberrant eIF4F-independent 48S initiation complexes that load at random locations on mRNA and prevent its translation (Figure 1C).
A previous study showed that high concentrations of PatA (10 μ M and higher) preventscopurification of eIF4A and eIF4G from cell lysates [4]. The recent data obtained by Bordeleau et al. [1] with pull-down and FRET assays showed that 10 μ M PatA did not affect the direct binding of eIF4A to eIF4G variants containing only one of the two eIF4A binding sites. By contrast, the same concentration of PatA prevented incorporation of recombinant eIF4A into eIF4F bound to m7GTP-Sepharose. Since the latter experiment used the ribosomal high-salt wash as a source of eIF4F, which may contain additional RNA binding proteins and mRNA, it is possible that PatA stabilization of either the eIF4A:mRNA or eIF4A:mRNA-protein complexes prevented the incorporation of eIF4A into eIF4F.
There is a discrepancy in the effect that high concentration of PatA has on the 48S complex stability. Low et al. [4] found that 100 μ M PatA stabilized or even increased the level of radiolabeled mRNA incorporated into the 48S complex and interpreted this as being the result of stalled 48S ribosome initiation complexes. In contrast, Bordeleau et al. [8] and [1] observed the reduction of 48S complexes upon treatment with 10 μ M PatA, which they interpret as being due to the sequestration of eIF4A from eIF4F. They suggest that the observed discrepancy is due to differences in the source of PatA.
Despite this discrepancy, the observation that the effects of PatA are mediated by the interaction between eIF4A and mRNA is important. A detailed understanding of the mechanism by which PatA inhibits translation should facilitate the development of chemotheraputic agents based on PatA. In particular, the observation that PatA acts through eIF4A suggests that it may be particularly useful in treating cancers in which eIF4A is either over-expressed or hyperactivated [16].
References
- 1.Bordeleau M, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J. Chem Biol. 2006;13:1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Hershey JWB, Merrick WC. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 3.Hood K, West L, Northcote P, Berridge M, Miller J. Apoptosis. 2001;6:207–219. doi: 10.1023/a:1011340827558. [DOI] [PubMed] [Google Scholar]
- 4.Low W, Dang Y, Schneider-Poetsch T, Shi Z, Choi N, Merrick W, Romo D, Liu J. Mol Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Romo D, Choi N, Li S, Buchler I, Shi Z, Liu J. J Am Chem Soc. 2004;126:10582–10588. doi: 10.1021/ja040065s. [DOI] [PubMed] [Google Scholar]
- 6.Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 7.Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. Proc Natl Acad Sci USA. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers GW, Jr, Komar AA, Merrick WC. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 10.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 11.Yoderhill J, Pause A, Sonenberg N, Merrick WC. J Biol Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]
- 12.Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Methot N, Pickett G, Keene JD, Sonenberg N. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 14.Rau M, Ohlmann T, Morley SJ, Pain VM. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 15.von der Haar T, McCarthy J. Mol Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 16.Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. Mol Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


