Abstract
Neonatal monocytes display immaturity of numerous functions compared with adult cells. Gene expression arrays provide a promising tool for elucidating mechanisms underlying neonatal immune function. We used a well-established microarray to analyze differences between LPS-stimulated human cord blood and adult monocytes to create dynamic models for interactions to elucidate observed deficiencies in neonatal immune responses.
We identified 168 genes that were differentially expressed between adult and cord monocytes after 45 min incubation with LPS. Of these genes, 95% (159 of 167) were over-expressed in adult relative to cord monocytes. Differentially expressed genes could be sorted into nine groups according to their kinetics of activation. Functional modelling suggested differences between adult and cord blood in the regulation of apoptosis, a finding confirmed using annexin binding assays. We conclude that kinetic studies of gene expression reveal potentially important differences in gene expression dynamics that may provide insight into neonatal innate immunity.
Background
The defects in neonatal adaptive immunity are relatively easy to understand a priori. Although there are complexities to be considered [1,2], experimental evidence demonstrates that newborns, lacking prior antigen exposure, must develop immunologic memory based on postnatal experience with phogens and environmental immunogens [3-5].
It is less clear why there should be defects in newborns' innate immunity, although these defects are well documented. For example, newborns have long been known to exhibit defects in phagocytosis [6], chemotaxis [7,8], and adherence [9], the latter possibly due to aberrant regulation of critical cell-surface proteins that mediate leukocyte-endothelial interactions [10]. Newborn monocytes also exhibit diminished secretion of numerous cytokines under both stimulated and basal conditions [11-13].
Elucidating the causes of these defects is a crucial question in neonatal medicine, since infection remains a major cause of morbidity and mortality in the newborn period. However, unravelling the complex events in monocyte and/or neutrophil activation, from ligand binding to activation of effector responses, is clearly a daunting challenge. Any one of numerous pathways from the earliest cell signalling events to protein synthesis or secretion could be relevant, and focusing on any one may overlook critical aspects of cellular regulation. In this context, genomic and/or proteomic approaches may offer some important advantages, at least in the initial phases of investigation, by allowing investigators to survey the panoply of biological processes that may be relevant to identifying critical biological distinctions.
Recently published work has documented differences in gene expression between adult and cord blood monocytes [14], although these studies did not elucidate the fundamental, functional differences between cord blood and adult cells. The studies we report here demonstrate how computational analyses, applied to microarray data, can elucidate critical biological functions when analysis extends beyond the identification of differentially-expressed genes.
Methods
Cells and cellular stimulation
Monocytes were purified from cord blood of healthy, term infants and from the peripheral blood of healthy adults by positive selection using anti-CD-14 mAb-coated magnetic beads (Miltenyi Biotec, Auburn, CA, USA) according to the manufacturer's instructions. Informed consent was obtained from adult volunteers; collection of cord blood was ruled exempt from consent after review by the Oklahoma Health Sciences Center IRB. In brief, blood was collected into sterile tubes containing sodium citrate as an anticoagulant (Becton Dickinson, Franklin Lakes, NJ). Peripheral blood mononuclear cells (PBMC) were prepared from the anti-coagulated blood using gradient separation on Histopaque-1077 performed directly in the blood collection tubes. Cells were washed three times in Ca2+ and Mg2+-free Hanks's balanced salt solution. PBMC were incubated for 20 min at 4°C with CD14 microbeads at 20 μl/1 × 107 cells. The cells were washed once, re-suspended in 500 μl Ca2+ and Mg2+-free PBS containing 5% FBS/1 × 108 cells. The suspension was then applied to a MACs column. After unlabeled cells passed through, the column was washed with 3 × 500 μl Ca2+ and Mg2+-free PBS. The column was removed from the separator and was put on a new collection tube. One ml of Ca2+ and Mg2+-free PBS was then added onto the column, which was immediately flushed by firmly applying the plunger supplied with the column.
Purified monocytes were incubated with LPS from Escherichia coli 0111:4B (Sigma, St. Louis, MO) at 10 ng/ml for 45 min and 2-hours in RPMI 1640 with 10% fetal bovine serum or studied in the absence of stimulation ("zero time"). It should be noted that this product is not "pure," and stimulates both TLR-4 and TRL-2 signaling pathways [15]. A smaller number of replicates (n = 5) was analyzed after 24 hr incubation. After the relevant time points, monocytes were lysed with TriZol (Invitrogen, Carlsbad, CA, USA) and RNA was isolated as recommended by the manufacturer. Cells from eight different term neonates and eight different healthy adults were used for these studies.
Gene microarrays
The microarrays used in these experiments were developed at the Oklahoma Medical Research Foundation Microarray Research Facility and contained probes for 21,329 human genes. Slides were produced using commercially available libraries of 70 nucleotide long DNA molecules whose length and sequence specificity were optimized to reduce the cross-hybridization problems encountered with cDNA-based microarrays (Qiagen-Operon). The oligonucleotides were derived from the UniGene and RefSeq databases. The RefSeq database is an effort by the NCBI to create a true reference database of genomic information for all genes of known function. All 11,000 human genes of known or suspected function were represented on these arrays. In addition, most undefined open reading frames were represented (approximately 10,000 additional genes).
Oligonucleotides were spotted onto Corning® UltraGAPS™ amino-silane coated slides, rehydrated with water vapor, snap dried at 90°C, and then covalently fixed to the surface of the glass using 300 mJ, 254 nm wavelength ultraviolet radiation. Unbound free amines on the glass surface were blocked for 15 min with moderate agitation in a 143 mM solution of succinic anhydride dissolved in 1-methyl-2-pyrolidinone, 20 mM sodium borate, pH 8.0. Slides were rinsed for 2 min in distilled water, immersed for 1 min in 95% ethanol, and dried with a stream of nitrogen gas.
Labeling, hybridization, and scanning
Fluorescently labeled cDNA was separately synthesized from 2.0 μg of total RNA using an oligo dT12–18 primer, PowerScript reverse transcriptase (Clontech, Palo Alto, CA), and Cy3-dUTP (Amersham Biosciences, Piscataway, NJ) for 1 hour at 42°C in a volume of 40 μl. Reactions were quenched with 0.5 M EDTA and the RNA was hydrolyzed by addition of 1 M NaOH for 1 hr at 65°C. The reaction was neutralized with 1 M Tris, pH 8.0, and cDNA was then purified with the Montage PCR96 Cleanup Kit (Millipore, Billerica, MA). cDNA was added to ChipHybe™ hybridization buffer (Ventana Medical Systems, Tucson, AZ) containing Cot-1 DNA (0.5 mg/ml final concentration), yeast tRNA (0.2 mg/ml), and poly(dA)40–60 (0.4 mg/ml). Hybridization was performed on a Ventana Discovery system for 6 hr at 42°C. Microarrays were washed to a final stringency of 0.1× SSC, and then scanned using a dual-color laser (Agilent Biotechnologies, Palo Alto, CA). Fluorescent intensity was measured by Imagene™ software (BioDiscovery, El Segundo, CA).
PCR validation of array data
Reverse transcription
Three cord blood samples (C1, C2, and C5) and three adult samples (A1, A5, and A6) from the 45 minute time point were used for PCR validation. First strand cDNA was generated from 3.6 μg of total RNA per sample using the OmniScript Reverse Transcriptase and buffer (Qiagen, Valencia, CA), 1 μl of 100 μM oligo dT primer (dT15) in a 40 μl volume. Reactions were incubated 60 min at 37° and inactivated at 93° for 5 min. cDNA was diluted 1:100 in water and stored at -20°C.
Quantitative PCR
Gene-specific primers for 10 genes (Erbb3, Tmod, Dscr1l1, Sp1, Scya4, Gro2, Cri1, Scya3, Scya3l1, and Il-1a) were designed with a 60°C melting temperature and a length of 19–25 bp for PCR products with a length of 90–140 bp, using Applied Biosystems Inc (ABI, Foster City, CA) Primer Express 1.5 software. PCR was run with 2 μl cDNA template in 15 μl reactions in triplicate on an ABI SDS 7700 using the ABI SYBR Green I Master Mix and gene specific primers at a concentration of 1 μM each. The temperature profile consisted of an initial 95°C step for 10 minutes (for Taq activation), followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min, and then a final melting curve analysis with a ramp from 60°C to 95°C over 20 min. Gene-specific amplification was confirmed by a single peak in the ABI Dissociation Curve software. No template controls were run for each primer pair. Since equal amounts of total RNA were used for cDNA synthesis, Ct values should reflect relative abundance [16]. These values were used to calculate the average group Ct (Cord vs. Adult) and the relative ΔCt was used to calculate fold change between the two groups [17].
Apoptosis assays
Exposed membrane phospholipids (a marker for early apoptosis) were detected in adult and neonatal monocytes after LPS stimulation using a commercially available annexin V binding assay. Monocytes from cord blood and adult peripheral blood were obtained as outlined above. Isolated monocytes were either labeled immediately with annexin V-FITC or were stimulated for 14 hours with LPS 10 ng/ml prior to labeling (this time point was derived empirically to maximize apoptosis). Annexin V-FITC staining was completed via the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences, San Jose, CA) using 5 μl of propidium iodine and 5 μl annexin V-FITC as recommended by the manufacturer. Analysis by flow cytometry was accomplished on a FACS Calibur automated benchtop flow cytometer. Data obtained by flow cytometry was analyzed by non-parametric t-test (Mann-Whitney test). An alpha level of 0.05 was considered statistically significant.
Statistical analysis
Microarrays were normalized and tested for differential expression using methods described previously [18]. Differential expression was concluded if the genes met the following criteria: a minimum expression level at least 10 times above background at one or more time points, a minimum 1.5-fold difference in the mean expression values between groups at one or more time points, and a minimum of 80% reproducibility using the jack-knife method. A jack-knife is the most common type of Leave-one-out-cross-validation (LOOCV); it is used here to cross-validate genes selected by differential analysis [19]. Time series analysis was performed using the hypervariable (HV) gene method previously described by our group [20].
After selection, HV genes are clustered and interrogated for gene-gene interactions. K-means clustering, an unsupervised technique, was performed on the HV genes to create unbiased clusters. Discriminate function analysis (DFA), a supervised technique, was used to determine and spatially map gene-to-gene interactions [21].
All statistical analysis was performed in Matlab R14 (Natick, MA) and Statistica v7 (Tulsa, OK, USA). An alpha level of 0.05 was considered statistically significant for all analyses.
Analysis of the apoptosis assays was undertaken using both parametric and non-parametric analysis methods. Parametric analysis was undertaken using the student's t-test; non-parametic analysis used the Mann-Whitney U-test. A p-value of > 0.05 was the threshold for rejecting the null hypothesis.
Discriminant function analysis
DFA is a method that identifies a subset of genes whose expression values can be linearly combined in an equation, denoted a root, whose overall value is distinct for a given characterized group. DFA therefore, allows the genes that maximally discriminate among the distinct groups analyzed to be identified. In the present work, a variant of the classical DFA, named the Forward Stepwise Analysis, was used to select the set of genes whose expression maximally discriminated among experimentally distinct groups. The Forward Stepwise Analysis was built systematically in an iterative manner. Specifically, at each step all variables were reviewed to identify the one that most contributes to the discrimination between groups. This variable was included in the model, and the process proceeded to the next iteration. The statistical significance of discriminative power of each gene was also characterized by partial Wilk's Lambda coefficients, which are equivalent to the partial correlation coefficient generated by multiple regression analyses. The Wilk's Lambda coefficient used a ratio of within-group differences and the sum of within-plus between-group differences. Its value ranged from 1.0 (no discriminatory power) to 0.0 (perfect discriminatory power).
Computer analysis of functional associations between differentially expressed genes
In addition to the above analyses, genes showing the most significant differences between neonatal and adult cells were characterized functionally using pre-existing databases such as PubMed, BIND, KEGG, and Ontoexpress. Biological associations of the differentially expressed genes were modelled using Ingenuity Pathways Analysis (Redwood City, CA). Data analyzed through this technique can then be resolved into cogent models of the specific biological pathways activated under the experimental conditions used in the microarray analyses.
Results
Differential gene expression analysis
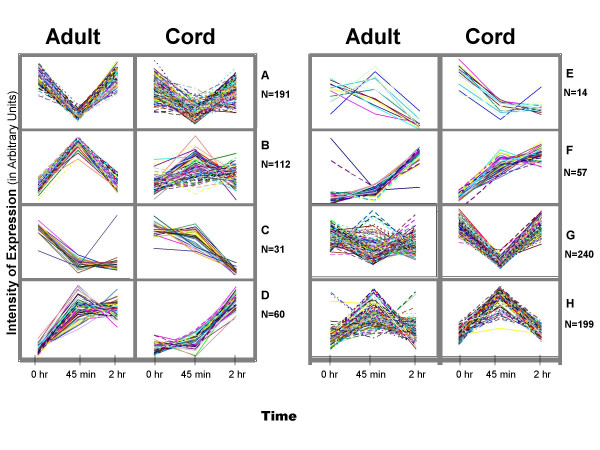
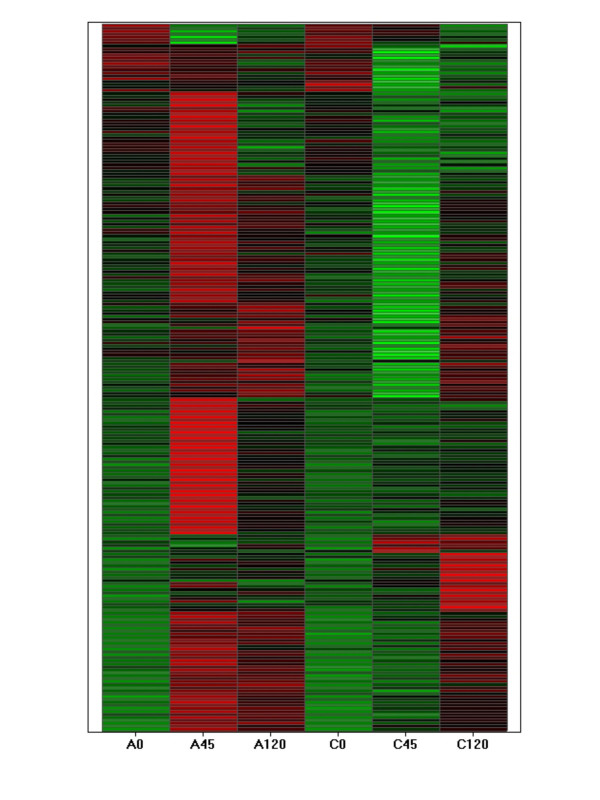
Table 1 lists genes determined to be differentially expressed between cord and adult peripheral blood monocytes, as described above. No genes were found to be statistically significantly differentially expressed between adult and cord monocytes in the absence of LPS exposure. 168 genes were differentially expressed between adult and cord monocytes after 45 min incubation with LPS. 95% of these genes (159 of 168) were over-expressed in adult relative to cord monocytes. After 120 minutes of LPS exposure, 24 genes were differentially expressed between adult and cord monocytes. Of the latter genes, 23 were more highly expressed in cord than adult monocytes. This pattern of differentially expressed genes suggested an initial delayed response to LPS followed by an enhanced transcription of genes in cord relative to adult monocytes. To test this hypothesis, k-means clustering was used to categorize differentially expressed genes based on their temporal profiles. Relative decreases in gene transcription by cord monocytes at 45 min were seen in 6 of the 9 clusters (Figure 1). Each of these clusters contained between 15 and 46 genes. Examination of the clusters showed that differences between groups after 45 minutes of LPS exposure were attributable to a) genes in certain clusters that were up-regulated in adult monocytes only, b) genes in other clusters that were down-regulated in cord monocytes only, or c) genes in yet other clusters that were up-regulated in adult and down-regulated in cord monocytes. These results, summarized in a heat map in Figure 2, indicated a high complexity of gene expression differences between adult monocytes and cord blood monocytes in response to LPS.
Table 1.
Differentially expressed genes between adult and cord monocytes at specific time points. T = time (min) at which the sample was taken. Numbers indicate corrected expression values.
| Adult | Adult | Adult | Cord | Cord | |||||
| Genbank # | Symbol | Gene Description | T = 0 | t = 45 | t = 120 | t = 0 | t = 45 | t = 120 | |
| Apoptosis | |||||||||
| NM_033423 | CTLA1 | Similar to granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | 317 | 419 | 299 | 199 | 193 | 264 | |
| AB037796 | PDCD6IP | Programmed cell death 6 interacting protein | 75 | 155 | 68 | 79 | 70 | 81 | |
| NM_024969 | TAIP-2 | TGFb-induced apoptosis protein 2 | 63 | 113 | 107 | 53 | 68 | 116 | |
| NM_003127 | SPTAN1 | Spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | 713 | 842 | 1171 | 724 | 824 | 2093 | |
| Protein synthesis, processing, degradation | |||||||||
| AK001313 | RPLP0 | Ribosomal protein, large, P0 | 704 | 1465 | 947 | 703 | 756 | 669 | |
| NM_006799 | PRSS21 | Protease, serine, 21 (testisin) | 204 | 789 | 457 | 169 | 360 | 400 | |
| NM_003774 | GALNT4 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) | 576 | 651 | 648 | 528 | 378 | 578 | |
| AK057790 | cDNA FLJ25061 fis, clone CBL04730 | 245 | 373 | 302 | 244 | 215 | 200 | ||
| NM_004223 | UBE2L6 | Ubiquitin-conjugating enzyme E2L 6 | 128 | 191 | 146 | 108 | 99 | 109 | |
| NM_014710 | GPRASP1 | KIAA0443 gene product | 122 | 182 | 106 | 113 | 119 | 95 | |
| NM_021090 | MTMR3 | Myotubularin related protein 3 | 109 | 171 | 137 | 108 | 87 | 138 | |
| AF339824 | HS6ST3 | Heparan sulfate 6-O-sulfotransferase 3 | 89 | 112 | 91 | 94 | 46 | 76 | |
| NM_012180 | FBXO8 | F-box only protein 8 | 40 | 67 | 42 | 45 | 33 | 43 | |
| U66589 | RPL5 | Ribosomal protein L5 | 34 | 48 | 37 | 30 | 26 | 36 | |
| NM_001870 | CPA3 | Carboxypeptidase A3 (mast cell) | 183 | 495 | 610 | 146 | 949 | 756 | |
| NM_006145 | DNAJB1 | DnaJ (Hsp40) homolog, subfmaily B, member 1 | 179 | 277 | 408 | 168 | 299 | 745 | |
| AK025547 | MRPL30 | Mitochondrial ribosomal protein L30 | 83 | 118 | 126 | 81 | 101 | 211 | |
| NM_000439 | PCSK1 | Proprotein convertase subtilisin/kexin type 1 | 39 | 55 | 53 | 40 | 78 | 88 | |
| Cell/Organism Movement | |||||||||
| NM_002067 | GNA11 | Guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | 555 | 870 | 607 | 540 | 468 | 664 | |
| NM_002465 | MYBPC1 | Myosin binding protein C, slow type | 81 | 140 | 154 | 88 | 80 | 161 | |
| NM_003275 | TMOD | Tropomodulin | 276 | 151 | 481 | 257 | 344 | 503 | |
| AK026164 | MYL6 | Myosin, light polypeptide 6, alkali, smooth muscle and non-muscle | 7 | 6 | 48 | 5 | 16 | 11 | |
| Small Molecule Interactions | |||||||||
| NM_006030 | CACNA2D2 | Calcium channel, voltage-dependent, alpha 2/delta subunit 2 | 670 | 1390 | 1021 | 641 | 639 | 946 | |
| AK025170 | SFXN5 | FLJ21517 fis, clone COL05829 | 431 | 537 | 437 | 405 | 295 | 374 | |
| NM_021097 | SLC8A1 | Solute carrier family 8 (sodium/calcium exchanger), member 1 | 396 | 456 | 458 | 412 | 276 | 369 | |
| Signal Transduction | |||||||||
| NM_032144 | RAB6C | RAB6C | 827 | 1658 | 1307 | 626 | 773 | 1251 | |
| NM_001982 | ERBB3 | V-erb-b2 erythroblastic leukemia viral oncogene homolog 3 | 603 | 1375 | 671 | 555 | 584 | 643 | |
| AK026479 | SNX14 | Sorting nexin 14 | 682 | 1207 | 879 | 624 | 567 | 883 | |
| NM_018979 | PRKWNK1 | Protein kinase, lysine deficient 1 | 451 | 813 | 782 | 516 | 480 | 792 | |
| NM_004811 | LPXN | Leupaxin | 329 | 539 | 445 | 323 | 298 | 503 | |
| BC005365 | clone IMAGE:3829438, mRNA, partial cds | 257 | 418 | 275 | 275 | 275 | 206 | ||
| NM_004723 | ARHGEF2 | Rho/rac guanine nucleotide exchange factor (GEF) 2 | 215 | 300 | 228 | 197 | 176 | 186 | |
| AF130093 | MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | 237 | 285 | 275 | 221 | 171 | 223 | |
| AK000383 | MKPX | Mitogen-activated protein kinase phosphatase x | 218 | 221 | 244 | 233 | 126 | 197 | |
| NM_022304 | HRH2 | Histamine receptor H2 | 45 | 121 | 86 | 42 | 74 | 79 | |
| NM_030753 | WNT3 | Wingless-type MMTV integration site family member 3 | 105 | 117 | 92 | 109 | 63 | 81 | |
| AB024574 | GTPBP2 | GTP binding protein 2 | 89 | 90 | 99 | 74 | 57 | 92 | |
| NM_002836 | PTPRA | Protein tyrosine phosphatase, receptor type, A | 8 | 6 | 80 | 6 | 16 | 28 | |
| NM_003656 | CAMK1 | Calcium/calmodulin-dependent protein kinase I | 4940 | 10131 | 4446 | 4785 | 4907 | 7190 | |
| Cellular Metabolism & Cell Division | |||||||||
| NM_006170 | NOL1 | Nucleolar protein 1 (120 kD) | 575 | 1815 | 1021 | 499 | 896 | 1093 | |
| AL133115 | COVA1 | Cytosolic ovarian carcinoma antigen 1 | 1381 | 1294 | 848 | 1309 | 658 | 808 | |
| D86962 | GRB10 | Growth factor receptor-bound protein 10 | 619 | 906 | 200 | 609 | 512 | 179 | |
| NM_005628 | SLC1A5 | Solute carrier family 1 (neutral amino acid transporter), member 5 | 338 | 801 | 600 | 311 | 397 | 524 | |
| D17525 | MASP1 | Mannan-binding lectin serine protease 1 (C4/C2 activating component of Ra-reactive factor) | 372 | 654 | 43 | 361 | 325 | 55 | |
| NM_016518 | PIPOX | Pipecolic acid oxidase | 240 | 545 | 330 | 221 | 293 | 286 | |
| NM_012157 | FBXL2 | F-box and leucine-rich repeat protein 2 | 274 | 501 | 374 | 249 | 277 | 298 | |
| NM_018446 | AD-017 | Glycosyltransferase AD-017 | 301 | 369 | 337 | 288 | 223 | 327 | |
| NM_001609 | ACADSB | Acyl-Coenzyme A dehydrogenase, short/branched chain | 354 | 368 | 325 | 273 | 211 | 276 | |
| NM_001647 | APOD | Apolipoprotein D | 259 | 358 | 289 | 261 | 202 | 205 | |
| NM_012113 | CA14 | Carbonic anhydrase XIV | 218 | 356 | 279 | 251 | 194 | 270 | |
| AB067472 | DKFZP434L1435 | KIAA1885 protein | 150 | 213 | 186 | 166 | 119 | 163 | |
| NM_002916 | RFC4 | Replication factor C (activator 1) 4 (37 kD) | 102 | 177 | 119 | 105 | 86 | 132 | |
| NM_004889 | ATP5J2 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit f, isoform 2 | 106 | 147 | 76 | 102 | 76 | 62 | |
| AK057066 | cDNA FLJ32504 fis, clone SMINT1000016, weakly similar to 2-hydroxyacylsphingosine 1b | 69 | 121 | 126 | 64 | 75 | 84 | ||
| AK021722 | AGPAT5 | Lysophosphatidic acid acyltransferase, epsilon | 37 | 71 | 48 | 42 | 39 | 46 | |
| NM_003664 | AP3B1 | Adaptor-related protein complex 3, beta 1 subunit | 34 | 52 | 29 | 37 | 24 | 30 | |
| AF146760 | Sept10 | Septin 10 | 22 | 36 | 23 | 26 | 16 | 28 | |
| NM_004910 | PITPNM | Phosphatidylinositol transfer protein, membrane-associated | 2611 | 2809 | 2410 | 2974 | 4590 | 2675 | |
| NM_018216 | FLJ10782 | Pantothenic acid kinase | 10 | 9 | 10 | 9 | 18 | 15 | |
| NM_001714 | BICD1 | Bicaudal D homolog 1 (Drosophila) | 230 | 562 | 407 | 197 | 447 | 691 | |
| AK054944 | LENG5 | Leukocyte receptor cluster (LRC) member 5 | 67 | 100 | 91 | 78 | 74 | 158 | |
| Gene Expression | |||||||||
| NM_005088 | DXYS155E | DNA segment on chromosome X and Y (unique) 155 expressed sequence | 4857 | 3489 | 3214 | 5177 | 2241 | 2725 | |
| NM_006298 | ZNF192 | Zinc finger protein 192 | 552 | 988 | 761 | 537 | 578 | 820 | |
| NM_004991 | MDS1 | Myelodysplasia syndrome 1 | 401 | 691 | 480 | 390 | 361 | 420 | |
| NM_021784 | HNF3B | Hepatocyte nuclear factor 3, beta | 320 | 632 | 367 | 347 | 361 | 391 | |
| AF153201 | LOC58502 | C2H2 (Kruppel-type) zinc finger protein | 288 | 532 | 335 | 244 | 297 | 324 | |
| NM_025212 | IDAX | Dvl-binding protein IDAX (inhibition of the Dvl and Axin complex) | 297 | 490 | 311 | 303 | 254 | 241 | |
| AK022962 | PBX1 | Pre-B-cell leukemia transcription factor 1 | 237 | 456 | 326 | 245 | 261 | 345 | |
| NM_017617 | NOTCH1 | Notch-1 homolog | 309 | 358 | 353 | 324 | 208 | 370 | |
| NM_001451 | FOXF1 | Forkhead box F1 | 165 | 347 | 306 | 177 | 208 | 328 | |
| NM_007136 | ZNF80 | Zinc finger protein 80 (pT17) | 199 | 269 | 203 | 205 | 143 | 177 | |
| NM_021975 | RELA | V-rel reticuloendotheliosis viral oncogene homolog A, nuclear factor of kappa light polypeptide gene | 184 | 221 | 139 | 150 | 124 | 122 | |
| NM_031214 | TARDBP | TAR DNA binding protein | 76 | 154 | 109 | 74 | 91 | 90 | |
| NM_014007 | ZNF297B | Zinc finger protein 297B | 109 | 137 | 122 | 109 | 77 | 111 | |
| NM_014938 | MONDOA | Mlx interactor | 74 | 90 | 92 | 69 | 53 | 86 | |
| NM_005822 | DSCR1L1 | Down syndrome critical region gene 1-like 1 | 45 | 80 | 30 | 40 | 27 | 26 | |
| NM_004289 | NFE2L3 | Nuclear factor (erythroid-derived 2)-like 3 | 73 | 63 | 41 | 64 | 39 | 38 | |
| NM_054023 | SCGB3A2 | Secretoglobin family 3a, member 2 | 37 | 59 | 45 | 43 | 34 | 49 | |
| NM_012107 | BP75 | Bromodomain containing protein 75 kDa human homolog | 44 | 51 | 34 | 37 | 22 | 30 | |
| NM_007212 | RNF2 | Ring finger protein 2 | 48 | 40 | 30 | 45 | 18 | 26 | |
| D89859 | ZFP161 | Zinc finger protein 161 homolog (mouse) | 500 | 596 | 4280 | 458 | 481 | 6699 | |
| NM_014335 | CRI1 | CREBBP/EP300 inhibitory protein 1 | 52 | 84 | 86 | 57 | 72 | 196 | |
| Immune Function | |||||||||
| NM_014889 | MP1 | Metalloprotease 1 (pitrilysin family) | 352 | 401 | 398 | 379 | 260 | 351 | |
| NM_014312 | CTXL | Cortical thymocyte receptor (X. laevis CTX) like | 386 | 370 | 375 | 392 | 224 | 299 | |
| NM_002053 | GBP1 | Guanylate binding protein 1, interferon-inducible, 67 kD | 259 | 369 | 334 | 245 | 214 | 251 | |
| NM_005356 | LCK | Lymphocyte-specific protein tyrosine kinase | 186 | 206 | 187 | 235 | 124 | 181 | |
| NM_000564 | IL5RA | Interleukin 5 receptor, alpha | 112 | 106 | 124 | 121 | 63 | 150 | |
| NM_001311 | CRIP1 | Cysteine-rich protein 1 (intestinal) | 45 | 31 | 39 | 49 | 60 | 43 | |
| NM_002984 | SCYA4 | Small inducible cytokine A4 MIP1B | 492 | 2001 | 2483 | 517 | 1523 | 3897 | |
| NM_002983 | SCYA3 | Small inducible cytokine A3 MIP1A | 248 | 1798 | 2207 | 185 | 1364 | 3673 | |
| NM_014443 | IL17B | Interleukin 17B | 663 | 696 | 681 | 706 | 703 | 1155 | |
| NM_006018 | HM74 | Putative chemokine receptor-GTP-binding protein | 13 | 25 | 19 | 15 | 26 | 34 | |
| Miscellaneous Functions | |||||||||
| AB033041 | VANGL2 | Vang, van gogh-like 2 (Drosophila) | 983 | 1246 | 1351 | 981 | 796 | 1304 | |
| AK021444 | POSTN | Periostin, osteoblast specific factor | 569 | 917 | 789 | 522 | 479 | 629 | |
| NM_003691 | STK16 | Serine/threonine kinase 16 | 403 | 777 | 458 | 395 | 348 | 393 | |
| NM_006438 | COLEC10 | Collectin sub-family member 10 (C-type lectin) | 284 | 762 | 500 | 260 | 351 | 528 | |
| AK057699 | FLJ33137 fis, clone UTERU1000077 | 375 | 637 | 613 | 369 | 392 | 616 | ||
| NM_017671 | C20orf42 | Chromosome 20 open reading frame 42 | 362 | 557 | 551 | 280 | 323 | 478 | |
| AK054683 | DCLRE1C | DNA cross-link repair 1C | 486 | 555 | 574 | 476 | 293 | 515 | |
| NM_033060 | KAP4.10 | Keratin associated protein 4.10 | 210 | 245 | 197 | 154 | 123 | 172 | |
| AF319045 | CNTNAP2 | Contactin associated protein-like 2 | 112 | 215 | 173 | 120 | 113 | 176 | |
| NM_001046 | SLC12A2 | Solute carrier family 12 (sodium/potassium/chloride transporters), member 2 | 158 | 148 | 184 | 146 | 86 | 161 | |
| NM_016279 | CDH9 | Cadherin 9, type 2 (T1-cadherin) | 77 | 112 | 69 | 65 | 51 | 64 | |
| NM_014208 | DSPP | Dentin sialophosphoprotein | 60 | 90 | 64 | 57 | 53 | 59 | |
| NM_015669 | PCDHB5 | Protocadherin beta 5 | 92 | 83 | 62 | 98 | 42 | 47 | |
| AK023198 | OPRK1 | Opioid receptor, kappa 1 | 58 | 76 | 41 | 48 | 46 | 38 | |
| NM_018240 | KIRREL | Kin of IRRE like (Drosophila) | 60 | 75 | 47 | 66 | 43 | 46 | |
| AK056781 | ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | 54 | 62 | 42 | 47 | 41 | 42 | |
| NM_022123 | NPAS3 | Basic-helix-loop-helix-PAS protein | 17 | 22 | 9 | 16 | 12 | 13 | |
| NM_001246 | ENTPD2 | Ectonucleoside triphosphate diphosphohydrolase 2 | 3438 | 3272 | 3731 | 3767 | 3590 | 6309 | |
| Unknown Function | |||||||||
| AK056884 | FLJ32322 fis, clone PROST2003577 | 2007 | 2878 | 2008 | 1825 | 1548 | 1958 | ||
| NM_017812 | FLJ20420 | Coiled-coil-helix-coiled-coil-helix domain containing 3 | 1105 | 1915 | 1370 | 1125 | 940 | 1358 | |
| AJ420459 | LOC51184 | Protein x 0004 | 661 | 1579 | 881 | 603 | 771 | 768 | |
| BC011575 | Similar to RIKEN cDNA 0610031J06 gene, clone IMAGE:4639306 | 974 | 1556 | 1412 | 1020 | 844 | 1261 | ||
| AK057357 | FLJ32926 | DKFZp434D2472 | 1188 | 1378 | 1159 | 1043 | 515 | 1136 | |
| NM_025019 | TUBA4 | tubulin, alpha 4 | 1446 | 1173 | 1330 | 1477 | 782 | 1366 | |
| AK023150 | FLJ13088 fis, clone NT2RP3002102 | 798 | 1087 | 905 | 845 | 564 | 785 | ||
| NM_017833 | C21orf55 | Chromosome 21 open reading frame 55 | 741 | 1079 | 799 | 687 | 508 | 665 | |
| BC001407 | Similar to cytochrome c-like antigen | 524 | 1004 | 629 | 506 | 502 | 577 | ||
| AK023104 | FLJ22648 fis, clone HSI07329 | 441 | 984 | 621 | 488 | 471 | 495 | ||
| AK024617 | FLJ20964 fis, clone ADSH00902 | 824 | 955 | 745 | 788 | 535 | 824 | ||
| BC009536 | IMAGE:3892368 | 553 | 924 | 775 | 597 | 498 | 671 | ||
| AK056287 | FLJ31725 fis, clone NT2RI2006716 | 435 | 862 | 907 | 405 | 459 | 893 | ||
| AK021611 | FLJ11549 fis, clone HEMBA1002968 | 535 | 812 | 675 | 545 | 392 | 630 | ||
| BC015119 | IMAGE:3951139 | 445 | 784 | 487 | 455 | 435 | 439 | ||
| AK056492 | FLJ31930 fis, clone NT2RP7006162 | 252 | 651 | 525 | 266 | 367 | 457 | ||
| AB058711 | KIAA1808 | KIAA1808 protein | 208 | 637 | 357 | 199 | 339 | 366 | |
| BC011266 | IMAGE:4156795 | 354 | 632 | 432 | 356 | 328 | 460 | ||
| AK023316 | FLJ13254 fis, clone OVARC1000787 | 416 | 596 | 357 | 400 | 290 | 352 | ||
| NM_024696 | FLJ23058 | Hypothetical protein FLJ23058 | 456 | 541 | 346 | 436 | 313 | 359 | |
| AF253316 | Pheromone receptor (PHRET) pseudogene | 136 | 520 | 425 | 128 | 301 | 347 | ||
| AK056007 | BICD1 | Bicaudal D homolog 1 (Drosophila) | 704 | 505 | 439 | 624 | 243 | 305 | |
| AB020632 | KIAA0825 | KIAA0825 protein | 249 | 498 | 353 | 246 | 272 | 339 | |
| NM_017609 | DKFZp434A1721 | Hypothetical protein DKFZp434A1721 | 182 | 485 | 319 | 190 | 298 | 304 | |
| NM_018190 | FLJ10715 | Hypothetical protein FLJ10715 | 202 | 483 | 310 | 174 | 206 | 266 | |
| AK057046 | FLJ32484 fis, clone SKNMC2001555 | 229 | 473 | 294 | 261 | 302 | 228 | ||
| NM_013395 | AD013 | Proteinx0008 | 448 | 461 | 496 | 403 | 304 | 378 | |
| BC008501 | MGC14839 | Similar to RIKEN cDNA 2310030G06 | 379 | 414 | 329 | 443 | 264 | 290 | |
| AK021988 | FLJ11926 fis, clone HEMBB1000374 | 321 | 411 | 399 | 280 | 218 | 288 | ||
| AF119872 | PRO2272 | 257 | 405 | 327 | 257 | 205 | 250 | ||
| NM_022744 | FLJ13868 | Hypothetical protein FLJ13868 | 267 | 376 | 239 | 270 | 212 | 172 | |
| AK022364 | FLJ12302 fis, clone MAMMA1001864 | 172 | 355 | 316 | 164 | 184 | 332 | ||
| BC002644 | MGC4859 | Hypothetical protein MGC4859 similar to HSPA8 | 282 | 335 | 382 | 257 | 223 | 331 | |
| AK022201 | FLJ12139 fis, clone MAMMA1000339 | 267 | 302 | 152 | 235 | 123 | 131 | ||
| NM_017953 | FLJ20729 | Hypothetical protein FLJ20729 | 170 | 290 | 258 | 138 | 170 | 218 | |
| AK057473 | FLJ32911 fis, clone TESTI2006210 | 160 | 268 | 265 | 163 | 123 | 247 | ||
| U50383 | RAI15 | Retinoic acid induced 15 | 206 | 265 | 236 | 198 | 159 | 186 | |
| AK027027 | FLJ23374 fis, clone HEP16126 | 134 | 261 | 170 | 134 | 152 | 141 | ||
| AK057288 | FLJ32726 fis, clone TESTI2000981 | 206 | 249 | 312 | 216 | 152 | 244 | ||
| U79280 | PIPPIN | Ortholog of rat pippin | 274 | 229 | 189 | 238 | 117 | 134 | |
| AK023628 | FLJ13566 fis, clone PLACE1008330 | 140 | 195 | 230 | 133 | 128 | 193 | ||
| NM_025263 | CAT56 | CAT56 protein | 126 | 194 | 147 | 127 | 101 | 130 | |
| AF311324 | Ubiquitin-like fusion protein | 191 | 189 | 179 | 190 | 106 | 138 | ||
| NM_005708 | GPC6 | Glypican 6 | 107 | 185 | 144 | 109 | 88 | 146 | |
| AB037778 | KIAA1357 | KIAA1357 protein | 153 | 180 | 156 | 149 | 118 | 146 | |
| AK055939 | FLJ31377 fis, clone NESOP1000087 | 152 | 167 | 179 | 136 | 105 | 173 | ||
| NM_018316 | FLJ11078 | Hypothetical protein FLJ11078 | 89 | 145 | 118 | 73 | 94 | 103 | |
| AF402776 | BIC | BIC noncoding mRNA | 82 | 136 | 171 | 96 | 88 | 153 | |
| BC003416 | IMAGE:3450973 | 64 | 133 | 93 | 83 | 73 | 111 | ||
| AL137491 | DKFZp434P1530 | 62 | 130 | 88 | 57 | 72 | 74 | ||
| AK057770 | FLJ25041 fis, clone CBL03194 | 110 | 130 | 114 | 108 | 83 | 84 | ||
| AB058769 | KIAA1866 | KIAA1866 protein | 89 | 126 | 122 | 102 | 83 | 91 | |
| AB058747 | WAC | WW domain-containing adapter with a coiled-coil region | 60 | 124 | 103 | 57 | 76 | 77 | |
| AK054885 | C6orf31 | Chromosome 6 open reading frame 31 | 51 | 119 | 108 | 41 | 68 | 119 | |
| AK022235 | FLJ12173 fis, clone MAMMA1000696 | 109 | 103 | 94 | 90 | 62 | 77 | ||
| AK026853 | AOAH | Acyloxyacyl hydrolase (neutrophil) | 59 | 98 | 64 | 59 | 61 | 56 | |
| AK024877 | FLJ21224 fis, clone COL00694 | 53 | 96 | 110 | 55 | 54 | 103 | ||
| NM_003171 | SUPV3L1 | Suppressor of var1, 3-like 1 (S. cerevisiae) | 65 | 93 | 60 | 60 | 55 | 58 | |
| NM_052933 | TSGA13 | Testis specific, 13 | 66 | 80 | 70 | 68 | 44 | 71 | |
| AK057907 | FLJ25178 fis, clone CBR09176 | 42 | 77 | 31 | 47 | 43 | 41 | ||
| AK055748 | FLJ31186 fis, clone KIDNE2000335 | 88 | 67 | 68 | 79 | 44 | 71 | ||
| BC013757 | IMAGE:4525041 | 40 | 54 | 39 | 43 | 33 | 32 | ||
| AL365511 | Novel human gene mapping to chomosome 22 | 19 | 48 | 29 | 20 | 27 | 37 | ||
| AK026889 | APRIN | Androgen-induced proliferation inhibitor | 31 | 35 | 42 | 34 | 21 | 34 | |
| AK057423 | FLJ32861 fis, clone TESTI2003589 | 36 | 32 | 34 | 30 | 18 | 31 | ||
| AK055543 | MLSTD1 | Male sterility domain containing 1 | 31 | 31 | 32 | 27 | 18 | 30 | |
| AK056513 | FLJ31951 fis, clone NT2RP7007177 | 33 | 29 | 20 | 22 | 13 | 20 | ||
| NM_013319 | TERE1 | Transitional epithelia response protein | 22 | 28 | 19 | 24 | 17 | 22 | |
| AK026456 | FLJ22803 fis, clone KAIA2685 | 15 | 26 | 14 | 16 | 13 | 17 | ||
| AK021610 | cDNA FLJ11548 fis, clone HEMBA1002944 | 34 | 26 | 29 | 31 | 15 | 28 | ||
| AK026823 | FLJ23170 fis, clone LNG09984 | 15 | 22 | 14 | 19 | 8 | 18 | ||
| AK056805 | FLJ32243 fis, clone PROST1000039 | 400 | 177 | 186 | 343 | 314 | 160 | ||
| NM_012238 | SIRT1 | Sirtuin silent mating type information regulation 2 homolog 1 (S. cerevisiae) | 149 | 156 | 170 | 178 | 134 | 109 | |
| NM_016099 | GOLGA7 | golgi autoantigen, golgin subfamily a, 7 | 10493 | 15165 | 9882 | 11947 | 11564 | 15698 | |
| AK022482 | FLJ12420 fis, clone MAMMA1003049 | 6052 | 9099 | 5803 | 6362 | 7620 | 9309 | ||
| AK026490 | RAB32 | RAB32, member RAS oncogene family | 3677 | 7044 | 4641 | 3671 | 5553 | 7561 | |
| NM_020684 | NPD007 | NPD007 protein | 674 | 794 | 764 | 630 | 720 | 1215 | |
| AL390158 | ATXN7L3 | Ataxin 7-like 3 | 319 | 460 | 378 | 339 | 403 | 598 | |
| NM_017752 | FLJ20298 | Hypothetical protein FLJ20298 | 146 | 237 | 282 | 133 | 233 | 493 | |
| AB037743 | KIAA1322 | KIAA1322 protein | 236 | 202 | 199 | 239 | 246 | 319 | |
| AF339819 | clone IMAGE:38177 | 77 | 111 | 110 | 96 | 125 | 174 | ||
| AK055215 | FLJ30653 fis, clone DFNES2000143 | 47 | 48 | 58 | 43 | 80 | 92 |
Figure 1.
LPS-stimulated genes in cord blood and adult monocytes can be differentiated on the basis of kinetics of expression. Expression level (in relative intensity units) is shown of the y-axis and time on the x-axis. At the 45 min time point, significant differences in expression level were seen between adult and neonatal monocytes for each of the gene groups A-H.
Figure 2.
Heat map representation of differences in gene expression of adult and cord blood monocytes in response to LPS. Z-transformed scores of the mean expression values for adult monocytes prior to (A0), after 45 min (A45), and after 120 min (A120) of LPS exposure are graphically shown to the left. Similar scores from cord blood monocytes prior to (C0), after 45 min (C45), and after 120 min C120) of LPS exposure, respectively. The heat map was produced using software from Spotfire Decision Site (Somerville, MA).
In addition to the above genes which differed in expression between groups following LPS exposure, 516 genes were also identified that were differentially expressed over time within a group. A supplementary table containing these data is available upon request. For these genes, a similar pattern of dynamic expression was seen as was observed in the other group. Therefore, these genes reflect common responses to LPS in monocytes from both sources.
A subset of genes that were differentially expressed either between adult and cord blood monocytes were selected for validation using the quantitative real-time polymerase chain reaction method (QRT-PCR). These included four genes that differed between groups after 45 min of LPS exposure (Erbb3, Tmod, Dscr1l1, and Sp1), and six genes that differed in expression after 2 hours of LPS exposure (Scya4, Gro2, Cri1, Scya3, Scya3l1, and Il-1a). Nine of the ten genes tested for QRT-PCR validation demonstrated similar levels of relative expression in QRT-PCR experiments as in the microarrays. Only CRI1 failed to corroborate the microarray data.
Hypervariable gene analysis
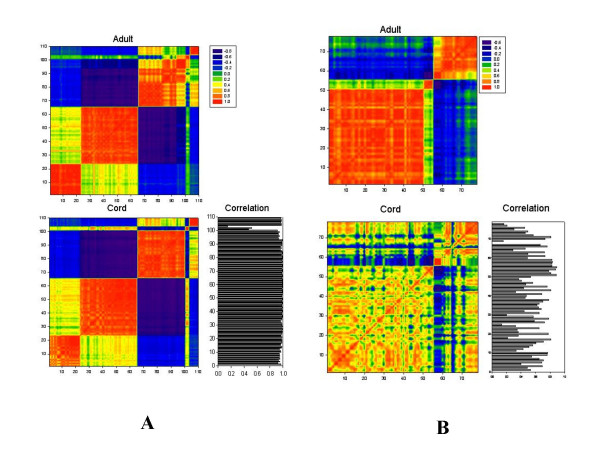
One hundred eighty-eight hypervariable (HV) genes were selected from expressed genes in adult and cord blood monocytes based on their changes across three time points. These genes exhibited significantly higher expression variation over time than the majority of genes. Differences in variation between two experimental sample sets, in this case adult and neonatal samples, can represent differences in homeostatic control mechanisms between these two sets [20]. The selected genes were hypervariable in both sample groups. HV genes with highly correlated expression levels in a given population are likely to share function [20]. A correlation based clustering procedure was carried out for these HV genes as described in the methods section. Genes belonging to the 5 largest clusters were used for creation of a graphical output, denoted a correlation mosaic. A correlation mosaic allows identification of the genes within clusters by visual inspection and subsequent functional analysis of genes within clusters (Figures 3A &3B). Figure 3A represents 110 genes of the same cluster allocation between adult and cord blood monocyte samples, demonstrating a very high similarity between cells from these two groups, as measured by the correlation coefficients between genes from adult and cord monocytes with value > 0.90 (figure 3A, black and white graph to the right). Three genes on this list (#101–103) were the exception: transcriptional regulator interacting with the PHS-bromodomain 2 (Trip-Br2), interleukin 1 beta (Il1b), and the GRO2 oncogene(Gro2). These genes may play a critical role in differentiation between adult and cord monocyte behaviour [22,23]. The high similarity of these mosaics presents evidence for the presence of fundamental processes in monocyte development that appear to be quite similar in both groups of samples. The details of the genes used in Figure 3A are presented as Table 2. Another group of 78 genes were found that have different cluster designations between adult and cord blood monocytes (Figure 3B). Details of these genes are listed in Table 3.
Figure 3.
Correlative mosaic for genes selected as HV-genes in cord blood and adult monocytes, belonging to five clusters of highest content. A. Genes of the same cluster in cord and adult; B. Genes of different cluster in cord and adult. Correlation coefficients are color-coded according to the key in the upper right. The correlation between the adult and cord blood monocyte profiles for each gene are shown in black and white, lower right.
Table 2.
Genes from which correlation mosaics in Figure 3A were derived. Genes in this table show the highest level of correlation by DFA analysis comparing adult and cord blood monocytes.
| Order in mosaic | Accession No. | Gene symbol | Description |
| 1 | NM_017614 | BHMT2 | Betaine-homocysteine methyltransferase 2 |
| 2 | NM_001651 | AQP5 | Aquaporin 5 |
| 3 | NM_020163 | LOC56920 | Semaphorin sem2 |
| 4 | NM_012343 | NNT | Nicotinamide nucleotide transhydrogenase |
| 5 | NM_000096 | CP | Ceruloplasmin (ferroxidase) |
| 6 | NM_005819 | STX6 | Syntaxin 6 |
| 7 | NM_052951 | C20orf167 | Chromosome 20 open reading frame 167 |
| 8 | NM_001348 | DAPK3 | Death-associated protein kinase 3 |
| 9 | X73502 | KRT20 | Cytokeratin 20 |
| 10 | NM_052887 | TIRAP | Toll-interleukin 1 receptor (TIR) domain-containing adapter protein |
| 11 | NM_019555 | ARHGEF3 | Rho guanine nucleotide exchange factor (GEF) 3 |
| 12 | NM_014380 | NGFRAP1 | Nerve growth factor receptor (TNFRSF16) associated protein 1 |
| 13 | NM_001272 | CHD3 | Chromodomain helicase DNA binding protein 3 |
| 14 | NM_005842 | SPRY2 | Sprouty homolog 2 (Drosophila) |
| 15 | NM_012332 | MT-ACT48 | Mitochondrial Acyl-CoA Thioesterase |
| 16 | BC015041 | VATI | Vesicle amine transport protein 1 |
| 17 | NM_003872 | NRP2 | Neuropilin 2 |
| 18 | NM_005849 | IGSF6 | Immunoglobulin superfamily, member 6 |
| 19 | NM_014323 | ZNF278 | Zinc finger protein 278 |
| 20 | NM_030674 | SLC38A1 | Solute carrier family 38, member 1 |
| 21 | NM_004153 | ORC1L | Origin recognition complex, subunit 1-like (yeast) |
| 22 | NM_005249 | FOXG1B | Forkhead box G1B |
| 23 | NM_021048 | MAGEA10 | Melanoma antigen, family A, 10 |
| 24 | M60502 | FLG | Filaggrin |
| 25 | NM_004997 | MYBPH | Myosin binding protein H |
| 26 | J05046 | INSRR | Insulin receptor-related receptor |
| 27 | M33987 | CA1 | Carbonic anhydrase I |
| 28 | D31886 | RAB3GAP | RAB3 GTPase-ACTIVATING PROTEIN |
| 29 | L24498 | GADD45A | Growth arrest and DNA-damage-inducible, alpha |
| 30 | L07590 | PPP2R3 | Protein phosphatase 2 (formerly 2A), regulatory subunit B" (PR 72), alpha isoform and (PR 130), bet |
| 31 | D87024 | IGLV4-3 | Immunoglobulin lambda variable 4-3 |
| 32 | L35848 | MS4A3 | Membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| 33 | M18216 | CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| 34 | M11952 | TRBV7–8 | T cell receptor beta variable 7–8 |
| 35 | D89094 | PDE5A | Phosphodiesterase 5A, cGMP-specific |
| 36 | M77140 | GAL | Galanin |
| 37 | D13628 | ANGPT1 | Angiopoietin 1 |
| 38 | M81635 | EPB72 | Erythrocyte membrane protein band 7.2 (stomatin) |
| 39 | D89859 | ZFP161 | Zinc finger protein 161 homolog (mouse) |
| 40 | D26069 | CENTB2 | Centaurin, beta 2 |
| 41 | L10717 | ITK | IL2-inducible T-cell kinase |
| 42 | L04282 | ZNF148 | Zinc finger protein 148 (pHZ-52) |
| 43 | L41944 | IFNAR2 | Interferon (alpha, beta and omega) receptor 2 |
| 44 | M82882 | ELF1 | E74-like factor 1 (ets domain transcription factor) |
| 45 | L26339 | RCD-8 | Autoantigen |
| 46 | D87328 | HLCS | Holocarboxylase synthetase (biotin-[proprionyl-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| 47 | D00943 | MYH6 | Myosin, heavy polypeptide 6, cardiac muscle, alpha (cardiomyopathy, hypertrophic 1) |
| 48 | D00099 | ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| 49 | L36531 | ITGA8 | Integrin, alpha 8 |
| 50 | D42084 | METAP1 | Methionyl aminopeptidase 1 |
| 51 | M76766 | GTF2B | General transcription factor IIB |
| 52 | J04621 | SDC2 | Syndecan 2 (heparan sulfate proteoglycan 1, cell surface-associated, fibroglycan) |
| 53 | D31888 | RCOR | REST corepressor |
| 54 | L32832 | ATBF1 | AT-binding transcription factor 1 |
| 55 | D86981 | APPBP2 | Amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| 56 | M94362 | LMNB2 | Lamin B2 |
| 57 | M54968 | KRAS2 | V-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene homolog |
| 58 | D42046 | DNA2L | DNA2 DNA replication helicase 2-like (yeast) |
| 59 | D86964 | DOCK2 | Dedicator of cyto-kinesis 2 |
| 60 | D50683 | TGFBR2 | Transforming growth factor, beta receptor II (70–80 kD) |
| 61 | M96843 | ID2B | Striated muscle contraction regulatory protein |
| 62 | M61906 | PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) |
| 63 | M12679 | HUMMHCW1A | Cw1 antigen |
| 64 | M63623 | OMG | Oligodendrocyte myelin glycoprotein |
| 65 | J04162 | FCGR3B | Fc fragment of IgG, low affinity IIIb, receptor for (CD16) |
| 66 | L48516 | PON3 | Paraoxonase 3 |
| 67 | M54927 | PLP1 | Proteolipid protein1 (Pelizaeus-Merzbacher disease, spastic paraplegia 2, uncomplicated) |
| 68 | D86973 | GCN1L1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) |
| 69 | D43968 | RUNX1 | Runt-related transcription factor 1 (acute myeloid leukemia 1-aml1 oncogene) |
| 70 | L05500 | ADCY1 | Adenylate cyclase 1 (brain) |
| 71 | D80010 | LPIN1 | Lipin 1 |
| 72 | D50918 | SEPT6 | Septin 6 |
| 73 | D86988 | RENT1 | Regulator of nonsense transcripts 1 |
| 74 | M90391 | IL16 | Interleukin 16 (lymphocyte chemoattractant factor) |
| 75 | M62324 | MRF-1 | Modulator recognition factor I |
| 76 | L77565 | DGS-H | DiGeorge syndrome gene H |
| 77 | D86970 | TIAF1 | TGFB1-induced anti-apoptotic factor 1 |
| 78 | D38169 | ITPKC | Inositol 1,4,5-trisphosphate 3-kinase C |
| 79 | D87684 | UBXD2 | UBX domain-containing 2 |
| 80 | D84454 | SLC35A2 | Solute carrier family 35 (UDP-galactose transporter), member 2 |
| 81 | M97496 | GUCA2A | Guanylate cyclase activator 2A (guanylin) |
| 82 | M95585 | HLF | Hepatic leukemia factor |
| 83 | L38517 | IHH | Indian hedgehog homolog (Drosophila) |
| 84 | L20860 | GP1BB | Glycoprotein Ib (platelet), beta polypeptide |
| 85 | M26880 | UBC | Ubiquitin C |
| 86 | D86962 | GRB10 | Growth factor receptor-bound protein 10 |
| 87 | D63481 | SCRIB | Scribble |
| 88 | D17525 | MASP1 | Mannan-binding lectin serine protease 1 (C4/C2 activating component of Ra-reactive factor) |
| 89 | L26584 | RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 |
| 90 | M65066 | PRKAR1B | Protein kinase, cAMP-dependent, regulatory, type I, beta |
| 91 | J05158 | CPN2 | Carboxypeptidase N, polypeptide 2, 83 kD |
| 92 | L36861 | GUCA1A | Guanylate cyclase activator 1A (retina) |
| 93 | L11239 | GBX1 | Gastrulation brain homeo box 1 |
| 94 | D90145 | SCYA3L1 | Small inducible cytokine A3-like 1 |
| 95 | M96739 | NHLH1 | Nescient helix loop helix 1 |
| 96 | M12959 | TRA@ | T cell receptor alpha locus |
| 97 | D80005 | C9orf10 | C9orf10 protein |
| 98 | M13231 | TRGC2 | T cell receptor gamma constant 2 |
| 99 | D28588 | SP2 | Sp2 transcription factor |
| 100 | M57732 | TCF1 | Transcription factor 1, hepatic-LF-B1, hepatic nuclear factor (HNF1), albumin proximal factor |
| 101 | NM_014755 | TRIP-Br2 | Transcriptional regulator interacting with the PHS-bromodomain 2 |
| 102 | NM_000576 | IL1B | Interleukin 1, beta |
| 103 | NM_002089 | GRO2 | GRO2 oncogene |
| 104 | NM_002089x | GPRC5D | G protein-coupled receptor, family C, group 5, member D |
| 105 | NM_002713 | PPP1R8 | Protein phosphatase 1, regulatory (inhibitor) subunit 8 |
| 106 | NM_014383 | TZFP | Testis zinc finger protein |
| 107 | NM_012248 | SPS2 | Selenophosphate synthetase 2 |
| 108 | AL137438 | SEC15L | SEC15 (S. cerevisiae)-like |
| 109 | NM_005387 | NUP98 | Nucleoporin 98 kD |
| 110 | NM_003476 | CSRP3 | Cysteine and glycine-rich protein 3 (cardiac LIM protein) |
Table 3.
Genes from which the mosaic in Figure 3B were derived. Genes from which correlation mosaics in Figure 3B were derived. Genes in this table show the greatest differences by DFA analysis comparing adult and cord blood monocytes.
| Order in Mosaic | Accession No. | Gene Symbol | Description |
| 1 | AK055855 | CLDN10 | Claudin 10 |
| 2 | NM_000565 | IL6R | Interleukin 6 receptor |
| 3 | NM_006150 | LMO6 | LIM domain only 6 |
| 4 | NM_022787 | NMNAT | NMN adenylyltransferase-nicotinamide mononucleotide adenylyl transferase |
| 5 | NM_002743 | PRKCSH | Protein kinase C substrate 80K-H |
| 6 | NM_004847 | AIF1 | Allograft inflammatory factor 1 |
| 7 | NM_021073 | BMP5 | Bone morphogenetic protein 5 |
| * 8 | AK025306 | CLK1 | CDC-like kinase 1 |
| 9 | NM_004280 | EEF1E1 | Eukaryotic translation elongation factor 1 epsilon 1 |
| * 10 | NM_004432 | ELAVL2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| 11 | NM_012181 | FKBP8 | FK506 binding protein 8 (38 kD) |
| 12 | NM_002091 | GRP | Gastrin-releasing peptide |
| 13 | NM_016355 | LOC51202 | Hqp0256 protein |
| 14 | NM_021204 | MASA | E-1 enzyme |
| 15 | NM_004204 | PIGQ | Phosphatidylinositol glycan, class Q |
| 16 | NM_002928 | RGS16 | Regulator of G-protein signalling 16 |
| 17 | NM_005839 | SRRM1 | Serine/arginine repetitive matrix 1 |
| 18 | NM_003166 | SULT1A3 | Sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| 19 | NM_000356 | TCOF1 | Treacher Collins-Franceschetti syndrome 1 |
| 20 | NM_016437 | TUBG2 | Tubulin, gamma 2 |
| * 21 | NM_022568 | ALDH8A1 | Aldehyde dehyrdogenase 8 family, member A1 |
| 22 | AF209930 | CHRD | Chordin |
| 23 | NM_005274 | GNG5 | Guanine nucleotide binding protein (G protein), gamma 5 |
| 24 | NM_018384 | IAN4L1 | Immune associated nucleotide 4 like 1 (mouse) |
| 25 | NM_000640 | IL13RA2 | Interleukin 13 receptor, alpha 2 |
| 26 | AK021692 | LOC51141 | Insulin induced protein 2 |
| 27 | NM_012443 | SPAG6 | Sperm associated antigen 6 |
| 28 | NM_003155 | STC1 | Stanniocalcin 1 |
| 29 | NM_022003 | FXYD6 | FXYD domain-containing ion transport regulator 6 |
| 30 | NM_002763 | PROX1 | Prospero-related homeobox 1 |
| 31 | NM_002836 | PTPRA | Protein tyrosine phosphatase, receptor type, A |
| 32 | AL136835 | TOLLIP | Toll-interacting protein |
| 33 | AB058691 | ALX4 | Aristaless-like homeobox 4 |
| 34 | AF112345 | ITGA10 | Integrin, alpha 10 |
| 35 | NM_022788 | P2RY12 | Purinergic receptor P2Y, G protein-coupled, 12 |
| 36 | NM_001213 | C1orf1 | Chromosome 1 open reading frame 1 |
| 37 | NM_005860 | FSTL3 | Follistatin-like 3 (secreted glycoprotein) |
| 38 | NM_013320 | HCF-2 | Host cell factor 2 |
| 39 | NM_058246 | LOC136442 | Similar to MRJ gene for a member of the DNAJ protein family |
| 40 | NM_020169 | LXN | Latexin protein |
| 41 | BC008993 | MGC17337 | Similar to RIKEN cDNA 5730528L13 gene |
| 42 | BC002712 | MYCN | V-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| 43 | AK026164 | MYL6 | Myosin, light polypeptide 6, alkali, smooth muscle and non-muscle |
| 44 | NM_006215 | SERPINA4 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 |
| 45 | NM_004790 | SLC22A6 | Solute carrier family 22 (organic anion transporter), member 6 |
| 46 | NM_022911 | SLC26A6 | Solute carrier family 26, member 6 |
| 47 | NM_003374 | VDAC1 | Voltage-dependent anion channel 1 |
| 48 | NM_017818 | WDR8 | WD repeat domain 8 |
| 49 | NM_003416 | ZNF7 | Zinc finger protein 7 (KOX 4, clone HF.16) |
| 50 | NM_002313 | ABLIM | Actin binding LIM protein |
| 51 | NM_012074 | CERD4 | Cer-d4 (mouse) homolog |
| 52 | NM_000787 | DBH | Dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| * 53 | NM_000561 | GSTM1 | Glutathione S-transferase M1 |
| 54 | BC014075 | GTPBP1 | GTP binding protein 1 |
| 55 | NM_033260 | HFH1 | Winged helix/forkhead transcription factor |
| 56 | NM_033033 | KRTHB2 | Keratin, hair, basic, 2 |
| 57 | NM_004789 | LHX2 | LIM homeobox protein 2 |
| 58 | NM_014106 | PRO1914 | PRO1914 protein |
| * 59 | NM_006799 | PRSS21 | Protease, serine, 21 (testisin) |
| * 60 | NM_002900 | RBP3 | Retinol binding protein 3, interstitial |
| 61 | NM_033022 | RPS24 | Ribosomal protein S24 |
| * 62 | AB029021 | TRIM35 | Tripartite motif-containing 35 |
| * 63 | NM_020989 | CRYGC | Crystallin, gamma C |
| * 64 | BI198124 | HMG1L10 | High-mobility group (nonhistone chromosomal) protein 1-like 10 |
| 65 | NM_014163 | HSPC073 | HSPC073 protein |
| 66 | AF181985 | JIK | STE20-like kinase |
| 67 | NM_017607 | PPP1R12C | Protein phosphatase 1, regulatory (inhibitor) subunit 12C |
| * 68 | NM_002873 | RAD17 | RAD17 homolog (S. pombe) |
| 69 | NM_022095 | ZNF335 | Zinc finger protein 335 |
| * 70 | M90355 | BTF3L2 | Basic transcription factor 3, like 2 |
| 71 | NM_002079 | GOT1 | Glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) |
| 72 | NM_004146 | NDUFB7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 (18 kD, B18) |
| 73 | L38486 | MFAP4 | Microfibrillar-associated protein 4 |
| * 74 | AF111848 | ACTB | Actin, beta |
| 75 | NM_001916 | CYC1 | Cytochrome c-1 |
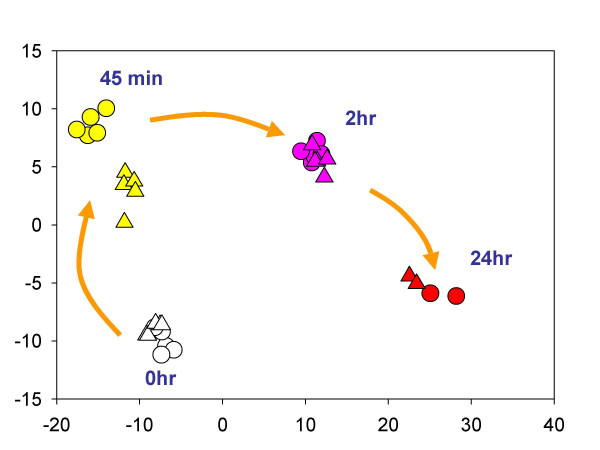
We analyzed these genes using DFA in order to find those genes most likely to highlight the differences between cord and adult monocytes. DFA identified genes having high discriminatory capabilities. The DFA software selected genes from Table 3 with highest discriminatory capabilities for this case. A total of 12 genes (marked with asterisk in Table 3) were used by the DFA program to differentiate dynamical changes in both cord and adult monocytes after LPS stimulation. Values of the roots obtained by DFA analysis were used to graphically depict the differences of the gene expression values obtained in cord and adult samples in different stages after stimulation (Fig. 4). The spatial organization of the elements in this representation provides a measure of the overall similarity of the dynamic behaviour of these samples. The greatest temporal changes in gene expression for cord and adult monocytes noted above after 45 min of LPS exposure were also observed in the analysis using these 12 genes. However, almost no differences occurred at the 2 hr time point between cord and adult cells suggesting that the global behavior of the cells is similar, but the kinetics of change differ i.e. many of the changes are the same in both groups, but they occur at different rates.
Figure 4.
DFA analysis of phases of monocyte activation comparing cord and adult cells. DFA identified a subset of genes (see Table 3) whose expression values can be linearly combined in an equation, denoted a root, whose overall value is distinct for a given characterized group. These roots used as coordinate for presentation of these groups of samples in scatterplot. Results from individual samples for adult monocyte (circles) and cord monocytes (triangles) are discussed in the text. Results from individual samples for adult monocyte (circles) and cord monocytes (triangles) are shown.
Apoptosis assays
The products of a subset of genes that were differentially expressed between groups after 45 min exposure to LPS are involved in apoptosis. We therefore performed a series of functional experiments comparing apoptosis in adult (n = 10) and neonatal (n = 10) cord bloods. Results of these assays are shown in Table 4. Annexin assays demonstrated that adult monocytes display different kinetics for both apoptosis and necrosis as compared with neonatal monocytes. Flow cytometry revealed that 43 ± 5% (mean + SD) of adult and 53 + 8% of neonatal monocytes are undergoing apoptosis after stimulation with LPS for 14 hours (p < 0.002), while 38 + 8% of adult and 25 + 9% of neonatal monocytes are necrotic after 14 hours of LPS stimulation (p < 0.003). The number of live monocytes after 14 hours of LPS stimulation was not statistically different between the two groups. There was also no statistically significant difference in the number of live, apoptotic, or necrotic monocytes between adult and neonatal samples prior to LPS stimulation (data not shown).
Table 4.
Results of Annexin Binding Assays
| Cell Type | Apoptotic Cells | Necrotic Cells | Significance |
| Adult monocytes | 43 ± 5% | 38 % ± 8% | P < 0.002 |
| Cord blood monocytes | 53 ± 8% | 25% ± 9% | P < 0.003 |
Discussion
Following a given physiologic stimulus, signalling kinase activation, transcription factor translocation, and gene transcription all occur in rapid order. However, like all biological processes, mRNA accumulation (or decreases) does not occur uniformly, and we hypothesized that examining the kinetics of mRNA accumulation or disappearance might provide clues into relevant cellular dynamics. We used a well-developed and validated gene expression microarray to examine the dynamics of mRNA accumulation and differences between adult and neonatal monocytes in that process.
Genes were found to be differentially expressed between adult and cord monocytes after either 45 or 120 minutes of LPS exposure, with little difference at 24 hr (see Figure 4). Interestingly, no statistically significant differences in gene expression were observed between these groups in untreated cells. Previous reports by others indicated altered functions of cord blood monocytes in cytokine secretion and cellular adhesion. Results from this study cast new light on these findings and add complexity to understanding such differences. In some cases, our data support previous speculations about neonatal immune function. For example, the increased expression of IL-17B in neonatal monocytes is consistent with the observations of Vanden Eijnden and colleagues that newborns compensate for their relative immune deficiency by over-expression of the IL23-IL-17 signalling pathway in dendritic cells [24]. Similarly, we found significant elevations in cord monocyte transcripts of the chemokines MIP1B and MIP1A after 2 hrs of LPS exposure, consistent with Sullivan and colleagues' report of higher amounts of MIPα in cord blood samples compared with adults [25]. On the other hand, transcripts for cadherin 9, Rock1, periostin, heparin sulfate 6-O-sulfotransferase 3, and C20orf42, whose products participate in various mechanisms that are associated with adhesion [26-28] were statistically significantly increased in adult monocytes after 45 min of LPS exposure, although no differences in expression for these genes between groups were detected at the later time point. These data suggest complex, dynamic relations for genes whose products are associated with cellular adhesion, and collectively highlight the importance of examining gene expression profiles (or related protein expression levels) over time.
The limits of gene expression profiling as a technique, albeit a very useful technique, must be acknowledged. The technique examines only RNA transcripts, not protein synthesis. Thus, alterations in other critical inflammatory mediators, such as eicosanoids, remain unobserved with this method. Furthermore, it is well known that there are many proteins, including critical inflammatory mediators, whose synthesis and secretion is not directly related in mRNA accumulation [29]. Thus, gene expression profiling should be complemented with other methods in order to maximize there potential.
In the final analysis, the utility of gene expression profiling will be demonstrated only if they provide insights into relevant physiologic or pathophysiologic function. For that reason, we elected to test the validity of the array data by examining a physiologic mechanism implicated by computer modelling of the array data. As noted in Table 1, adult monocytes over-expressed a small number of genes associated with the regulation of apoptosis. Since monocyte activation is a "balancing act" between signals inducing apoptosis and those inducing activation and differentiation [30,31], differences in the kinetics of expression or activation of enzymes or transcription factors that regulate apoptosis could have a crucial outcome on whether monocyte responses are pro- or anti-inflammatory. Annexin assays confirmed that there are significant differences in the appearance of apoptotic cells between adults and newborn monocytes (Table 4). Since apoptotic cells dampen the inflammatory response, it is interesting to speculate that the related blunted neonatal response to inflammatory stimuli (including infection) may result, at least in part, from the excessive production of apoptotic cells during monocyte activation.
There has been, to our knowledge, one previously published paper using gene expression arrays to study neonatal monocyte function [14]. Our findings differ somewhat from those described by these authors. The most obvious difference was our finding of no statistically significant differences between adult and cord blood samples in the resting state. We should note, however, that it is otherwise difficult to compare the two studies. Jiang and colleagues used a 1000-fold greater dose of LPS to stimulate the monocytes, and RNA was prepared after 18 hr of stimulation. Thus, it is difficult to determine which of the effects observed by these authors were the direct result of LPS activation or were mediated through autocrine activation by proteins secreted in response to LPS. Furthermore, the non-physiologic dose of LPS used by those authors makes the biological/pathological relevance of that study difficult to interpret. Finally, we should note that the study by Jiang and colleagues used different methodologies for purifying monocytes. While our method, positive selection using CD14-coated microbeads, carries the theoretical risk of activating the cells through TLR-4/CD14 signaling pathways, adherence procedures carry the greater risk of activating the cells, as β2 integrins are activated during the adherence process.
From the bioinformatics standpoint, our data demonstrate how gene microarray experiments can quickly move from the generation of gene lists to the development of plausible and testable models of relevant biology and physiology. Specifically, they demonstrate that computer-assisted, physiologic modelling is another means of corroborating array findings and provides the advantage of providing an approach for immediately testing the biological relevance of microarray data before embarking on the sometimes laborious task of confirming differential expression of dozens or even hundreds of genes identified in a microarray experiment. As described in the results section, the differences between groups in gene expression at 45 min were attributable to a unique up-regulation of specific genes in adult monocytes, a unique down-regulation of other genes in cord monocytes, or a combination of both processes for other genes. We have searched for mechanisms that account for these patterns. Specifically, we have analyzed the genes within derived k-means clusters to determine if a large number of genes within a cluster are related to overlapping functions using Ingenuity Pathway Assist software, or alternatively to shared transcriptional response elements upstream of these genes. However, these strategies have failed to elucidate reasons to explain these findings.
Our studies also suggest that, while expensive and time-consuming to undertake, studying the kinetics of gene expression using microarrays can be highly informative. The previously reported study [14] examining gene expression differences between adult and cord blood monocytes was performed at only a single time point (18 hr after activation with a non-physiologic dose of LPS). Our studies suggest that the relevant biology may lie not in the specific genes that are differentially expressed at one particular time point, but, as one would predict with a dynamic system, which genes are expressed when. Timing of mRNA accumulation could determine, among other things, whether pro-apoptotic signals are processed in monocytes before cellular necrosis ensues.
The validity of the dynamic/kinetic approach is further supported by the correlation analyses (Figures 3 and 4). These analyses demonstrate clearly that the accumulation of a specific mRNA is not an independent event. Gene transcription and mRNA degradation are dynamic processes closely tied to the accumulation or degradation of other mRNAs and the transcription of their cognate proteins. We contend that, without this dynamic view of cellular activity, investigators attempting to use microarray data to elucidate relevant biological or pathological processes will encounter unnecessary obstacles in attempts to move from the generation of gene lists to testing specific hypotheses.
Abbreviations
LPS – Lipopolysaccharide
DFA – Discriminant function analysis
HV – Hypervariable
Acknowledgments
Acknowledgements
Supported in part by the National Institutes of Health (NIH), National Center for Research Resources, a component of the NIH, General Clinical Research Center Grant MO1 RR-14467, NIH grants P20 RR020143-01, P20 RR15577, P20 RR17703, and P20 R016478-04 and by the Oklahoma Center for Science and Technology (OCAST).
The authors also wish to extend their thanks to Julie McGhee, M.D., for her review and thoughtful comments on this manuscript.
Contributor Information
Shelley Lawrence, Email: lawrence@pediatrix.com.
Yuhong Tang, Email: yuhong-tang@omrf.ouhsc.edu.
M Barton Frank, Email: Bart-Frank@omrf.ouhsc.edu.
Igor Dozmorov, Email: igor-dozmorov@omrf.ouhsc.edu.
Kaiyu Jiang, Email: kaiyu-jiang@ouhsc.edu.
Yanmin Chen, Email: yanmin-Chen@ouhsc.edu.
Craig Cadwell, Email: craig-cadwell@omrf.ouhsc.edu.
Sean Turner, Email: sean-turner@omrf.ouhsc.edu.
Michael Centola, Email: michael-centola@omrf.ouhsc.edu.
James N Jarvis, Email: james-jarvis@ouhsc.edu.
References
- Kobayashi S, Ohnuma K, Uchiyama M, Iino K, Iwata S, Dang NH, Morimoto C. Association of CD26 with CD45RA outside lipid rafts attenuates cord blood T-cell activation. Blood. 2004;103:1002–1010. doi: 10.1182/blood-2003-08-2691. [DOI] [PubMed] [Google Scholar]
- Adkins B, LeClerc C, Marshall-Clark S. Neonatal adaptive immunity comes of age. Nature Rev Immunol. 2004;4:553–564. doi: 10.1038/nri1394. [DOI] [PubMed] [Google Scholar]
- Garcia AM, Fadel SA, Cao S, Sarzotti M. T cell immunity in neonates. Immunol Res. 2000;22:177–190. doi: 10.1385/IR:22:2-3:177. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai ZP, Lv P, Gao XM. Phenotypic and functional analysis of human T lymphocytes in early second- and third-trimester fetuses. Clin Exp Immunol. 2002;129:302–308. doi: 10.1046/j.1365-2249.2002.01920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zola H, Fusco M, Weedon H, Macardle PJ, Ridings J, Roberton DM. Reduced expression of the interleukin-2-receptor gamma chain on cord blood lymphocytes: relationship to functional immaturity of the neonatal immune response. Immunol. 1996;87:86–91. [PMC free article] [PubMed] [Google Scholar]
- Schuit KE, Powell DA. Phagocytic dysfunction in monocytes of normal newborn infants. Pediatrics. 1980;65:501–504. [PubMed] [Google Scholar]
- Tan ND, Davidson D. Comparative differences and combined effects of interleukin-8, leukotriene B4, and platelet-activating factor on neutrophil chemotaxis of the newborn. Pediatr Res. 1995;38:11–16. doi: 10.1203/00006450-199507000-00003. [DOI] [PubMed] [Google Scholar]
- Anderson DC, Hughes BJ, Smith CW. Abnormal mobility of neonatal polymorphonuclear leukocytes. J Clin Invest. 1981;68:863–874. doi: 10.1172/JCI110341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DC, Freeman KLB, Heerdt B, Hughes BJ, Jack RM, Smith CW. Abnormal stimulated adherence of neonatal granulocytes: impaired induction of surface Mac-1 by chemotactic factors or secretagogues. Blood. 1987;70:740–750. [PubMed] [Google Scholar]
- Torok C, Lundahl J, Hed J, Lagercrantz H. Diversity of regulation of adhesion molecules (Mac-1 and L-selectin) in monocytes and neutrophils from neonates and adults. Arch Dis Child. 1993;68:561–565. doi: 10.1136/adc.68.5_spec_no.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan D, Ho W, Cutilli J, Campbell DE, Douglas SD. C-C chemokine profile of cord blood mononuclear cells: selective defect in RANTES production. Blood. 2000;95:715–718. [PubMed] [Google Scholar]
- Bessler H, Mendel C, Straussberg R, Gurary N, Aloni D, Sirota L. Effects of dexamethasone on IL-1beta, IL-6, and TNF-alpha production by mononuclear cells of newborns and adults. Biol Neonate. 1999;75:225–233. doi: 10.1159/000014099. [DOI] [PubMed] [Google Scholar]
- Kotiranta-Ainamo A, Rautonen J, Rautonen N. Interleukin-10 production by cord blood mononuclear cells. Pediatric Res. 1997;41:110–113. doi: 10.1203/00006450-199701000-00017. [DOI] [PubMed] [Google Scholar]
- Jiang H, Van de Ven C, Satwani P, Baxi LV, Cairo MS. Differential gene expression patterns by oligonucleotide microarray of basal versus lipopolysaccharide-activated monocytes from cord blood versus adult peripheral blood. J Immunol. 2004;172:5870–5879. doi: 10.4049/jimmunol.172.10.5870. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Busin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalisation to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Analytical Biochem. 2002;309:293–300. doi: 10.1016/S0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Knowlton N, Dozmorov IM, Centola M. Microarray Data Analysis Toolbox (MDAT): for normalization, adjustment and analysis of gene expression data. Bioinformatics. 2004;20:3687–3690. doi: 10.1093/bioinformatics/bth424. [DOI] [PubMed] [Google Scholar]
- Efron B, Gong G. A leisurely look at the bootstrap, the jackknife, and cross-validation. American Statistician. 1983;37:36–48. doi: 10.2307/2685844. [DOI] [Google Scholar]
- Dozmorov I, Knowlton N, Tang Y, Shields A, Pathipvanich P, Jarvis JN, Centola M. Hypervariable genes – experimental error or hidden dynamics. Nucleic Acids Res. 2004;32:e147. doi: 10.1093/nar/gnh146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Wichern D. Applied Multivariate Statistics. Prentice Hall; 2002. [Google Scholar]
- Peters AM, Bertram P, Gahr M, Speer CP. Reduced secretion of interleukin-1 and tumor necrosis factor-alpha by neonatal monocytes. Biol Neonate. 1993;63:157–62. doi: 10.1159/000243926. [DOI] [PubMed] [Google Scholar]
- Marwitz PA, Tenbergen-Meekes AJ, Heijnen CJ, Rijkers GT, Zegers BJ. Interleukin 1 in the in vitro antigen-induced antibody response in the human adult and newborn. Scand J Immunol. 1990;32:451–459. doi: 10.1111/j.1365-3083.1990.tb03185.x. [DOI] [PubMed] [Google Scholar]
- Vanden Eijnden S, Goriely S, De Wit D, Goldman M, Willems F. Preferential production of the IL-12(p40)/IL-23(p19) heterodimer by dendritic cells from human newborns. Eur J Immunol. 2006;36:21–26. doi: 10.1002/eji.200535467. [DOI] [PubMed] [Google Scholar]
- Sullivan SE, Staba SL, Gersting JA, Hutson AD, Theriaque D, Christensen RD, Calhoun DA. Circulating concentrations of chemokines in cord blood, neonates, and adults. Pediatr Res. 2002;51:653–657. doi: 10.1203/00006450-200205000-00018. [DOI] [PubMed] [Google Scholar]
- Juliano RL. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu Rev Pharmacol Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- Swetman CA, Leverrier Y, Garg R, Gan CH, Ridley AJ, Katz DR, Chain BM. Extension, retraction and contraction in the formation of a dendritic cell dendrite: distinct roles for Rho GTPases. Eur J Immunol. 2002;32:2074–2083. doi: 10.1002/1521-4141(200207)32:7<2074::AID-IMMU2074>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- Hamilton BJ, Burns CM, Nichols RC, Rigby WF. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J Biol Chem. 1997;272:28732–28741. doi: 10.1074/jbc.272.45.28732. [DOI] [PubMed] [Google Scholar]
- Morand EF, Bucala R, Leech M. Macrophage inhibitory factor. Arthritis Rheum. 2003;48:291–299. doi: 10.1002/art.10728. [DOI] [PubMed] [Google Scholar]
- Roger T, Glauser MP, Calandra T. Macrophage migration inhibitory factor (MIF) modulates innate immune responses induced by endotoxin and gram-negative bacteria. J Endotoxin Res. 2001;7:456–460. doi: 10.1179/096805101101533089. [DOI] [PubMed] [Google Scholar]