Abstract
We have molecularly characterized a proteolytic cleavage in conserved nuclear pore complex proteins. This cleavage, previously demonstrated to be essential for the biogenesis of two nuclear pore complex proteins in mammals (Nup98 and Nup96) and yeast (Nup145-N and Nup145-C), occurs between Phe and Ser residues within a highly conserved domain in a polyprotein precursor. Here, we show that a protease is not involved in the cleavage event. By using a combination of domain mapping and site-directed mutagenesis, we demonstrate that the human nuclear pore complex protein Nup98 specifically cleaves itself between F863 and S864. A region of Nup98, amino acids 715–920, is able to cleave, whereas a smaller region, amino acids 772–920, does not cleave. In addition, we have generated a Nup98 mutant that cleaves under defined conditions in vitro. Further, the two cleaved fragments of Nup98 form a complex, providing a possible mechanism whereby specific, yet low-affinity, binding between Nup98 and Nup96 is responsible for the nuclear targeting of Nup96. Although apparently unrelated evolutionarily, Nup98 has converged on an autoproteolytic biogenesis mechanism similar to that of hedgehog proteins, the inteins, and the N-terminal nucleophile proteins.
Exchange of protein and RNA between the nucleoplasm and cytosol occurs exclusively through the nuclear pore complex (NPC) (reviewed in ref. 1). As such, the NPC has a central role in numerous aspects of cellular function, including all signal transduction pathways, transcription, and translation. A superfamily of receptors, the karyopherins (also termed importins/exportins, or transportins) recognize signals for nuclear import and nuclear export of proteins and nucleic acids (for review, see refs. 2–4). In an incompletely understood sequence of events, karyopherins recognize cognate transport substrates, move to and dock at the NPC by virtue of specific interactions with a subset of NPC proteins (nucleoporins, or nups), pass through the NPC, and release the transport substrates. The nucleotide-bound state of the GTPase Ran is a determining factor in the directionality of transport (5).
A subset of nups containing degenerate peptide repeats (GLFG, FG, FXFG) is thought to serve as a docking site for karyopherin/substrate complexes as they traverse the NPC (6, 7). Several vertebrate FXFG-containing nups have been localized by immunoelectron microscopy. For example, Nup214/CAN is at the cytoplasmic face of the NPC, p62 is in the central region, and Nup98 is at the nuclear face (for review, see ref. 8). As such, regulated docking and undocking of karyopherin/substrate complexes to asymmetrically disposed nups could be a mechanism for traversing the depth of the NPC. The NPC is probably a dynamic structure, for example, the central channel is gated to preclude diffusion of moderately sized proteins, yet appears to dilate to allow transport of large ribonucleoprotein complexes (9).
Two of the repeat-containing nups, Nup214/CAN and Nup98, have been shown to be involved in leukemia when they are mutated. Chromosomal translocations resulting in in-frame fusions between these nups and several DNA-binding proteins, including DEK and HOXA9, cause aberrant transcription, apparently by virtue of the interaction between FG repeats and transcriptional coactivators (10–12). Additionally, a Nup214/CAN-SET translocation has been implicated in leukemia, perhaps through the impairment of the protein phosphatase 2A inhibitory activity of SET (13, 14).
Herein, we describe experiments to molecularly characterize the basis for an unusual posttranslational cleavage event in the biogenesis of Nup98 and Nup96. Nup98 and Nup98–Nup96, the polyprotein product of an alternatively spliced mRNA, have both been recently shown to be proteolytically cleaved in a fashion similar to the Saccharomyces cerevisiae Nup98–Nup96 homologue Nup145p (15–18). The Nup98 cleavage occurs 6 kDa from the C terminus of the protein, generating two fragments, one approximately 92 kDa (here termed Nup98-N) and one 6 kDa (here termed Nup98-C) (Fig. 1A). Similarly, cleavage of the Nup98–Nup96 polyprotein generates two bona fide nups, consisting of Nup98-N and a 96-kDa C-terminal fragment (15). Likewise, the cleavage of Nup145p in S. cerevisiae generates two nups, a 65-kDa N-terminal fragment, Nup145-N, and an 80-kDa C-terminal fragment, Nup145-C (16). Interestingly, the Nup98–Nup96 cleavage has been shown to be essential for the assembly of both Nup98-N and Nup96 into the NPC (15). We investigated the basis of the cleavage event to gain insight into this unusual chemical step that appears to be involved in NPC assembly. Surprisingly, we have determined that no protease is required for the specific cleavage of Nup98. Instead, Nup98 is able to cleave itself through the unique chemical reactivity of an enzyme-like active site that includes the tripeptide HFS at amino acids 862–864. As such, Nup98 is the founder of a new subset in the class of proteins able to carry out autoproteolysis, which also includes hedgehog, the inteins, and the N-terminal nucleophile (ntn) proteins.
Figure 1.
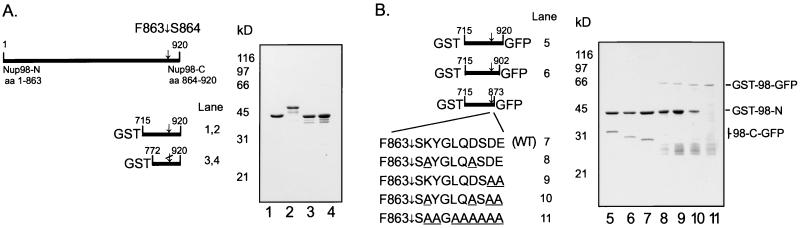
A minimal domain for Nup98 cleavage. (A) Full-length Nup98 and N-terminal deletions are represented at left, the cleavage site is indicated (↓), as are the N and C termini. The deletions were made in either wild-type Nup98 or a mutant Nup98 that is unable to cleave. Purified proteins were separated by SDS/12.5% PAGE and visualized with Coomassie blue R-250. The wild-type and mutant constructs of Nup98 (715–920) are in lanes 1 and 2, and the wild-type and mutant constructs of Nup98 (772–920) are in lanes 3 and 4. (B) N-terminal deletions (lanes 5–7) and site-directed mutants (lanes 8–11) are represented at left. The cleavage site and termini are represented, and the region of the C terminus that was subjected to mutagenesis is expanded. Mutagenized amino acids are underlined. Proteins were purified from similar culture volumes, separated by SDS/12.5% PAGE, and visualized with Coomassie blue R-250. GST, glutathione S-transferase; GFP, green fluorescent protein.
MATERIALS AND METHODS
Molecular Biology Methods.
Standard protocols were followed for PCR and cloning (19). Wild-type Nup98 and a cleavage-deficient mutant (Nup98 (F863S/Y866R)) (15) were subcloned by using unique SalI/NotI sites from myc-pAlter-MAX into pGEX 4T-2 (Amersham Pharmacia Biotech). Deletion mutants were cloned into pGEX 5X-1 (Amersham Pharmacia Biotech). The GFP gene was subcloned from pYX242-GFP (19) into pGEX 5X-1. Site-directed mutants were generated by overlap extension PCR (20). Plasmids were sequenced at the Rockefeller University Protein/DNA Technology Center (New York, NY) to ensure that they were free of spurious mutations.
Protein Methods.
Protein expression from the pGEX plasmids was performed following the manufacturer’s instructions with minor modifications. For immunoblotting, whole cell extracts were prepared by centrifugation of the induced culture and resuspension of cell pellets in urea/SDS sample buffer followed by sonication. Alternatively, for protein purification, cell pellets were resuspended in 0.05 culture volumes of 50 mM Hepes, 150 mM NaCl (pH 8) followed by sonication. Cleared lysates containing overexpressed proteins were incubated in batch with 0.01 vol of packed glutathione (GSH)-Sepharose (Amersham Pharmacia Biotech). Following 15 min at room temperature, or 1 hr at 4°C, the resin was washed in batch with resuspension buffer and eluted with 10 mM reduced GSH in resuspension buffer. SDS/PAGE gels were run, and proteins were visualized with Coomassie blue R-250 or transferred to nitrocellulose and probed with Abs following standard protocols (19). For the immunodetection of GST-containing fusion proteins, the monoclonal anti-GST antibody B14 (Santa Cruz Biotechnology) was used at 0.04 μg/ml. Protein sequencing and mass spectral analysis were performed at the Rockefeller University Protein/DNA Technology Center (21, 22).
In Vitro Cleavage of Nup98 (H862Q/S864C).
Nup98 (H862Q/S864C) was purified at 4°C as described above. Directly after elution with GSH, free GSH was removed by using a microspin G-50 column (Amersham Pharmacia). To initiate cleavage, 0.1 vol of a 10× concentrated solution of either DTT or hydroxylamine was added, and the mixture was shifted to room temperature.
RESULTS
Heterologous Expression and Minimal Domain for Nup98 Cleavage.
Previously, the cleavage of Nup145p was shown to occur in vivo in S. cerevisiae (18). Bacterial expression of a fragment of Nup145p predicted to be 97 kDa instead resulted in the production of a 70-kDa protein, possibly indicating that the cleavage of Nup145p may also occur in bacteria (17). In addition, the cleavage sites of both Nup98 and Nup145p have been determined (15, 16). No previous investigations have directly addressed which protease, if any, was involved in the cleavage of Nup98 or Nup145p. As a first step to determining whether a protease was responsible for the observed cleavage of Nup98, wild-type Nup98 and an uncleavable mutant (15) were expressed as fusion proteins with GST in Escherichia coli. A slight migration difference on SDS/PAGE between these proteins suggested that both the cleavage of Nup98 between residues 863 and 864 (see schematic, Fig. 1) and its abrogation by mutation were maintained in bacteria (data not shown). In addition, the removal of residues 1–714 from both of these proteins preserved the approximately 6-kDa migration difference (Fig. 1A, lanes 1 and 2). Surprisingly, removal of an additional 57 N-terminal residues from each protein produced fusion proteins with identical mobilities (Fig. 1A, lanes 3 and 4). The identical mobilities of the wild-type and mutant proteins suggested that the wild-type protein was no longer able to cleave, even though it contained 92 aa N-terminal to the cleavage site. Such a protein, with at least 57 aa on either side of the cleavage site would be expected to contain sufficient information for any bacterial protease. As this protein was not cleaved, it did not appear likely that a protease was involved.
To ensure that Nup98 was indeed being cleaved in E. coli and to map the required region of Nup98-C, Nup98 (amino acids 715–920) was cloned into an expression vector in-frame with GST upstream and green fluorescent protein (GFP) downstream. In this manner, the cleaved C-terminal domain would be over 30 kDa, as opposed to the 6 kDa of Nup98-C alone, and would be identifiable by immunoblotting for GFP. As expected, both fragments of Nup98 (715–920) could be identified by probing whole cell lysates blotted to nitrocellulose with Abs to the respective N- and C-terminal fusion partners, whereas almost no full-length product was observed (data not shown). Surprisingly, GSH-Sepharose retained not just the N-terminal, GST-containing fragment, but also a protein with the expected mobility of the cleaved C-terminal fragment (Fig. 1B, lane 5). That this 32-kDa protein was indeed Nup98-C-GFP was confirmed by N-terminal sequencing. The sequence thus generated, SKYGL, matched the N terminus of Nup98-C. Importantly, the sequencing data also confirmed that the cleavage observed in the mammalian protein (15) was precisely recapitulated in E. coli. The binding of the two fragments of Nup98 was confirmed by cloning Nup98 (715–920) downstream of maltose-binding protein. In this case, both fragments were retained on an amylose column. We therefore conclude that Nup98-C binds to Nup98-N. Additionally, amino acids 715–863 of Nup98-N are sufficient to bind Nup98-C. Further, because the N-terminal 51 (out of a total of 57) amino acids of Nup98-C are identical to the corresponding residues of Nup96, Nup98-N most likely binds Nup96 as well.
We then endeavored to more finely map Nup98-C. As shown schematically in Fig. 1B and in lanes 5–7, Nup98-C could be reduced from 57 aa and 6 kDa to 10 aa and 1 kDa with no evident decrease in cleavage activity or Nup98-N binding. Mutants were then made to examine the contributions of individual, highly conserved amino acids (for sequence alignment, see Fig. 2). The combined K865A/D871A double mutant was still able to cleave, although with less efficiency as indicated by the purification of uncleaved GST-Nup98-GFP (Fig. 1B, compare lane 8 to lane 7). Similarly, D873A/E874A was also able to cleave, though not as efficiently as wild type (Fig. 1B, compare lane 9 to lane 7). In addition, although the Nup98-C-GFP still copurified with GST-Nup98-N, its affinity was evidently reduced for both of these mutants and could be clearly detected only by immunoblotting (data not shown). When these two double mutants were combined, the resulting mutant, with four of the nine amino acids following S864 changed to Ala, was still able to cleave, though again with reduced efficiency (Fig. 1B, lane 10). Finally, when all of the non-Gly resides following S864 were changed to Ala, no cleavage was observed (Fig. 1B, lane 11). As such, it appears that the nine residues following S864 are coordinately involved in both the cleavage and the binding of the two fragments.
Figure 2.
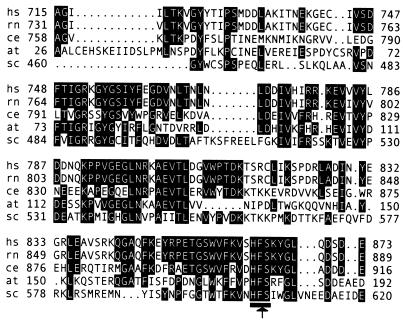
The minimal cleavage domain of human Nup98 is evolutionarily conserved. The corresponding regions of the human (hs), rat (rn), Caenorhabditis elegans, A. thaliana, and S. cerevisiae proteins are aligned (GenBank accession numbers U41815, P49793, AAA91249, AAD32891, and P49687). Residues common to at least three of the organisms are highlighted. The cleavage site and HFS tripeptide are indicated.
The Minimal Domain for Cleavage Is Highly Conserved.
A BLAST search (23) was performed to identify proteins with similarity to the minimal cleavage domain of Nup98, amino acids 715–873. Homologous proteins, all predicted to be nups, were identified in several species. In Fig. 2, the cleavage domain from the Nup98 homologues from human, rat, Caenorhabditis elegans, Arabidopsis thaliana, and S. cerevisiae are aligned, with residues common among at least three of the five proteins highlighted. These proteins share a high degree of sequence similarity throughout this domain, including absolute conservation of the HFS tripeptide around the cleavage site (Fig. 2, underlined). Relative to the human protein, the domains from rat, C. elegans, A. thaliana, and S. cerevisiae are 98%, 43%, 40%, and 36% identical, respectively. Other nucleoporins with similarity to this domain only upstream of the cleavage site were also identified, for example Nup116p of S. cerevisiae, which is 29% identical and 50% similar to the Nup98 cleavage domain over 143 aa.
The absolute conservation of the HFS tripeptide suggests that it is essential for some aspect of the common functions of this protein class, and is consistent with the conserved biogenesis mechanism that leads to scission of the F/S peptide bond. In this light, it was recognized that, although no overall sequence similarity could be seen between these nucleoporins and other proteins that are processed autoproteolytically, HX(S/C/T) motifs, where cleavage occurs between X and the hydroxyl- or thiol-containing amino acids, are present at C-terminal cleavage sites of many inteins and at the cleavage sites of several ntn proteins (for reviews, see refs. 24 and 25). As such, we characterized mutants in this region to further establish the autoproteolytic mechanism and explore the mechanistic similarities between Nup98 and previously described autoproteolytic proteins.
Mutants in the HFS Motif: Nup98 Amino Acids 862–864.
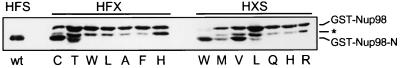
Peptide bond hydrolysis in hedgehog proteins, the inteins, and ntn proteins is, in large part, the result of unique reactivity of the S, C, or T following the scissile amide bond (for review see refs. 24 and 25). In a key early step in the cleavage of these proteins, the hydroxyl- or thiol-containing amino acid is involved in a reversible N→O (or N→S) acyl shift, forming an intermediate with a (thio)ester replacing the X-S (or C/T) peptide bond. Such acyl shifts have been well characterized and, in many cases, are quite facile (26–28). The (thio)ester intermediate is then attacked by water (or, in the hedgehog case, cholesterol), leading to hydrolysis (and, for hedgehog, the addition of cholesterol). If Nup98 hydrolysis shares a mechanistic framework with that of inteins, ntn proteins, and hedgehog proteins, it would be expected that the HFS tripeptide could be changed to HFC or HFT without completely blocking hydrolysis. Conversely, non-hydroxyl- or non-thiol-containing amino acids at position 864 would not be expected to undergo cleavage. To test the requirements of position 864, we randomized this amino acid in the context of the GST-Nup98 (643–920) fusion protein by overlap extension PCR. Position 864 was randomized with an NNS codon (where N can be any nucleotide, S is either G or C), allowing all 20 amino acids, but only the amber stop codon that is suppressed in the strain used for screening, XL1-Blue. Resulting mutants were screened for SDS/PAGE migration differences, and presumably cleavage differences, by blotting whole cell lysates with Abs specific for GST. Of the 36 mutants screened, 2 cleaved, 6 partially cleaved, and 28 did not cleave. The 8 cleavers and partial cleavers and 8 of the noncleavers were sequenced. The 2 cleavers had S at position 864, whereas the partial cleavers additionally had C or T (or a stop codon, because of incomplete suppression) at this position (Fig. 3). As in other autoproteolytic proteins, replacement of the wild-type nucleophilic residue with other possible nucleophiles decreased the efficiency of cleavage of Nup98 (compare Fig. 3, lanes wt with C and T).
Figure 3.
Mutations in the amino acids immediately preceding and following the Nup98 cleavage site. The wild-type protein is in the first lane, followed by seven mutants in amino acid 864 and seven mutants in amino acid 863. The cleavage-site tripeptide is above the lanes, where X is the amino acid directly below the lane. A whole cell extract was prepared, and proteins were separated by SDS/10% PAGE. GST-containing fusion proteins were visualized by immunoblotting. The band between the uncleaved and cleaved proteins (∗) most likely corresponds to a faster migrating conformation of the uncleaved protein.
We analyzed the purified wild-type, S864C, and S864L proteins by matrix-assisted laser desorption ionization time-of-flight MS, taking advantage of our observation that Nup98-C binds to Nup98-N (Fig. 1B). The wild-type C terminus gave a peak at m/z 6165.82, comparing favorably with the predicted Nup98-C mass of 6168.88. The S864C mutant gave a peak at m/z 6183.71, in accordance with the predicted mass of 6184.95. In this manner, we confirmed the sequence of the mutant and that the mutant cleaves in the same location as the wild-type protein. Further, as no other peaks in this area were observed, it appears that the band between the uncleaved and cleaved proteins (Fig. 3, ∗) corresponds to a faster migrating, uncleaved species. This band also decreased in intensity, in concert with the uncleaved protein, under prolonged incubation of the S864C mutant, also indicating that it is a conformational or covalent isomer of the uncleaved protein. As expected, the S864L mutant gave no peaks in the region of m/z 6000, agreeing with its observed slower migration on SDS/PAGE and the conclusion that it is unable to cleave.
Next, the importance of the conserved F863 was probed by randomization and screening in a manner analogous to that for S864. As was the case for amino acid 864, only a minority of the amino acid 863 mutants were able to cleave, some of them only slightly (Fig. 3). As seen in Fig. 3, F863 could be replaced by W with only a small decrease, or by M, L, or V with a greater decrease in cleavage efficiency. The phenyl side chain of F863 is not likely to participate in the chemistry of the cleavage, yet the cleavage activity of Nup98 seems to require a large hydrophobic residue at this position. This circumstance is reminiscent of that in Flavobacterium glycosylasparaginase, which contains an HDT cleavage site. In that case, the interaction between D151 and R180 may act to twist the D151–T152 amide bond into a configuration that favors attack by the T152 side chain (29). Another example of amide bond twist in autoproteolysis is found at the N-terminal cleavage site of the intein GyrA. The amide bond between A0 and S1, corresponding to the scissile bond in the wild-type protein, was in the cis conformation in the recently determined crystal structure of a GyrA mutant (30).
In Vitro Cleavage of a Nup98 Mutant.
Our results with regard to a minimal domain needed for cleavage and activities of mutants in F863 and S864 are consistent with an autoproteolytic mechanism for Nup98 cleavage; however, it was possible that we were characterizing the substrate preferences of a highly unusual protease. To completely rule out the involvement of a protease in the cleavage of Nup98, we sought to generate either a slow-cleaving mutant or one whose cleavage would be triggerable under defined conditions. In either case, the goal was to construct a mutant whose cleavage properties were completely incompatible with cleavage by an external protease. In hedgehog proteins and the inteins, a His distant from the cleavage site is completely conserved throughout the class whose mutagenesis disrupts cleavage (31, 32). In the case of the GyrA intein and hedgehog from Drosophila, this His, H75 and H329, respectively, is hydrogen bonded to the amide nitrogen of the scissile peptide bond (30, 33). In the case of Nup98, examination of closely related proteins does not readily identify completely conserved amino acids that are likely to function as general acids or bases (see Fig. 2). Indeed, we were unable to identify mutants of highly conserved amino acids distant from the cleavage site that led to complete loss of cleavage activity. Instead, mutations in highly conserved amino acids led to either mostly insoluble, uncleaved protein, and small amounts of soluble, cleaved protein (S729A/D731A, R752A/K753A, Y755A/S757A, D772A/D773A), or no apparent decrease in cleavage (S746A/D747A) (data not shown).
The uncleaved S864C and S864T mutants (Fig. 3) were stable enough to be purified, but only at low temperature when the purification was completed as quickly as possible. Even so, the uncleaved proteins were not readily purified away from the cleaved proteins. As a result, it was difficult to study the cleavage of the precursor in the presence of a large quantity of cleaved protein.
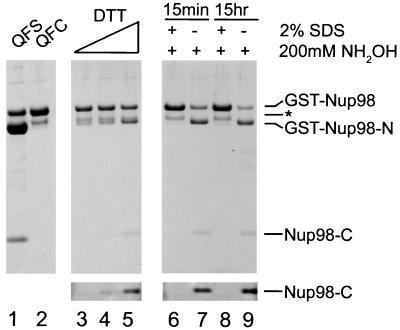
We therefore sought additional mutants that might allow us to purify uncleaved protein. It had been previously reported that mutation of the HNC motif of the sce VMA intein to QNC leads to a protein that cleaves very slowly in the absence of exogenously added thiols (34). We therefore made both the H862Q and the combined H862Q/S864C mutants of Nup98. As shown in Fig. 4, the mutation of the conserved His to Gln only slightly decreased the cleavage activity of the otherwise wild-type protein (lane 1). This is somewhat remarkable given the proposed role of this residue, in analogy to that in the Flavobacterium glycosylasparaginase, which is to deprotonate the nucleophilic Thr (29). The mutability of this motif seems to be evidence for a multifactorial basis of the cleavage activity. Such a multifactorial basis for catalysis is common in proteases; for example, subtilisin is still able to catalyze the hydrolysis of anilide substrates by a factor of over 103, even after all of the catalytic triad residues are mutated to Ala (35). When the H862Q mutation was combined with S864C, the intrinsic rate of cleavage was slow relative to the time scale of protein purification. As a result, we could purify the uncleaved protein without a substantial quantity of cleaved protein (Fig. 4, lane 2). The purified double mutant was subjected to an increasing concentration of DTT. As expected, more cleavage was observed with an increasing concentration of DTT (Fig. 4, lanes 3–5). DTT-mediated cleavage occurred at both pH 6.2 and pH 8 (data not shown).
Figure 4.
Cleavage of H862Q-containing mutants. Purified Nup98(H862Q) and Nup98(H862Q/S864C) are in lanes 1 and 2. Nup98(H862Q/S864C) was incubated at ambient temperature for 20 hr with 25, 50, and 200 mM DTT (lanes 3, 4, and 5, respectively). SDS-denatured and native Nup98(H862Q/S864C) were incubated with 200 mM NH2OH at room temperature for 15 min (lanes 6 and 7) and 15 hr (lanes 8 and 9). Additionally, the contrast of the lower region of lanes 3–9 was enhanced (Lower) to facilitate detection of Nup98-C. Proteins were separated by SDS/4–12% NuPAGE (NOVEX, San Diego) and visualized with Coomassie blue R-250.
Most striking is the cleavage of the QFC mutant mediated by hydroxylamine, a nucleophilic reagent too mild to attack amides, but very reactive with thioesters (36). Purified QFC mutant was first incubated at room temperature in the presence or absence of 2% SDS for 10 min. Next, hydroxylamine was added to 200 mM to both the SDS-denatured and untreated samples. Within 15 min, the untreated sample was mostly cleaved, whereas the denatured sample appeared unchanged (Fig. 4, lanes 6 and 7). After overnight incubation, neither the native nor the previously denatured sample was cleaved appreciably more than after 15 min (Fig. 4, lanes 8 and 9). To confirm that the hydroxylaminolysis occurred at the corresponding cleavage site in the wild-type protein, the 6-kDa product was sequenced. As expected, the N-terminal sequence generated, XKYGLQ, did correspond to amino acids 864–869 of Nup98. Taken together, these results indicate that the QFC mutant is still able to efficiently complete the first step of cleavage, namely the N→S acyl shift, but the rate of hydrolysis of the resulting thioester is slow relative to the competing (unproductive) S→N acyl shift. Hydroxylamine therefore acts to trap the transient thioester intermediate, as hydroxylaminolysis is irreversible under the conditions used. Additionally, consistent with the case in other autocleaving proteins (29), no accumulation of thioester intermediate is seen, as indicated from the inability of hydroxylamine to cleave SDS-denatured protein. These results further exclude the involvement of a protease in the cleavage reaction.
Cleavage of the hedgehog proteins occurs not via hydrolysis of the thioester intermediate, but by alcoholysis (37). We cannot currently rule out such a reaction in the processing of Nup98 in situ, but the efficiency of wild-type Nup98 cleavage in bacteria would seem to indicate that competing reactions would be unlikely to compete kinetically with hydrolysis. This situation can be compared with the case of Drosophila hedgehog, which, when expressed in E. coli, slowly cleaves in the absence of cholesterol or high concentrations of DTT (33).
An Additional Motif for Autoproteolysis.
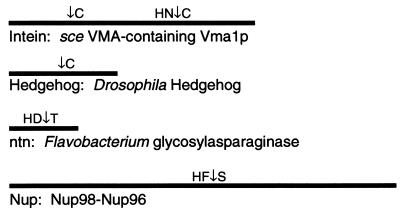
Previously, three classes of proteins have been shown to be capable of proximal or cis autoproteolysis (for reviews, see refs. 24 and 25). None of these proteins have been shown to be capable of acting on cleavage site-containing peptides added in trans. By virtue of the similarities we have observed between these proteins and Nup98, with regard to cleavage in heterologous systems, domains required for cleavage, cleavage site sequences, and behavior of mutants in these cleavage-site amino acids, we suggest that Nup98 is an additional member of this class of proteins. Proteins previously shown to have autoproteolysis as an essential step in their biogenesis, and the Nup98–Nup96 polyprotein described here, are represented in Fig. 5. Several classes of proteins capable of autoproteolysis by divergent mechanisms exist, but will not be discussed here (see for example refs. 38 and 39).
Figure 5.
Proteins capable of cis-autoproteolysis. One protein from each autoproteolytic subgroup is schematically represented. Cleavage sites (↓) and adjacent amino acids are indicated.
The inteins are unique in that they undergo two sequential cleavages involving a branched intermediate and a ligation that fuses the previously uncontiguous domains. The C-terminal domain of the hedgehog proteins bears sequence and structural similarity to the inteins and contains all of the machinery needed for cleavage (30, 33, 40). As a result, the cholesterol modification/cleavage of hedgehog proteins is mechanistically similar to the N-terminal cleavage of the inteins. This cleavage site does not have an analogously conserved His, as observed in the Nup98 case (see Fig. 5). However, the C-terminal cleavage site of inteins does have such a His, and this cleavage may more closely resemble that of Nup98, except that the intein C-terminal residue is almost always Asn, which has been shown to cyclize to form a succinimide as part of the cleavage mechanism (41). No analogous residue exists in the Nup98 homologues, and no succinimide formation is predicted to occur on Nup98 cleavage. The Nup98 motif would therefore seem to most closely resemble the ntn motif. Not only are these two classes of proteins similar in their cleavage site and presumed mechanism (as discussed above; see also Fig. 6), but they also share the property that amino acids N- and C-terminal to the cleavage site are necessary for processing. This is in stark contrast to the case for hedgehog C termini and the inteins. Perhaps by virtue of their nature as mobile elements (42) or lipophilic modification factors (37), they do not generally require involvement of residues of hedgehog N termini or the exteins.
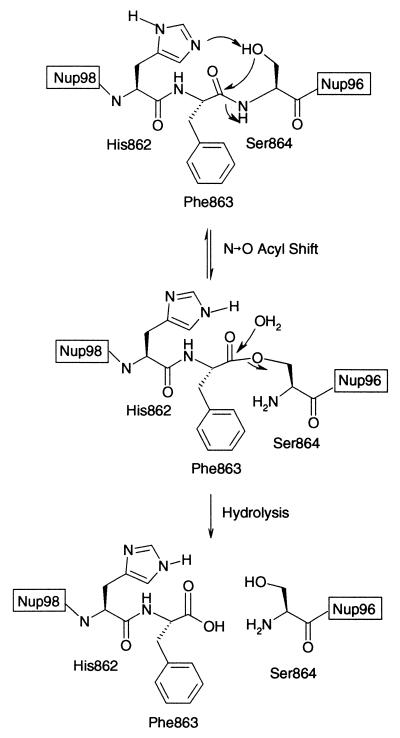
Figure 6.
The proposed mechanism for the autoproteolysis of Nup98 and Nup98–Nup96. (The tetrahedral hydroxyoxazolidine intermediate between the precursor and the ester has been omitted for clarity.)
The proposed Nup98 cleavage mechanism, based on our observations and by analogy to the cleavage mechanism for other autoproteolytic proteins, is summarized in Fig. 6. In the first step, the penultimate residue of Nup98-N, H862, is involved in the deprotonation of the N-terminal residue of Nup96 (or Nup98-C) S864. By virtue of interactions of F863 with a hydrophobic binding site in Nup98, the F863–S864 amide bond is twisted, allowing the attack of the S864 hydroxide on the preceding carbonyl group. The outcome of this attack is an N→O acyl shift, resulting in an ester, rather than an amide, between Nup98-N and Nup96 (or Nup98-C). This ester is then hydrolyzed, with H862 possibly acting as a general base.
The Importance of Autoprocessing for Nup98 Function.
Previously, it was observed that an uncleavable Nup98–Nup96 precursor was imported into the nucleus but not properly assembled into NPCs, whereas Nup96 expressed alone was localized in the cytoplasm (15). Also, when Nup98-N was expressed, it was able to assemble into NPCs, even though it was not the product of cleavage of the full-length precursor (15). These observations could be interpreted to implicate a localized proteolysis in the processing of Nup98–Nup96. Proteolysis may occur in the nucleus, where processing of the Nup98–Nup96 precursor could lead to the assembly of both nups into the NPC. Alternatively, a cytoplasmically localized proteolysis may be thought to be consistent with the proper localization and assembly of Nup98-N and the inability to observe an uncleaved precursor in vivo in a pulse–chase experiment (15). In any case, if a protease were involved in the cleavage, it may have been expected to be differentially localized.
Our results strongly indicate that there is no protease (nuclear or otherwise) involved in the Nup98 or Nup98–Nup96 cleavage. What then is the purpose of this processing event? It is highly unlikely that the autoprocessing is used in a manner analogous to a differentially localized protease. Whereas it should not be difficult to localize a protease, it would seem to be very difficult to compartmentalize the autoproteolysis activity that we have observed. The autoproteolysis is very rapid for the wild-type protein both in E. coli and in situ (15). Rapid cleavage is common throughout the class of autoproteolytic proteins, where uncleaved wild-type proteins are seen only in the most unusual cases, for example when an intein from a thermophilic organism was expressed and characterized far below its optimal temperature (43). It remains possible that a negative regulator of autoproteolysis exists, but such a regulator would be restricted to functioning in a very narrow time window.
It is more likely that the autoproteolysis mechanism is used for the twofold purposes of precisely regulating Nup98-N/Nup96 stoichiometry and delivering both Nup98-N and Nup96 to the NPC. The stoichiometry of Nup98-N/Nup96 is strictly controlled, as no Nup96 is synthesized unless it is fused to Nup98-N. Why might it be important to control Nup98-N/Nup96 stoichiometry? One possibility is that they form a functionally relevant, yet low-affinity complex, analogous to that seen here, that forms only at high local concentrations of the components (see Fig. 1B, lanes 5–7, and Fig. 4, lane 1). As Nup96 is not able to localize to the nucleus on its own, it is possible that it is targeted to the nucleus by virtue of its interaction with Nup98-N. In the case of the alternatively spliced mRNA that does not contain Nup96, it is then possible that Nup98-C has a function distinct from Nup98-N either in the nucleus or at the NPC.
A subcomplex of the NPC containing Nup96, Nup107, and at least two Sec13-related proteins has been isolated from rat liver; similarly, a subcomplex of Nup145-C, Nup120p, Nup84p, Nup85p, Seh1p, and Sec13p has been isolated from yeast (15, 16). Neither of these subcomplexes contains Nup98-N/Nup145-N. Therefore, it would appear that the interactions between Nup98-N and Nup96 (or Nup145-C and -N) are stable enough for targeting the complex of nups to the NPC, yet labile enough to allow different complexes containing these nups to be formed at the NPC. This may be a general theme in NPC assembly, accentuating the dynamic nature of this supramolecular structure. In addition, the complex formation and targeting that appear to depend on cleavage likely require properly folded nups. In this light, the cleavage event could additionally be seen as a mechanism for quality control, ensuring that only properly folded nups are cleaved and targeted to the NPC.
Acknowledgments
We thank T. Muir, B. Ayers, and members of the Blobel lab for helpful discussions; E. Ellison for technical assistance; and B. Fontoura for the myc-pAlter-MAX Nup98 constructs. We also thank B. Cravatt, B. Fontoura, T. Muir, L. Pemberton, and M. Rout for constructive comments on the manuscript. J.S.R. was supported by a National Institutes of Health postdoctoral fellowship. Protein sequence analysis was provided by the Rockefeller University Protein/DNA Technology center, which is supported in part by National Institutes of Health shared instrumentation grants and by funds provided by the U.S. Army and Navy for purchase of equipment.
ABBREVIATIONS
- NPC
nuclear pore complex
- nup
nucleoporin
- ntn
N-terminal nucleophile
- GFP
green fluorescent protein
- GSH
glutathione
- GST
glutathione S-transferase
References
- 1.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 2.Pemberton L F, Blobel G, Rosenblum J S. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 3.Wozniak R W, Rout M P, Aitchison J D. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 4.Adam S A. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- 5.Moore M S. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 6.Rexach M, Blobel G. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 7.Radu A, Moore M S, Blobel G. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 8.Gant T M, Goldberg M W, Allen T D. Curr Opin Cell Biol. 1998;10:409–415. doi: 10.1016/s0955-0674(98)80018-5. [DOI] [PubMed] [Google Scholar]
- 9.Kiseleva E, Goldberg M W, Allen T D, Akey C W. J Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T, Yamazaki Y, Hatano Y, Miura I. Blood. 1999;94:741–747. [PubMed] [Google Scholar]
- 11.Kasper L H, Brindle P K, Schnabel C A, Pritchard C E, Cleary M L, van Deursen J M. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu G K, Grosveld G, Markovitz D M. Proc Natl Acad Sci USA. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Makkinje A, Damuni Z. J Biol Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 15.Fontoura B M, Blobel G, Matunis M J. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teixeira M T, Siniossoglou S, Podtelejnikov S, Benichou J C, Mann M, Dujon B, Hurt E, Fabre E. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabre E, Boelens W C, Wimmer C, Mattaj I W, Hurt E C. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 18.Emtage J L, Bucci M, Watkins J L, Wente S R. J Cell Sci. 1997;110:911–925. doi: 10.1242/jcs.110.7.911. [DOI] [PubMed] [Google Scholar]
- 19.Rosenblum J S, Pemberton L F, Bonifaci N, Blobel G. J Cell Biol. 1998;143:887–899. doi: 10.1083/jcb.143.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi N, Welschof M, Zewe M, Braunagel M, Dubel S, Breitling F, Little M. BioTechniques. 1994;17:310. , 312, 314–315. [PubMed] [Google Scholar]
- 21.Gharahdaghi F, Kirchner M, Fernandez J, Mische S M. Anal Biochem. 1996;233:94–99. doi: 10.1006/abio.1996.0012. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez J, Andrews L, Mische S M. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Schaffer A A, Miller W, Madden T L, Lipman D J, Koonin E V, Altschul S F. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao Y, Kent S B. Chem Biol. 1997;4:187–194. doi: 10.1016/s1074-5521(97)90287-8. [DOI] [PubMed] [Google Scholar]
- 25.Perler F B, Xu M Q, Paulus H. Curr Opin Chem Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 26.Iwai K, Ando T. Methods Enzymol. 1967;11:263–282. [Google Scholar]
- 27.Dawson P E, Muir T W, Clark-Lewis I, Kent S B. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 28.Tam J P, Lu Y A, Liu C F, Shao J. Proc Natl Acad Sci USA. 1995;92:12485–12489. doi: 10.1073/pnas.92.26.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan C, Liu Y, Shao Y, Cui T, Liao W, Ewel A, Whitaker R, Paulus H. J Biol Chem. 1998;273:9695–9702. doi: 10.1074/jbc.273.16.9695. [DOI] [PubMed] [Google Scholar]
- 30.Klabunde T, Sharma S, Telenti A, Jacobs W R, Jr, Sacchettini J C. Nat Struct Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 31.Pietrokovski S. Protein Sci. 1998;7:64–71. doi: 10.1002/pro.5560070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J J, Ekker S C, von Kessler D P, Porter J A, Sun B I, Beachy P A. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka Hall T M, Porter J A, Young K E, Koonin E V, Beachy P A, Leahy D J. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 34.Chong S, Williams K S, Wotkowicz C, Xu M Q. J Biol Chem. 1998;273:10567–10577. doi: 10.1074/jbc.273.17.10567. [DOI] [PubMed] [Google Scholar]
- 35.Carter P, Wells J A. Nature (London) 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 36.Jencks W, Cordes S, Carriulo J. J Biol Chem. 1960;235:3608–3614. [PubMed] [Google Scholar]
- 37.Porter J A, Ekker S C, Park W J, von Kessler D P, Young K E, Chen C H, Ma Y, Woods A S, Cotter R J, Koonin E V, Beachy P A. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 38.van Poelje P D, Snell E. Annu Rev Biochem. 1990;59:29–59. doi: 10.1146/annurev.bi.59.070190.000333. [DOI] [PubMed] [Google Scholar]
- 39.Blair W S, Semler B L. Curr Opin Cell Biol. 1991;3:1039–1045. doi: 10.1016/0955-0674(91)90126-j. [DOI] [PubMed] [Google Scholar]
- 40.Perler F B. Cell. 1998;92:1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 41.Xu M Q, Comb D G, Paulus H, Noren C J, Shao Y, Perler F B. EMBO J. 1994;13:5517–5522. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gimble F S, Thorner J. Nature (London) 1992;357:301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- 43.Xu M Q, Southworth M W, Mersha F B, Hornstra L J, Perler F B. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]