Abstract
The bacterial envelope stress response (ESR) is triggered by the accumulation of misfolded outer membrane proteins (OMPs) upon envelope damage or excessive OMP synthesis, and is mediated by the alternative sigma factor, σE. Activation of the σE pathway causes a rapid downregulation of major omp mRNAs, which prevents further build-up of unassembled OMPs and liberates the translocation and folding apparatus under conditions that require envelope remodelling. The factors that facilitate the rapid removal of the unusually stable omp mRNAs in the ESR were previously unknown. We report that in Salmonella the ESR relies upon two highly conserved, σE-controlled small non-coding RNAs, RybB and MicA. By using a transcriptomic approach and kinetic analyses of target mRNA decay in vivo, RybB was identified as the factor that selectively accelerates the decay of multiple major omp mRNAs upon induction of the ESR, while MicA is proposed to facilitate rapid decay of the single ompA mRNA. In unstressed bacterial cells, the two σE-dependent small RNAs function within a surveillance loop to maintain envelope homeostasis and to achieve autoregulation of σE.
Introduction
Bacteria respond to unfavourable changes in their environment by inducing specific stress regulons. Alternative sigma (σ) factors play a key role in many stress responses by redirecting RNA polymerase to the promoters of particular stress regulons. Many bacteria possess a specialized sigma factor, σE, that controls aspects of pathogenesis and the development of maximal resistance to various environmental stresses (Rowley et al., 2006). In Escherichia coli and Salmonella, the rpoE-encoded σE protein is present as an inactive membrane-bound precursor in unstressed cells; upon envelope damage, controlled proteolysis cleaves the σE precursor from the membrane to yield cytoplasmic active σE (Ades et al., 1999; Ruiz and Silhavy, 2005). While diverse stresses (e.g. temperature shock, exposure to ethanol and antimicrobial peptides, hyperosmolarity, and entry into stationary phase) are known to induce σE (Rowley et al., 2006), it has been widely assumed that the actual σE-activating signal is the accumulation of misfolded outer membrane proteins (OMPs) in the periplasmic space, as has been reported in exponentially growing bacteria (Mecsas et al., 1993; Missiakas et al., 1996; Raivio and Silhavy, 1999).
The σE-controlled envelope stress response (ESR, also known as extracytoplasmic stress response) involves expression of > 80 transcription units of the E. coli and Salmonella genomes (Rhodius et al., 2006; Skovierova et al., 2006). While some of these genes are species-specific, most members of the σE core regulon act to synthesize and correctly assemble lipopolysaccharides and OMPs, which must be balanced to maintain envelope homeostasis.
Time-course experiments of global transcript changes upon RpoE expression identified the rapid disappearance of multiple omp mRNAs as a hallmark of the σE response (Rhodius et al., 2006). The rapid reduction of synthesis of the major OMPs represents an obvious solution to restore envelope homeostasis, as the decreased flow of OMPs to the envelope prevents further build-up of unassembled OMPs.
The factors that facilitate the rapid shut-off of OMP synthesis upon envelope stress remained unknown but we reasoned that these must involve the acceleration of omp mRNA decay. First, none of the hitherto identified σE-controlled genes (Rhodius et al., 2006; Skovierova et al., 2006; Wade et al., 2006) encodes a known transcriptional repressor of omp genes. Second, transcriptional repression of omp genes would not be an effective way to mediate this rapid response, because many omp mRNAs are unusually stable. For example, the ompA message decays with a half-life of ∼ 17 min in normally growing cells (von Gabain et al., 1983); RpoE expression reduces its half-life to ∼ 5 min (as calculated from Fig. 1 in Rhodius et al., 2006). This level of stability would normally lead to the continuation of OMP synthesis for many minutes from the existing omp mRNAs.
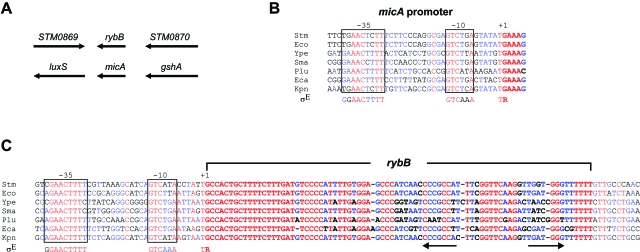
Fig. 1.
Conservation of σE consensus motifs in the promoters of rybB and micA sRNA genes. A. The S. typhimurium micA and rybB genes are located (counterclockwise) in the luxS-gshA and STM0869-STM0870 (ybjK-ybkL) intergenic regions as previously described in E. coli. The black arrows indicate the orientation of the genes. B. Alignment of micA promoter sequences, including the first five nucleotides of the MicA-encoding sequence (in bold) of diverse γ-proteobacteria (Stm: Salmonella typhimurium; Eco: Escherichia coli K12; Ype: Yersinia pestis; Sma: Serratia marcescens; Plu: Photorhabdus luminescens; Eca: Erwinia carotovora; Kpn: Klebsiella pneumoniae). The −10/−35 consensus motifs of σE-controlled promoters are shown below the alignment. C. Alignment of rybB genes, including σE motifs, of the same set of bacterial species as above. The RybB-encoding sequence is shown in bold. Arrows indicate a putative ρ-independent transcription terminator.
How bacterial cells actively degrade a specific set of mRNAs under stressful conditions is poorly understood. However, small non-coding RNAs (sRNAs) have recently emerged as a new class of auxiliary factors that facilitate the recognition of specific mRNAs by the general RNA decay machinery to mediate accelerated turn-over in response to adverse growth conditions. For example, iron starvation induces expression of the 90 nt RyhB sRNA, which then acts on the trans-encoded sodB mRNA to trigger its decay in a RNase E-dependent fashion (Massé and Gottesman, 2002; Masséet al., 2003). Similarly, the phosphosugar stress-induced 200 nt SgrS sRNA accelerates the turn-over of ptsG mRNA (Vanderpool and Gottesman, 2004; Morita et al., 2005).
About a third of the hitherto characterized enterobacterial sRNAs have been shown to target individual omp mRNAs (Guillier et al., 2006; Vogel and Papenfort, 2006), but this observation had not been linked to the σE-controlled ESR. We began to make this connection because mutations of the bacterial RNA chaperone, Hfq, caused misregulation of major OMPs (Vytvytska et al., 1998; 2000; Ding et al., 2004; Sittka et al., 2006) and led to chronic induction of the ESR (Ding et al., 2004; Sittka et al., 2006). Hfq binds a variety of sRNAs and promotes their interaction with their target mRNAs (Valentin-Hansen et al., 2004), and we predicted that one or more σE-regulated sRNAs would target multiple major omp mRNAs.
We have now identified two sRNAs, RybB and MicA, which act within the ESR of Salmonella typhimurium. Transcription of the RybB and MicA genes is stringently controlled by the availability of active σE, and the two sRNAs are proposed to serve dual functions. First, RybB facilitates the rapid removal of an unprecedented number of major omp mRNAs upon induction of the σE pathway. Second, in unstressed cells, RybB and MicA form an autoregulatory loop with the σE regulon that limits OMP biogenesis to prevent the accumulation of misfolded intermediates. Collectively, the two sRNAs are likely to facilitate the remodelling of the outer membrane upon environmental challenges that require adjustments to the bacterial envelope.
Results
σE controls the expression of RybB and MicA sRNAs
To identify σE-regulated sRNAs, we searched for σE binding motifs (Rhodius et al., 2006; Skovierova et al., 2006) in the promoter regions of the more than 50 sRNA genes predicted in the genome of S. typhimurium (Hershberg et al., 2003; Vogel and Sharma, 2005). This search yielded two candidate genes, rybB and micA. The intergenic location of these sRNA genes is conserved between Salmonella and E. coli with respect to the flanking protein-coding genes (Fig. 1A; Hershberg et al., 2003). The rybB and micA upstream regions each contain almost perfect matches to the −35 (GGAACTTTT) and −10 (GTCAAA) motifs of σE-controlled promoters, and these elements are strongly conserved in rybB and micA genes of other γ-proteobacteria (Fig. 1B and C). In Salmonella the putative rybB−35 and −10 promoter elements each differ from the σE consensus at one position; the putative micA promoter has one (−35) or two (−10) mismatches with the σE consensus boxes. In both genes, the −1 and +1 positions perfectly match the σE consensus.
MicA (also known as SraD) and RybB were previously identified in E. coli as ∼70 nt and ∼80 nt, respectively, RNAs that are detected upon entry into stationary phase (Argaman et al., 2001; Wassarman et al., 2001; Vogel et al., 2003). Similarly, we found that the two sRNAs are only slightly expressed in fast-growing Salmonella enterica serovar Typhimurium cells and strongly accumulate at stationary phase (Fig. 2A). This is consistent with a previous observation that stationary phase Salmonella display an elevated σE response (Testerman et al., 2002). In line with our prediction, RybB and MicA are not detected in a Salmonella rpoE mutant strain that lacks σE (Fig. 2B). However, the sRNAs rapidly accumulate following RpoE expression from the arabinose-inducible plasmid, pBAD-RpoE (Fig. 2C). In addition, envelope stress triggered by exposure to the antimicrobial peptide, polymyxin B (Humphreys et al., 1999), activates the rybB and micA genes in wild-type but not in rpoE mutant Salmonella (Fig. 2D).
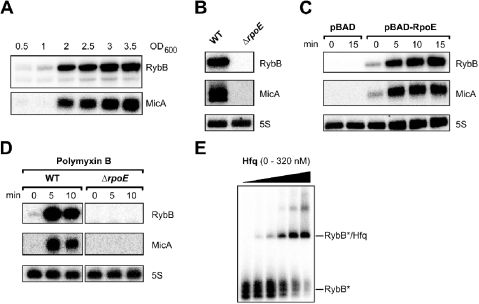
Fig. 2.
Growth phase- and σE-dependent expression of Salmonella rybB and micA. A. Northern blots of total RNA probed for RybB and MicA show that the two RNAs accumulate in stationary phase cells. Samples were taken from Salmonella wild-type cultures throughout growth from exponential to stationary phase at the OD600 values indicated above the panels. RNA extraction, Northern blotting and hybridization were done as described in the Experimental procedures section. B–D. Northern blots show that Salmonella rybB and micA expression is strictly dependent on σE. All blots were probed for 5S rRNA as loading control. B. Wild-type and isogenic rpoE mutant Salmonella were grown into stationary phase (6 h after cells had reached an OD600 of 2). The sRNA signals are lost in the absence of σE. C. The sRNAs rapidly accumulate following RpoE expression. Salmonella carrying either a pBAD control plasmid (pBAD33) or pBAD-RpoE (pAC-rpoEST4) expression plasmid, in which the rpoE gene is cloned under an arabinose-inducible promoter, were grown to late exponential phase (OD600 of 1), that is when rybB and micA are not expressed. Cultures were treated with l-arabinose (0.2% final concentration) to induce RpoE expression. Aliquotes were withdrawn for total RNA preparation prior to (0 min) and at the indicated time-points (5, 10 and 15 min) following induction. The slightly elevated RybB and MicA levels at the 0 min time-point in pBAD-RpoE cells may result from leaky PBAD-rpoE transcription in the absence of l-arabinose. D. The sRNAs are rapidly induced by treatment with the antimicrobial peptide, polymyxin B, in wild-type but not ΔrpoE cells in late exponential phase (OD600 of 1.0). Total RNA was prepared prior to (0 min) and after 5 and 10 min of polymyxin B addition. E. RybB shows high affinity to Hfq protein in vitro. 32P-labelled RybB (4 nM) was incubated in the presence of 1000-fold excess of unlabelled yeast tRNA for 10 min at 37°C with increasing concentrations of purified Salmonella Hfq protein (from left to right: 0, 20, 40, 80, 160, 320 nM), followed by electrophoresis on a native gel. Shown is an autoradiograph of the gel.
MicA sRNA was recently shown to repress OmpA synthesis in E. coli in an Hfq-dependent manner (Rasmussen et al., 2005; Udekwu et al., 2005). RybB is a strongly conserved sRNA (Fig. 1C) of hitherto unknown function that was the second most abundant species in an E. coli sRNA library cloning screen (Vogel et al., 2003). E. coli RybB was also among the highest-scoring sRNAs in Hfq immunoprecipitation experiments (Zhang et al., 2003), suggesting that it acts to regulate mRNAs. We thus tested Salmonella RybB binding to Hfq protein in vitro by gel mobility shift experiments. As shown in Fig. 2E, in vitro synthesized RybB RNA displayed high affinity to purified Salmonella Hfq (Sittka et al., 2006), binding to the protein with an apparent kD of 100 nM, which is similar to the level of Hfq binding of E. coli MicA RNA (Rasmussen et al., 2005). We also found that the in vivo stability of RybB in Salmonella greatly depends on Hfq. RybB decays with a half-life of ∼ 8 min in wild-type Salmonella, which is identical to its E. coli counterpart (Vogel et al., 2003), but its half-life is reduced to less than 1 min in Salmonella deleted for hfq (data not shown). Taken together, these results identified RybB and MicA as excellent candidates for the predicted σE-regulated, Hfq-dependent sRNAs that repress major OMP synthesis upon induction of the ESR.
RybB targets multiple OMP-encoding mRNAs
To determine the putative RybB target mRNAs, the sRNA was cloned on a plasmid under control of an arabinose-inducible PBAD promoter, yielding plasmid pBAD-RybB. Salmonella carrying pBAD-RybB or a pBAD control vector were treated with arabinose in late exponential phase for 10 min, and the resulting global changes in transcript abundance were scored with whole-genome S. typhimurium microarrays. We chose this short pulse expression to only cause changes of those mRNAs with which RybB directly interacts; a similar approach has been used successfully to identify primary targets of several E. coli sRNAs (Masséet al., 2005; Tjaden et al., 2006).
Of the 4716 open reading frames represented on the Salmonella SALSA microarray, 14 transcripts were reduced > threefold, while three mRNAs showed > threefold elevated levels (Fig. 3A and Table S2). On average, the repressed mRNAs exhibited a far higher degree of regulation, and ∼80% of these encoded OMPs. These proteins included the most abundant OMPs of Salmonella, i.e. OmpA, OmpC, OmpD and OmpF (Lee and Schnaitman, 1980), with the ompC and ompD mRNAs showing the strongest repression (22- and 14-fold respectively).
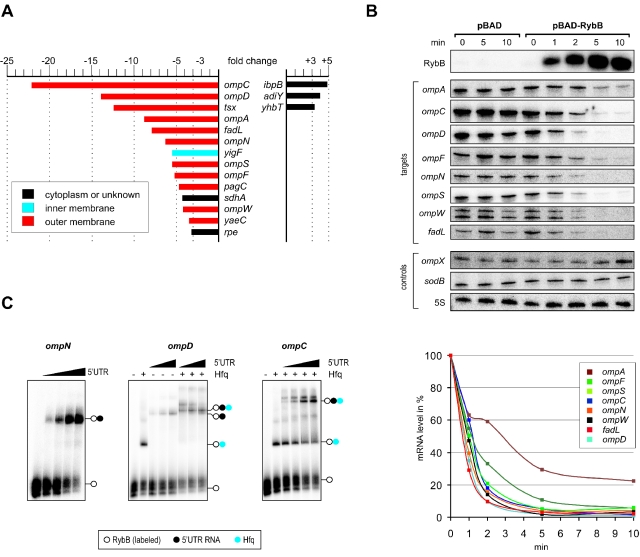
Fig. 3.
RybB targets a large set of mRNAs that encode OMPs. A. Fold changes of mRNA levels after RybB pulse overexpression. Salmonella carrying expression plasmid pBAD-RybB were cultured to an OD600 of 1.5, and RybB expression was induced with l-arabinose for 10 min. Total RNA was extracted and probed on Salmonella SALSA whole-genome microarrays, and data were analysed as described (Nagy et al., 2006). The fold changes given here are normalized to the mRNA expression changes of induced cells that carried a pBAD control vector, to correct for the effects of l-arabinose on global mRNA expression. The chart shows all mRNAs whose levels changed by > threefold, after statistical filtering (see Supplementary material for the entire data set). Bar colours indicate the predicted cellular localization of the encoded proteins. B. Northern blot validation of RybB-induced target mRNA decay. Salmonella carrying either the control pBAD vector or the pBAD-RybB expression vector were arabinose-induced at an OD of 1.5, and total RNA extracted at the time-points indicated above the panels. Northern hybridization with gene-specific probes (indicated to the left) confirmed rapid induction of RybB expression (upper panel), and a concomitant drop in the steady-state levels of eight target mRNAs (ompA/C/D/F/N/S/W, and fadL) in pBAD-RybB cells. The ompX and sodB mRNAs, whose expression did not change in the aforementioned microarray experiments, were probed as controls (panels below). Probing for 5S rRNA confirmed equal RNA loading (lower panel). A quantification of the blot signals obtained for the eight RybB target mRNAs in pBAD-RybB cells is given below. For each mRNA, the signal obtained at the 0 min time-point (prior to induction) was set to 100%. RybB expression reduces the half-life of these targets (except ompA) to ∼1 min. C. Gel-mobility shift assays show that RybB binds to RNA fragments derived from 5′ UTRs of three target mRNAs in vitro. RybB (32P-labelled; 5 nM final concentrations) binding assays with 5′ UTR RNAs were performed in the presence of a 1000-fold excess of yeast tRNA for 10 min at 37°C, followed by electrophoresis on a native gel, of which autoradiographs are shown. (Left panel) RybB was incubated with increasing concentrations of an unlabelled RNA fragment derived from the ompN 5′ UTR (from left to right: 0, 15, 30, 60, 120 nM). The positions of RybB (open circle) or the RybB/ompN complex (filled circle) are indicated to the right. (Middle panel) Gel mobility shifts with an unlabelled RNA derived from the ompD 5′ UTR (final concentration from left to right: 0, 0, 125, 250, 500, 125, 250, 500 nM). Here, Hfq was added to the individual reactions where indicated (+) at a final concentration of 30 nM; a blue circle indicates Hfq-specific mobility changes. (Right panel) Complex formation of RybB with an unlabelled RNA derived from the ompC 5′ UTR (final concentrations from left to right: 0, 0, 12.5, 25, 50, 100 nM). Addition of Hfq and complex formation is indicated as in the other two panels.
To confirm the transcriptomic data, we determined the kinetics of the RybB-mediated downregulation of eight omp target mRNAs on Northern blots (Fig. 3B). RybB expression from plasmid pBAD-RybB resulted in approximately twofold or greater reduction of these target mRNAs within 1 min, and in ≥ 10-fold reduction within 5 min (except ompA, which was reduced by threefold). In contrast, two RybB-independent mRNAs, sodB and ompX, that were probed as controls remained stable or increased. Likewise, arabinose induction of a strain carrying a pBAD control plasmid did not affect the abundance of the RybB targets.
Importantly, the observed downregulation is most likely to result from active mRNA degradation rather than transcriptional repression. Generally, OMP-encoding mRNAs are known to be unusually stable, e.g. the usual half-lives of the ompC and ompD mRNAs under this growth condition are 10 min and > 20 min respectively (Sittka et al., 2006); the action of RybB reduced these half-lives to ∼1 min (Fig. 3B). We hypothesized that RybB binds to 5′ UTR of its targets, which could block translation initiation and destabilize these mRNAs, as it was previously established for other bacterial antisense RNAs (Storz et al., 2004). To test this, RybB/omp 5′ UTR complex formation was assayed in vitro. Figure 3C shows that RybB readily forms complexes with the ompN 5′ UTR fragment, resulting in a nearly complete shift of RybB with ∼60 nM ompN 5′ UTR RNA. RybB complex formation was considerably weaker with the ompC and ompD 5′ UTRs, but was greatly enhanced when RybB was pre-incubated with 30 nM Hfq. While the precise RybB binding sites on its targets remain unknown, these experiments suggest that RybB promotes the decay of multiple major omp mRNAs by direct interaction with their 5′ UTRs, and that this regulation is Hfq-dependent.
Limited role of other OMP-regulatory sRNAs in the ESR
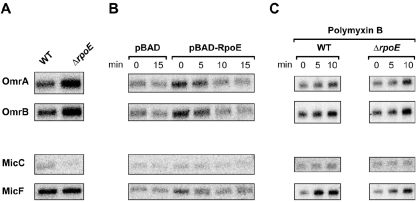
MicA, the other σE-regulated sRNA we had identified, was known to repress ompA mRNA in E. coli (Rasmussen et al., 2005; Udekwu et al., 2005). We used our transcriptomic approach to identify MicA mRNA targets in Salmonella and confirmed ompA repression, as well as showing that no other omp mRNA is regulated by MicA (K. Papenfort et al., unpubl. results). In addition to MicA, five sRNAs – MicC/F, OmrA/B and RseX – were previously shown to regulate omp mRNAs in E. coli (Mizuno et al., 1984; Chen et al., 2004; Douchin et al., 2006; Guillier and Gottesman, 2006). Several of these sRNAs repressed omp mRNAs that we found here to be targets of RybB. Specifically, both MicC and RseX were shown to act on ompC (Chen et al., 2004; Douchin et al., 2006), while MicF was known to target ompF in E. coli (Mizuno et al., 1984). By testing their Salmonella homologues for σE-dependent expression as described above, we found that these sRNAs were not members of the σE regulon (Fig. 4). Collectively, this pointed to RybB as the major facilitator of omp mRNA repression in the ESR.
Fig. 4.
The Salmonella omrA, omrB, micC and micF sRNA genes are not members of the σE regulon. The same Northern blots as in Fig. 2B– D were probed for the four SalmonellasRNAs. A. Wild-type and isogenic rpoE mutant Salmonella were grown into stationary phase (6 h after cells had reached an OD600 of 2). B. Salmonella carrying either a pBAD control plasmid (pBAD33) or pBAD-RpoE (pAC-rpoEST4) expression plasmid, in which the rpoE gene is cloned under an arabinose-inducible promoter, were grown to late exponential phase (OD600 of 1). Cultures were treated with l-arabinose (0.2% final concentration) to induce RpoE expression. Aliquots were withdrawn for total RNA preparation prior to (0 min) and at the indicated time-points (5, 10 and 15 min) following induction. C. RNA samples were taken from polymyxin B-treated wild-type and ΔrpoE cells in late exponential phase (OD600 of 1). Total RNA was prepared prior to (0 min) and after 5 and 10 min of polymyxin B addition. In none of these samples, we were able to detect an RseX-specific signal.
RybB accelerates global omp mRNA decay in the ESR
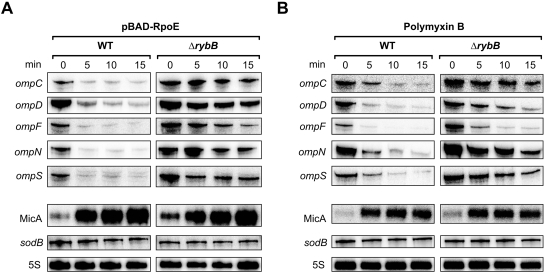
To determine the impact of the chromosomal rybB gene on omp mRNA decay in the ESR, we followed the changes of five RybB target mRNAs in wild-type and ΔrybB cells over time upon pBAD-RpoE expression. In wild-type Salmonella, RpoE expression caused a rapid decay of all these targets (Fig. 5A). Quantification of the Northern blot signals revealed a > 80% decrease within 5 min of RpoE induction, consistent with the fast disappearance of E. coli omp mRNAs observed by Rhodius et al. (2006). In contrast, target mRNA decay was delayed and incomplete in the rybB mutant, and exhibited some aberrant patterns, e.g. ompC and ompN mRNA levels increased within the first 5 min. The same pattern was observed when the σE pathway was activated with polymyxin B (Fig. 5B). The delayed omp mRNA decay in ΔrybB cells cannot be explained by an altered σE induction, because upregulation of the σE-dependent MicA is unabated (Fig. 5). To confirm that polymyxin B activates σE, which then transcribes rybB and micA to destabilize porin mRNAs, we also treated the ΔrpoE strain with polymyxin B and observed that downregulation of ompD mRNA was abrogated by the absence of σE activation (Fig. S1).
Fig. 5.
RybB facilitates omp mRNA decay in the ESR. A. Northern analysis of the decay of five RybB target mRNAs (ompC, ompD, ompF, ompN, ompS) in Salmonella wild-type and ΔrybB cells carrying plasmid pBAD-RpoE. Bacteria were grown to late exponential phase (OD600 of 1.5), and RpoE expression was induced with arabinose. Aliquots were withdrawn for total RNA preparation prior to (0 min) and at the indicated time-points (5, 10 and 15 min) following induction. The wild-type and ΔrybB samples were probed in parallel on the same blot. MicA probing shows that the ESR induction is not affected in ΔrybB. The blots were also probed for the ESR-independent sodB mRNA and 5S rRNA (loading control). B. Northern blots of Salmonella wild-type and ΔrybB cells grown to OD600 of 1.5 in which the ESR was induced by treatment with polymyxin B for the time indicated above the panels. Probing of the same mRNAs as in (A) shows a similar delay in the decay of the RybB-target mRNAs in rybB mutant Salmonella.
RybB and MicA maintain envelope homeostasis
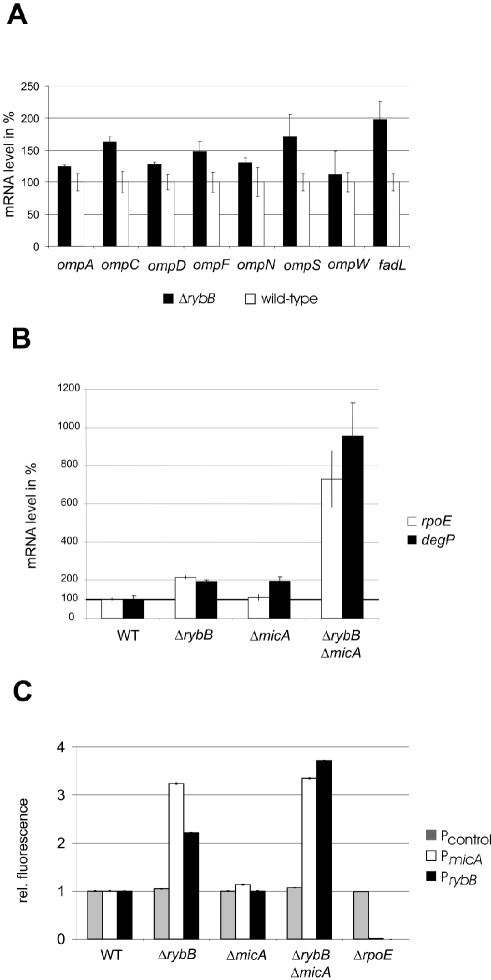
What are the functions of the σE-dependent sRNAs, RybB and MicA, under normal growth? In the experiments described above, we observed that ΔrybB cells had elevated levels of several RybB target mRNAs, i.e. at the zero time point prior to σE induction (Fig. 5). This observation was confirmed by determining the levels of eight RybB target mRNAs in wild-type and rybB mutant Salmonella by quantitative real-time PCR (Fig. 6A). We predicted that absence of RybB may increase major OMP synthesis, to impact upon envelope homeostasis, and could lead to chronic ESR activation in otherwise unstressed ΔrybB cells. To assay the ESR status in rybB and/or micA deletion strains, we determined the levels of rpoE and degP mRNA (Fig. 6B), two sensitive markers of the σE response (Dartigalongue et al., 2001; Rhodius et al., 2006; Rowley et al., 2006). In the ΔrybB strain, approximately twofold higher levels of these σE-dependent transcripts were observed. ΔmicA cells also had elevated rpoE and degP levels, albeit not as pronounced as with ΔrybB. However, rybB/micA double deletion activated σE even further as judged by approximately sevenfold and approximately ninefold higher levels of rpoE and degP mRNAs as compared with wild-type cells. To corroborate these results, we assayed the activity of the σE-dependent rybB and micA promoters in the sRNA deletion strains (Fig. 6C). To this end, the sRNA promoter regions were transcriptionally fused to a plasmid-borne gfp reporter gene, and the resulting plasmids were introduced into the wild-type, ΔrybB and/or ΔmicA, and ΔrpoE strains. A reporter plasmid in which gfp is under control of a constitutive PLtetO promoter (Lutz and Bujard, 1997) and which should not be affected by σE availability, served as control. By measuring the GFP activity of these promoter fusions, we found that the ΔrybB mutation – alone or in combination with ΔmicA– resulted in several-fold activation of the micA and rybB promoters, while the micA deletion alone had a lesser effect. In ΔrpoE cells, the transcriptional activity of both the rybB and the micA promoters were reduced to near background levels, which confirmed that these sRNA genes are strictly σE-dependent.
Fig. 6.
Loss of RybB and MicA functions induces the ESR. A. The ΔrybB mutation causes accumulation of major omp mRNAs in early stationary phase cultures (OD600 of 1.5). Shown is a comparison of major omp mRNA levels between wild-type and ΔrybB cells as determined by real-time PCR. The wild-type signals of each mRNA were set to 100%. B. Higher steady-state levels of the rpoE and degP mRNAs indicate an elevated ESR status in rybB, micA and sRNA double deletion strains. Wild-type Salmonella and isogenic mutant strains (as indicated) were grown to stationary phase (OD600 of 3), and rpoE and degP mRNA levels were determined by real-time PCR. The wild-type signals of the two mRNAs were set to 100%. C. The elevated ESR status in rybB and micA mutant cells is also apparent from a higher transcriptional activity of σE-dependent promoters of these genes. Transcriptional GFP fusions to the rybB and the micA promoters were introduced in Salmonella wild-type and the indicated mutant strains. A GFP fusion to the constitutive PLtetO promoter served as a control. Reporter activity (GFP fluorescence) was determined in cultures grown to stationary phase (OD600 of 3). Given are relative values, with the wild-type signals set to 1 in each case.
Discussion
Small non-coding RNAs have become important players in bacterial gene regulation (Gottesman, 2004; Storz et al., 2005). To date, systematic genome-wide searches for these regulatory molecules have led to the identification of ∼80 sRNAs in E. coli (reviewed in Vogel and Sharma, 2005), the majority of which are conserved in Salmonella and other closely related bacterial species (Hershberg et al., 2003). The precise role of most of the sRNAs is still unknown, though many are only expressed under specific conditions such as stationary phase and slow growth (Argaman et al., 2001; Wassarman et al., 2001; Vogel et al., 2003). It has long been argued that stationary phase expression of sRNA genes may actually reflect transcriptional control by distinct stress regulons which are gradually activated upon the cessation of growth. Our new findings place two such stationary phase-specific sRNAs, RybB and MicA, at the centre of an intensely investigated stress response, based upon the alternative sigma factor, σE.
σE is widespread among a diverse set of pathogenic and non-pathogenic bacteria, and becomes activated when bacterial envelope homeostasis is disturbed (reviewed in Rowley et al., 2006). The ESR plays fundamental roles in bacterial virulence and survival; it is induced when (i) elevated OMP production causes accumulation of misfolded OMPs in the periplasm, and (ii) the envelope requires remodelling following damage by external stresses. In both cases, the cell must avoid synthesis of the major OMPs because the ongoing translocation of these proteins cause membrane stress and so impact upon bacterial fitness. This negative link between the ESR and OMP protein production was first reported in 1993 (Mecsas et al., 1993), and was further highlighted by the observation of rapid removal of omp mRNAs upon σE induction (Rhodius et al., 2006), but it remained a mystery how this rapid omp mRNA decay could be reconciled with the extraordinarily stability of these messengers.
We examined this contradiction in S. typhimurium, and discovered that RybB and MicA solve this problem. We show that the σE-dependent sRNA, RybB, achieves the active degradation of the bulk of omp mRNAs upon activation of the ESR. Consequently, a chromosomal rybB deletion causes the accumulation of aberrant levels of omp mRNAs and chronic activation of the ESR, which is accentuated by a micA deletion.
This is the first example of such a complex system in γ-proteobacteria, where an alternative stress Sigma factor drives the expression of regulatory sRNAs which have functionally related target mRNAs. To our knowledge, the only other case are the σ54-controlled, highly homologous Qrr sRNAs of Vibrio species, which all act on a single target mRNA that encodes a quorum-sensing regulator (Lenz et al., 2004). However, it is unknown whether the action of the Qrr sRNAs also regulates σ54 activity, as reported here for the regulation of σE activity by RybB and MicA.
The conservation of rybB (Fig. 1C), micA (Udekwu et al., 2005) and rpoE (Rowley et al., 2006) genes within a large group of enterobacterial species suggests that an sRNA-mediated global omp mRNA decay is a fundamental ESR function in many bacteria. Homology is apparent at both the transcribed and the σE-dependent promoter regions, suggesting an evolutionarily conserved mechanism based upon the regulation of bacterial envelope homeostasis by RybB and MicA. A conserved regulatory pathway had been hypothesized for E. coli MicA when it was identified as a post-transcriptional regulator of ompA mRNA, but it was never suspected that a related pathway would target multiple omp mRNAs (Valentin-Hansen et al., 2004; Udekwu et al., 2005). Indeed, while this manuscript was in preparation, σE-dependent functions of the two sRNAs have been identified in E. coli (Johansen et al., 2006; E.G. Wagner and S. Gottesman, pers. comm.). In addition, loss of Hfq which causes a dramatic activation of the ESR in Salmonella (Figueroa-Bossi et al., 2006; Sittka et al., 2006) was also shown to activate the Salmonella micA gene in a σE-dependent fashion (Figueroa-Bossi et al., 2006). As MicA requires Hfq to repress ompA mRNA in E. coli (Rasmussen et al., 2005; Udekwu et al., 2005), and RybB is shown here both to display high affinity to Hfq and to be dependent on Hfq for binding to some of its target mRNAs in vitro, we speculate that the chronic envelope stress experienced by Salmonella hfq mutants may result from the loss of function of these two OMP-regulatory sRNAs.
Our results support the model shown in Fig. 7. First, in normally growing cells, the two σE-dependent sRNAs function within a surveillance loop to maintain envelope homeostasis. The constant flux of OMPs to the periplasm inevitably results in a fraction of misfolded OMPs, which will induce the σE pathway. Under this condition, RybB and MicA become activated and limit synthesis of OMP proteins. Of the two, RybB is the major facilitator of OMP repression, while MicA acts to repress the abundant OmpA protein. Second, if the ESR is triggered by an external stimulus, RybB and MicA function to rapidly shut off OMP synthesis. The downregulation of omp mRNA must be pivotal for the counteraction of envelope stress; it also occurs in the absence of RybB, albeit at a slower rate (Fig. 5), and may in part be mediated by transcriptional repression of the omp genes. However, it is also possible that other σE-regulated sRNAs remain to be identified. We note that polymyxin B treatment mediates a faster repression of some RybB target mRNAs, e.g. ompD and ompF, than arabinose-induced σE expression from plasmid pBAD-RpoE, and that this repression is less dependent on an intact rybB gene. Interestingly, we found that expression of MicF, the antisense regulator of ompF mRNA, is induced by polymyxin B in an rpoE-independent manner (Fig. 4) and could contribute to downregulation of ompF mRNA under this condition.
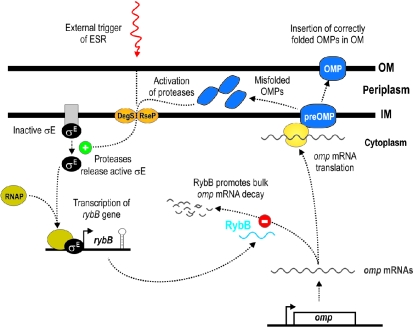
Fig. 7.
Proposed model of RybB and MicA functions in the ESR. σE activates expression of a range of phenotypes under conditions of envelope stress (Rowley et al., 2006). One of the key components of the ESR is a rapid decrease in mRNA levels of major OMPs (Rhodius et al., 2006), caused by a previously unknown mechanism. We have discovered a unique role for a sRNA, RybB, which is responsible for destabilizing many omp mRNA species in a σE-dependent fashion. The initial triggering of the ESR is initiated by DegS which releases functional σE to activate RybB, MicA and the rest of the σE regulon. RybB is proposed to bind to many omp mRNAs to accelerate their decay. This halts the synthesis of bulk OMPs to protect the cell from misfolded proteins accumulating in the periplasm. We propose a similar role for the σE-dependent MicA sRNA, i.e. ESR-induced ompA mRNA decay (not shown).
These experiments clearly identified RybB as a major accelerator of bulk omp mRNA decay. Why do the omp mRNAs need to be removed so quickly? The passage of enteric pathogens such as Salmonella through their host continuously requires these bacteria to remodel their envelope, and the speed of stress adaptation is one secret of their success as mammalian pathogens. However, the high stability of omp mRNAs threatens the successful adaptation processes required for envelope remodelling. We propose that the RybB-mediated omp mRNA decay selectively halts OMP production to free the membrane-associated translocation and the periplasmic folding machinery for the subsequent process of cell surface remodelling. The role of such a sRNA-controlled mechanism may allow Salmonella to react rapidly to environmental stress encountered during infection. We note that bacteriophages commonly use OMPs as receptors for docking (Mock and Pugsley, 1982; Koebnik et al., 2000; Nikaido, 2003); it is possible that a rapid shut-off of receptor expression via post-transcriptional control could become a matter of survival for bacteria under phage attack.
Why did such a mechanism evolve? The answer to this question probably lies in the purpose of the σE regulon. As σE is always present in the cell, but kept inactive by the anti-sigma factor RseA, induction of this regulon is quick and flexible. We suggest that the role of a small non-coding RNA is to facilitate a similarly rapid response: unlike protease-mediated degradatory systems, sRNA-based regulation does not necessitate the time-consuming process of translation. The design of the RybB-mediated surveillance loop indicates an unexpected ‘need for speed’ in the degradation of omp mRNAs by enteric bacteria in response to environmental stress.
Furthermore, our finding that RybB alters the kinetics of omp mRNA removal in the ESR may have general implications for our understanding of regulatory RNAs. Although stress-related targets have been identified for numerous sRNAs (Majdalani et al., 2005; Storz et al., 2005), physiological phenotypes of sRNA deletion strains have rarely been found. This lack of phenotypes, accompanied by small effects on target abundance in the absence of the regulatory sRNA, has led to the assumption that sRNAs primarily act to fine-tune stress responses. It is clear that bacteria show an enormous robustness in the control of their most important metabolic pathways and stress responses, mediated by redundancy in regulatory, catabolic and dissimilatory pathways. Therefore, in end-point assays that measure the ability to survive a certain stress under otherwise favourable conditions, the specific contribution of a regulatory sRNA can be masked by that of a more global protein-mediated response. In this study we have used kinetic analyses of omp mRNA repression to pinpoint a cellular function. Future experiments will be directed at determining the consequences of RybB and MicA action for bacterial physiology and its impact on OMP biogenesis.
What is the mechanism of RybB function? Our identification of so many functionally related mRNAs as the targets of RybB is unprecedented in bacteria. The closest example of a regulatory sRNA with multiple targets is the iron starvation-induced RyhB of E. coli, which negatively regulates mRNAs of iron-binding proteins when iron becomes scarce (Massé and Gottesman, 2002). Structural probing of a RyhB complex with one of its targets, sodB mRNA, showed RyhB to base-pair to the sodB 5′ UTR (Geissmann and Touati, 2004), and numerous other E. coli sRNAs that act as repressors have been shown by structural probing and/or compensatory base-pair changes to anneal to the 5′ UTRs of their respective target mRNAs (e.g. Schmidt et al., 1995; Argaman and Altuvia, 2000; Chen et al., 2004; Vogel et al., 2004; Rasmussen et al., 2005; Udekwu et al., 2005; Kawamoto et al., 2006). In most of these cases, sRNA pairing will sequester the ribosome binding site of the target mRNA, inhibit translation, and so accelerate RNase E-mediated degradation of the target (Masséet al., 2003; Morita et al., 2005). In other words, the rapid decay of a repressed mRNA appears to be an indirect effect of translation inhibition because the half-life of bacterial mRNA is strongly affected by its association with ribosomes (Deana and Belasco, 2005). Furthermore, the SgrS and RyhB sRNAs silence translation of pts6 and sodB mRNA, even in the absence of RNase E-mediated mRNA destabilization (Morita et al., 2006). Thus, the RybB-mediated shut-off of OMP synthesis is likely to be initiated by a block at the translational level.
Generally, trans-encoded antisense RNAs typically have short, imperfect sequence complementarity with their target(s) (for examples, see Wagner and Darfeuille, 2006), and are thus difficult to predict with statistical significance. The extremely rapid target decay upon pBAD-RybB expression, however, argues for direct RNA interactions. Using the TargetRNA algorithm (Tjaden et al., 2006) for bacterial sRNAs, Johansen et al. (2006) have predicted interactions of E. coli RybB with the 5′ UTRs of ompC and ompW, which appear to be conserved in Salmonella. Our preliminary results on structure probing of the RybB/ompN complex suggests that RybB forms an almost perfect 16 bp duplex with the 5′-coding region of ompN mRNA (F. Mika and J. Vogel, unpublished). However, the precise interaction sites of RybB with its many target mRNAs, as well as the contributions of Hfq and RNase E to RybB action, need to be identified on a case-to-case basis, which is the current focus of our work.
Intriguingly, the RybB/MicA functions described here bear striking similarity to the selective removal of membrane protein-encoding mRNAs within the eukaryotic unfolded protein response (UPR) (Hollien and Weissman, 2006). The UPR allows the endocytoplasmic reticulum (ER) to recover from the accumulation of misfolded proteins, when the folding capacity of the ER is exceeded. In this situation, IRE1 nuclease is activated to promote the rapid decay of a specific subset of mRNAs that are targeted to the ER (Hollien and Weissman, 2006), thus relieving the burden of misfolded proteins. By employing a sRNA (RybB) or a protein (IRE1), bacteria and eukaryotes have evolved different ways to cope with a similar problem; two distinct mechanisms that result in selective mRNA decay.
Experimental procedures
Oligodeoxynucleotides
Table S1 in the Supplementary material lists all oligodeoxynucleotides used in this study.
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. The Salmonella enterica serovar Typhimurium SL1344 strain was used as wild-type strain. Its ΔrpoE, ΔrybB and ΔmicA derivates were constructed using the lambda-red recombinase method (Datsenko and Wanner, 2000), and primer pairs JVO-1074/JVO-1075, JVO-0279/JVO-0280 and JVO-0019/JVO-0020 respectively. All chromosomal mutations were subsequently transferred to a fresh SL1344 background strain via P22 HT105/1 int-201 transduction (Schmieger, 1971). In strain JVS-00127, the kanamycin-resistance cassette of plasmid pKD4 replaces nucleotides 1–40 of the rybB gene. In strain JVS-00026, the kanamycin-resistance cassette of plasmid pKD4 replaces nucleotides 1–60 of the micA gene. In strain JVS-01028, the chloramphenicol-resistance cassette of plasmid pKD3 replaces nucleotides 100–1133 of the rpoE gene. All gene deletions were verified by PCR with JVO-0021/0023 for micA, JVO-1076/1077 for rpoE, JVO-0281/0282 for rybB. To construct the ΔrybB/ΔmicA strain JVS-01109, the kanamycin-resistance gene of JVS-00127, flanked by Flip recombinase target sites, was first removed with FLP recombinase as described in Datsenko and Wanner (2000), yielding strain JVS-01104. Subsequently, JVS-01104 served as a recipient for P22 transduction of the micA::KmR locus from JVS-00026.
Table 1.
Strains used in this study.
| Strain | Relevant markers/genotype | Reference/source |
|---|---|---|
| S. typhimurium | ||
| SL1344 | StrRhisG rpsL xyl | Hoiseth and Stocker (1981), provided by Dirk Bumann, MPI-IB Berlin |
| JVS-00026 | SL1344 ΔmicA::KmR | This study |
| JVS-01028 | SL1344 ΔrpoE::CmR | This study |
| JVS-00127 | SL1344 ΔrybB::KmR | This study |
| JVS-01104 | SL1344 ΔrybB | This study |
| JVS-01109 | SL1344 ΔrybBΔmicA::KmR | This study |
| E. coli | ||
| TOP10F′ | F′{lacIq Tn10 (TetR)} mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
Table 2.
Plasmids used in this study.
| Name | Relevant fragment | Comment | Origin/marker | Reference |
|---|---|---|---|---|
| pJV861-9 | PLtetO-lacZ::gfp | lacZ transcriptional GFP fusion plasmid | pSC101*/CmR | Urban and Vogel (2006) |
| pJV783-1 | PmicA-gfp | micA transcriptional GFP fusion plasmid | pSC101*/CmR | This study |
| pJV784-25 | PrybB-gfp | rybB transcriptional GFP fusion plasmid | pSC101*/CmR | This study |
| pJV990 | luc | pZE12-luc derivate, ColE1 origin exchanged pSC101* of pZS*24-MCS-1 | pSC101*/AmpR | This study and Lutz and Bujard (1997) |
| pAS0046 | gfp | Transcriptional fusion vector, based on pJV859-8 | pSC101*/CmR | Sittka et al. (2006) |
| pJV790 | PLtetO-lacZ::gfp | lacZ transcriptional GFP fusion plasmid | pSC101*/AmpR | This study |
| pJV791 | PmicA-gfp | micA transcriptional GFP fusion plasmid | pSC101*/AmpR | This study |
| pJV792 | PrybB-gfp | rybB transcriptional GFP fusion plasmid | pSC101*/AmpR | This study |
| pBAD/Myc-His A | pBAD expression plasmid | pBR322/AmpR | Invitrogen | |
| pKP8-35 (pBAD) | pBAD control plasmid, expresses ∼50 nt nonsense RNA derived from rrnB terminator | pBR322/AmpR | This study | |
| pKP17-1 (pBAD-RybB) | rybB | RybB expression plasmid, rybB is controlled by the plasmid-borne PBAD promoter | pBR322/AmpR | This study |
| pBAD33 | pBAD expression plasmid | pBR322/CmR | Guzman et al. (1995) | |
| pAC-rpoEST4 (pBAD-RpoE) | rpoE | RpoE expression plasmid, rpoE is controlled by the plasmid-borne PBAD promoter | pBR322/CmR | Miticka et al. (2003) |
| pKD3 | Template for mutant construction; carries chloramphenicol-resistance cassette | oriRγ/AmpR | Datsenko and Wanner (2000) | |
| pKD4 | Template for mutant construction; carries kanamycin-resistance cassette | oriRγ/AmpR | Datsenko and Wanner (2000) | |
| pKD46 | ParaB-γ-β-exo | Temperature-sensitive lambda-red recombinase expression plasmid | oriR101/AmpR | Datsenko and Wanner (2000) |
| pCP20 | Temperature-sensitive FLP recombinase expression plasmid | oriR101/AmpR, CmR | Datsenko and Wanner (2000) |
Plasmid construction
Plasmids for l-arabinose-inducible expression of rybB and micA genes were constructed by amplification of plasmid pBAD-His-myc [cycling parameters: 95°C/30′, 25× (95°C/10′, 59°C/30′, 72°C/2′), 72°C/10′] with primers JVO-0900/0901 (JVO-0901 introduces an XbaI restriction site upstream of the rrnB terminator sequence). The PCR product was digested with XbaI and DpnI. For amplification of the rybB insert, the sense primer (JVO-0906) starts with the sRNA +1 site (as previously mapped in E. coli; Argaman et al., 2001; Vogel et al., 2003) and carries a 5′ phosphate modification. The antisense primer (JVO-0282) binds close to the 3′ end of the rybB terminator, and adds an XbaI site to its sequence. Following amplification and PCR product digestion with XbaI, the vector- and sRNA-derived PCR products were ligated with T4 DNA ligase (5′ blunt end/3′ XbaI site), yielding plasmids pKP17-1 (pBAD-RybB). Correct inserts were confirmed by sequencing of the plasmids with vector primers, pBad-FW and pBad-REV.
Amplification of plasmid pBAD-His-myc using primers JVO-0900/JVO-0901, PCR product digestion with DpnI, and religation with T4 DNA ligase yielded the pBAD control plasmid, pKP8-35. PBAD expression in pKP8-35 results in a ∼50 nt nonsense transcript derived from the rrnB terminator sequence.
The transcriptional PmicA-gfp fusion plasmid (pJV783-1) was constructed by digestion of pAS0046 (Sittka et al., 2006) with AatII/NheI and ligation with a PCR product amplified with JVO-1230/-1231. To generate the PrybB-gfp transcriptional fusion plasmid (pJV784-25), pAS0046 was digested as described above and ligated with a PCR fragment amplified with JVO-1232/-1233. To replace the cat (chloramphenicol)-resistance cassette in both plasmids, an AatII/AvrII-generated fragment was replaced by the amp (ampicillin)-resistance cassette from pJV990, yielding plasmid pJV791 (transcriptional PmicA-gfp fusion) and pJV792 (transcriptional PrybB-gfp fusion). For construction of the control plasmid, the cat gene was replaced in the same way in plasmid pJV861-9 (encoding a short E. coli lacZ fragment fused to gfp under a PLtetO promoter; Urban and Vogel, 2006) resulting in pJV790. Competent E. coli TOP10 F′ cells (Invitrogen) were used for all cloning procedures.
Bacterial growth, l -arabinose induction and polymyxin B treatment
Growth in Luria–Bertani (LB) broth (220 rpm, 37°C) or on LB plates at 37°C was used throughout this study. SOC medium was used to recover transformants after heat-shock or electroporation and prior to plating. Antibiotics (where appropriate) were used at the following concentrations: 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 20 μg ml−1 chloramphenicol. For RybB, MicA and RpoE expression from pBAD-derived plasmids, cultures were treated with l-arabinose (final concentration of 0.2%). Polymyxin B (Sigma-Aldrich; #P9602–1VL) was used at a final concentration of 1 μg ml−1 (Fig. 2D) or 5 μg ml−1 (Fig. 5B).
Microarray experiments
Strain SL1344 was transformed with plasmids pKP8-35 (control) and pKP17-1 (pBAD-RybB), and grown in liquid culture from single colonies to an OD600 of 1.5, at which sRNA expression was induced with l-arabinose for 10 min. RNA extraction and data generation are described in the Supplementary material.
Northern blot analysis
Overnight cultures were diluted 1/100 in fresh medium and grown to the indicated cell densities (OD600). Culture aliquots were removed and mixed with 0.2 vol. of stop solution (5% water-saturated phenol, 95% ethanol), and snap-frozen in liquid nitrogen. After thawing on ice, bacteria were pelleted by centrifugation (2 min, 16 000 rcf, 4°C), and RNA was isolated using the Promega SV total RNA purification kit as described at http://www.ifr.ac.uk/safety/microarrays/protocols.html(Kelly et al., 2004) or using the Trizol method (in the case of the RpoE overexpression and polymyxin B experiments shown in Figs 2 and 4). The purified RNA was quantified on a Nanodrop machine (NanoDrop Technologies).
RNA samples (∼5 μg) were denatured for 5 min at 95°C in RNA loading buffer (95% [v/v] formamide, 0.1% [w/v] xylene cyanole, 0.1% [w/v] bromphenol blue, 10 mM EDTA), separated on 8.3 M urea/5% polyacrylamide gels, and transferred to Hybond-XL membranes (GE Healthcare) by electroblotting (1 h, 50 V, 4°C) in a tank blotter (Peqlab, Germany). Following pre-hybridization of the membranes in Rapid-hyb Buffer (GE Healthcare), [32P]-labelled gene-specific probes (Table S3) were added and hybridization was performed at the temperatures given in Table S3. After hybridization for 2 h, membranes were rinsed at room temperature in 2× SSC/0.1% SDS, followed by washing in three subsequent 15 min steps in SSC (2×, 1× or 0.5× respectively)/0.1% SDS solutions (at the hybridization temperature). Membranes hybridized with end-labelled oligodeoxyribonucleotides were rinsed in 5× SSC followed by three wash steps at 42°C in SSC (5×, 1× and 0.5× respectively). Signals were visualized on a phosphorimager (FLA-3000 Series, Fuji), and band intensities quantified with AIDA software (Raytest, Germany).
Hybridization probe generation
Primers for template amplification are listed in Table S3. Standard polymerase chain reactions were carried out on genomic DNA. Double-stranded DNA probes (PCR products) were random-labelled in the presence of [32P]-α-dCTP using the Rediprime II labelling kit (GE Healthcare). Single-stranded RNA probes (riboprobes) were generated from PCR fragments (a T7 RNA polymerase promoter sequence was added by the antisense primer) in the presence of an excess of [32P]-α-UTP over unlabelled UTP using the Ambion T7 polymerase Maxiscript kit. DNA oligonucleotides were labelled with [32P]-γ-ATP using T4 polynucleotide kinase (Fermentas). All labelled probes were purified over G50 columns (GE Healthcare) to remove unincorporated nucleotides prior to hybridization.
Synthesis, purification and labelling of RNA for in vitro binding assays
DNA templates carrying a T7 promoter sequence were generated by PCR using genomic DNA and primers as listed in Table S1. For RybB oligonucleotides JVO-1242 (adds T7 promoter)/-1243 (binds in the terminator region) were used. For the 5′ UTRs of ompN primer pair JVO-1244/-1245 [the fragment covers the ompN region from positions −73 to +89 (30th aa) relative to the start codon], ompC primer pair JVO-1246/JVO-1247 [the fragment covers the ompC region from positions −78 to +100 (33rd aa) relative to the start codon], and ompD primer pair JVO-1186/JVO-1058 [the fragment covers the ompD region from positions −69 to +118 (39th aa) relative to the start codon]. Primers JVO-1244, JVO-1246 and JVO-1186 add a T7 promoter sequence to the 5′ ends of the respective PCR fragments.
In vitro transcription was performed using the MEGAscript High Yield Transcription Kit (Ambion, #1333), followed by DNase I digestion (1 unit, 15 min, 37°C). Following extraction with phenol : chloroform : isopropanol (25:24:1 v/v), the RNA was precipitated overnight at −20°C with 1 vol. of isopropanol. RNA integrity was checked on a denaturing polyacrylamide gel.
To label in vitro synthesized RybB RNA, 20 pmol RNA was dephosphorylated with 10 units of calf intestine alkaline phosphatase (New England Biolabs) in a 20 μl reaction at 37°C for 1 h. Following phenol extraction, the RNA was precipitated overnight with ethanol/sodium acetate and 20 μg glycogen. The dephosphorylated RNA was 5′-end-labelled with 32P-γATP (20 μCi) and 1 unit of polynucleotide kinase (New England Biolabs) for 30 min at 37°C in a 20 μl reaction. Unincorporated nucleotides were removed using Microspin G-50 Colums (GE Healthcare), followed by purification of the labelled RNA on a denaturing polyacrylamide gel (6%/7 M urea). Upon visualization of the labelled RNA by exposure on a phosphorimager, the RNA was cut from the gel and eluted with RNA elution buffer (0.1 M sodium acetate, 0.1% SDS, 10 mM EDTA) at 4°C overnight, followed by phenol extraction and precipitation as before.
Electrophoretic mobility shift assays
RybB/Hfq binding assays were performed in 1× structure buffer [100 mM Tris pH 7, 1 M KCl, 100 mM MgCl2, provided along with RNase T1 (#2283) from Ambion, USA] as follows. 5′-labelled RybB RNA (∼4 nM final concentration in binding reaction) and 1 μg of yeast RNA (final concentration: 4.3 μM) were incubated in the presence of Hfq (concentrations as given in the figure legend) in 10 μl reactions at 37°C for 10 min. The Hfq dilutions, calculated for the Hfq hexamer were prepared in 1× dilution buffer (1× structure buffer with 1% glycerol, 0.1% Triton X-100). Salmonella Hfq protein was prepared as outlined in Sittka et al. (2006).
RybB/5′ UTR binding assays were performed as above in the absence of Hfq (ompN) or the presence of 30 nM Hfq (ompC, ompD). The final concentrations of 5′ UTR RNAs are given in legend to Fig. 3G. Prior to gel loading, the binding reactions were mixed with 3 μl of loading buffer (50% glycerol, 0.5× TBE, 0.2% bromphenol blue), and electrophoresed on native 6% polyacrylamide gels in 0.5× TBE buffer at 250 V at 4°C for 3 h. Gels were dried, and analysed using a phosphorimager (see above).
Quantitative RT-PCR
To determine the mRNA levels of wild-type and ΔrybB (JVS-0127) cells overnight cultures were diluted 1:100 and cells were grown to an OD600 of 3 [rpoE and degP (htrA)] or 1.5 (ompA, ompF, ompC, ompD, fadL, ompS, ompW, ompN). Two millilitre aliquots were removed and treated with 0.2 vol. of stop solution (95% EtOH; 5% water-saturated phenol). Cells were snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction. RNA extraction was carried out as described above (Promega SV total RNA purification kit) and RNA concentrations were determined on a Nanodrop machine (NanoDrop Technologies). The relative amount of target mRNA was determined by quantitative real-time PCR using Quantitect™ SYBR® Green RT-PCR Kit following manufacturer's instructions (Qiagen) and calculating standard deviations as outlined in Paland et al. (2006). Specific primer pairs for rpoE (JVO-1236/1237), degP (JVO-1234/1235), ompA (JVO-1090/1091), ompF (JVO-1328/1329), ompC (JVO-1092/1093), ompD (JVO-1094/1095), fadL (JVO-1393/1394), ompS (JVO-1332/1333), ompW (JVO-1334/1335) and ompN (JVO-1330/1331) were designed using the PRIMER EXPRESS™ software (Applied Biosystems). rfaH (JVO-1117/1118) was used as an internal standard.
Fluorescence measurements
Strains carrying the GFP fusion plasmids were inoculated 1:100 in LB medium. At the indicated cell density, 3 × 100 μl of culture was transferred to a 96 well plate, and fluorescence was measured at 37°C using a VICTOR3™ machine (1420 Multilabel Counter, Perkin Elmer). All experiments were done in triplicates. Plasmid pJV859-8, which expresses GFP from a constitutive PLtetO promoter, served as a control. Strains without a plasmid served as background control (autofluorescence). The detailed protocol of fluorescence measurement is described in Urban and Vogel (2006).
Acknowledgments
We are grateful to Mark Achtman, Thomas F. Meyer and Arturo Zychlinsky for helpful comments on the manuscript, Jan Kormanec for providing material, and Fabien Darfeuille for discussions of a possible link between sRNAs and σE. This work was supported by the core strategic grant of the BBSRC (Hinton lab).
Supplementary material
The following supplementary material is available for this article online:
An rpoE deletion abrogates the rapid downregulation of ompD mRNA upon polymyxin B treatment.
Oligodeoxynucleotides used in this study
Microarray results of pBAD-RybB expression.
Probes for Northern detection and hybridization conditions.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli sigma E regulon. J Biol Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J Biol Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Loss of Hfq activates the sigma-dependent envelope stress response in Salmonella enterica. Mol Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- von Gabain A, Belasco JG, Schottel JL, Chang AC, Cohen SN. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigma(E) regulon: role in down-regulation of outer membrane proteins. J Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JC, Dorman CJ. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- Lee DR, Schnaitman CA. Comparison of outer membrane porin proteins produced by Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980;142:1019–1022. doi: 10.1128/jb.142.3.1019-1022.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- Missiakas D, Betton JM, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- Miticka H, Rowley G, Rezuchova B, Homerova D, Humphreys S, Farn J, et al. Transcriptional analysis of the rpoE gene encoding extracytoplasmic stress response sigma factor sigmaE in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2003;226:307–314. doi: 10.1016/S0378-1097(03)00600-1. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock M, Pugsley AP. The BtuB group col plasmids and homology between the colicins they encode. J Bacteriol. 1982;150:1069–1076. doi: 10.1128/jb.150.3.1069-1076.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emody L, et al. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun. 2006;74:5914–5925. doi: 10.1128/IAI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paland N, Rajalingam K, Machuy N, Szczepek A, Wehrl W, Rudel T. NF-kappaB and inhibitor of apoptosis proteins are required for apoptosis resistance of epithelial cells persistently infected with Chlamydophila pneumoniae. Cell Microbiol. 2006;8:1643–1655. doi: 10.1111/j.1462-5822.2006.00739.x. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Zheng P, Delihas N. Secondary structures of Escherichia coli antisense micF RNA, the 5′-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry. 1995;34:3621–3631. doi: 10.1021/bi00011a017. [DOI] [PubMed] [Google Scholar]
- Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2006. [DOI] [PMC free article] [PubMed]
- Skovierova H, Rowley G, Rezuchova B, Homerova D, Lewis C, Roberts M, Kormanec J. Identification of the sigmaE regulon of Salmonella enterica serovar Typhimurium. Microbiology. 2006;152:1347–1359. doi: 10.1099/mic.0.28744-0. [DOI] [PubMed] [Google Scholar]
- Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol. 2002;43:771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2006. in press. [DOI] [PMC free article] [PubMed]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006. [DOI] [PubMed]
- Vogel J, Sharma CS. How to find small non-coding RNAs in bacteria. Biol Chem. 2005;386:1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Hüttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Vytvytska O, Jakobsen JS, Balcunaite G, Andersen JS, Baccarini M, von Gabain A. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc Natl Acad Sci USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Bläsi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- Wagner EG, Darfeuille F. Nellen W. Small RNAs: Analysis and Regulatory Functions. Springer Verlag: Berlin; 2006. Small regulatory RNAs in bacteria; pp. 1–30. [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An rpoE deletion abrogates the rapid downregulation of ompD mRNA upon polymyxin B treatment.
Oligodeoxynucleotides used in this study
Microarray results of pBAD-RybB expression.
Probes for Northern detection and hybridization conditions.