Abstract
The import of protein into chloroplasts is mediated by translocon components located in the chloroplast outer (the Toc proteins) and inner (the Tic proteins) envelope membranes. To identify intermediate steps during active import, we used sucrose density gradient centrifugation and blue-native polyacrylamide gel electrophoresis (BN-PAGE) to identify complexes of translocon components associated with precursor proteins under active import conditions instead of arrested binding conditions. Importing precursor proteins in solubilized chloroplast membranes formed a two-peak distribution in the sucrose density gradient. The heavier peak was in a similar position as the previously reported Tic/Toc supercomplex and was too large to be analyzed by BN-PAGE. The BN-PAGE analyses of the lighter peak revealed that precursors accumulated in at least two complexes. The first complex migrated at a position close to the ferritin dimer (approximately 880 kDa) and contained only the Toc components. Kinetic analyses suggested that this Toc complex represented an earlier step in the import process than the Tic/Toc supercomplex. The second complex in the lighter peak migrated at the position of the ferritin trimer (approximately 1320 kDa). It contained, in addition to the Toc components, Tic110, Hsp93, and an hsp70 homolog, but not Tic40. Two different precursor proteins were shown to associate with the same complexes. Processed mature proteins first appeared in the membranes at the same fractions as the Tic/Toc supercomplex, suggesting that processing of transit peptides occurs while precursors are still associated with the supercomplex.
Keywords: chloroplast, protein import, translocon complex, blue-native polyacrylamide gel electrophoresis
Introduction
Most proteins in chloroplasts are encoded by the nuclear genome and synthesized in the cytosol as precursors with N-terminal targeting signals called transit peptides. The import of precursor proteins into chloroplasts is mediated by a translocon complex. Many components of the translocon complex have been identified. They include the Toc (translocon at the outer envelope membrane of chloroplasts) and Tic (translocon at the inner envelope membrane of chloroplasts) proteins and at least two chaperones located in the stroma (for reviews, see Kessler and Schnell, 2006; Soll and Schleiff, 2004.)
Toc75, Toc159, and Toc34 are the major Toc components identified and form a core Toc complex (Schleiff et al., 2003b). Toc75 most likely functions as the protein-translocating channel in the outer membrane (Hinnah et al., 1997; Reumann et al., 1999). Toc159 and Toc34 are homologous GTPases (Kessler et al., 1994). Both proteins are essential for import of protein into chloroplasts (Bauer et al., 2000; Constan et al., 2004; Kubis et al., 2004) and both have been shown to bind transit peptides directly (Smith et al., 2004; Sveshnikova et al., 2000). However, which of these two GTPases is the initial receptor for precursors and the exact function of each protein remain in dispute (Kessler and Schnell, 2006). Tic110 is the major Tic component identified. It has an N-terminal membrane anchor followed by a large hydrophilic domain located in the stroma (Inaba et al., 2003; Jackson et al., 1998). It most likely functions as the stroma-side receptor for transit peptides and the scaffold for translocation of precursor across the inner envelope membrane into the stroma (Inaba et al., 2003, 2005). Translocation across the inner envelope membrane requires at least two additional proteins. Chloroplast stromal Hsp93 (ClpC) is most likely the motor that drives translocation (Nielsen et al., 1997), although direct evidence for this function is still lacking. Tic40 has a similar topology to Tic110 and is a co-chaperone that coordinates Tic110 and Hsp93 (Chou et al., 2003).
Despite some general ideas about their functions, how individual translocon components associate with one another during the import process, i.e. the number and composition of complexes and the temporal order of precursor association with these complexes, is much less clear.
Almost all studies analyzing interactions of translocon components with precursors use conditions that arrest precursors at a ‘binding’, or ‘docking’, stage (Olsen and Keegstra, 1992) by artificially removing ATP from chloroplast and precursor preparations and then controlling the exogenous ATP at below 100 μm. Under these conditions, precursors stably insert into the chloroplast envelope but remain partially exposed on the chloroplast surface. The transit peptide is still inaccessible by the stromal processing peptidase (Olsen et al., 1989). Schnell et al. (1994) have shown that, under a binding condition of 50 μm ATP at room temperature in the dark, precursors can be co-purified with Toc159, Toc75, and Toc34. When further chased with 5 mm ATP for 2.5 min, precursors can be co-purified with at least five proteins: Toc159, Toc75, Toc34, Tic110, and an hsp70 homolog of as yet unknown identity. This hsp70 homolog is an integral membrane protein most likely located in the intermembrane space side of the outer membrane (Marshall et al., 1990; Schnell et al., 1994). These results suggest that precursors are mostly associated with the Toc components under the arrested binding conditions and are associated with both Toc and Tic components under the high-ATP active import conditions. However, co-purification does not provide information about the number and size of the translocon complexes involved. It is also not clear whether interaction with the Toc components is an obligatory first step during normal import, or if the interaction results from artificially arresting import at the binding stage. It is possible that during active import, precursors may interact with a complete Tic/Toc complex directly.
Using various cross-linkers and then analyzing the cross-linked complexes on denaturing SDS-PAGE, Akita et al. (1997) have shown that when import was arrested under 75 μm ATP on ice in the dark, precursors are associated with at least four groups of complexes. When analyzed for the presence of the three Toc components, Tic110 and Hsp93 by immunoprecipitation, the first group contained all five components and was too large to enter the gel. The second group had an estimated molecular mass of 500–700 kDa. The lighter portion of this group contained only the Toc components and the heavier portion contained all five components. The third group contained only the three Toc components and had an estimated molecular mass of 250–350 kDa and the fourth contained only Toc75 and Toc159 and had a molecular mass of 110 kDa. It is not clear whether all of these complexes are required during active import. The temporal order of association of these complexes with precursors is also not known. Because the cross-linked products were analyzed on SDS-PAGE, the possibility of multiple conformations for a single complex also could not be investigated.
Blue-native polyacrylamide gel electrophoresis (BN-PAGE) has been used successfully in analyzing precursor import and assembly of the translocon complex in the mitochondrial membranes (Chacinska et al., 2005; Ishikawa et al., 2004) and the chloroplast thylakoid membrane (Cline and Mori, 2001). Possibly due to the complexity of the precursor-associated complexes, it has been more difficult to use this technique to analyze precursor transport across the chloroplast envelope membranes. In the current report, we first used sucrose density gradient centrifugation to fractionate chloroplast membranes isolated from chloroplasts under active import conditions, instead of the arrested binding conditions. We then analyzed the precursor-containing fractions from the sucrose density gradients using BN-PAGE. We identified multiple forms of native Toc and Tic/Toc complexes associated with importing precursors. Our results suggest that during active import, precursors interact with an 880-kDa Toc complex before associating with a very high-molecular-weight Tic/Toc supercomplex. Processing of transit peptides then takes place while precursors are still associated with the Tic/Toc supercomplex.
Results
Importing precursors form a two-peak distribution pattern in the sucrose density gradient
To find a suitable time point during active import for identification of the translocon complex, import time-course experiments were performed with isolated pea chloroplasts and in vitro-translated [35S]-labeled precursors to the small subunit of Rubisco (prRBCS). To maximize the signal intensity for further analyses, one volume of chloroplasts (at a concentration of 1 mg chlorophyll ml−1) was mixed with one volume of [35S]prRBCS. To prolong the time span of active import in order to increase the chance of observing intermediates, import was performed at 20°C instead of 25°C. Under these conditions, the amount of mature RBCS imported continued to increase for about 60 min (Figure 1a). The 20-min time point was chosen for further analyses because chloroplasts at this time point contained a large amount of prRBCS and the amount of RBCS imported was increasing linearly. When chloroplasts from a 20-min import reaction were isolated and further incubated in a buffer containing 3 mm ATP but without additional prRBCS, the prRBCS molecules associated with chloroplasts were gradually chased into mature RBCS (Figure 1b), indicating that the prRBCS molecules were on an active course of import.
Figure 1. Identifying a suitable time point during active import for identification of the translocon complex.
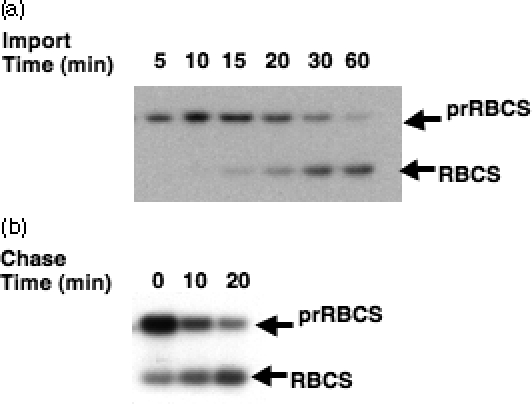
(a) [35S]prRBCS was incubated with isolated chloroplasts under import conditions at 20°C. At each time point, a portion of the import mixture was removed and the import reaction terminated by diluting with cold import buffer. An equal amount of protein was loaded in each lane.
(b) Chase after 20-min import. [35S]prRBCS was incubated with isolated chloroplasts under import conditions at 20°C for 20 min. Chloroplasts were isolated and resuspended in import buffer containing 3 mm ATP. A portion of the chloroplasts were immediately re-isolated (chase time 0 min), or the reaction was incubated further at 20°C for 10 or 20 min (chase time 10 and 20 min). An equal amount of protein was loaded in each lane.
To identify translocon complexes associated with precursors, chloroplasts from samples of the 20-min import (chase 0 min) and from samples that have been further chased for 10–20 min were re-isolated, treated with 0.5 mm dithiobis-9-succinimidylpropionate (DSP) cross-linker to preserve complex integrity and then hypotonically lysed. Total membranes from lysed chloroplasts were solubilized with 2%n-decyl-β-d-maltopyranoside and loaded onto a 15–55% sucrose density gradient. The [35S]prRBCS formed a two-peak distribution pattern in the sucrose density gradient (Figure 2a, top panel). The heavier peak (represented by fraction 19) was in a similar position to the previously reported Tic/Toc supercomplex (Nielsen et al., 1997).
Figure 2. Sucrose density gradient analyses of solubilized chloroplast membranes after prRBCS import and chase.
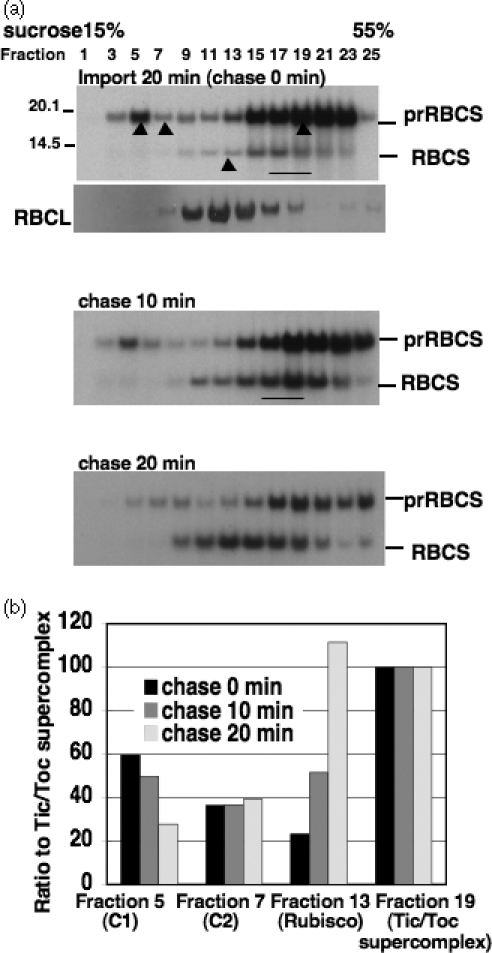
(a) Chloroplasts were incubated with prRBCS for 20 min under import conditions, then re-isolated and further chased for 0–20 min. Membrane fractions from chloroplasts of each time point were solubilized and analyzed by sucrose density gradients. Filled triangles indicate the four fractions quantified in (b). An immunoblot of the large subunit of Rubisco (RBCL) is shown to indicate the position of endogenous Rubisco holoenzyme. Underlines mark the mature RBCS at the supercomplex fractions.
(b) Quantification of fractions representing distinct complexes from gels shown in (a). For each gel, the amount of prRBCS in fraction 19 (supercomplex) was set to 100% and the relative amounts of prRBCS in fractions 5 and 7 and mature RBCS in fraction 13 were then plotted.
The lighter peak (represented by fraction 5) quickly diminished in intensity during the chase and disappeared before the disappearance of the supercomplex. Quantification of the data indicated that the ratio of the lighter peak to the supercomplex peak gradually decreased during the chase incubation (Figure 2b, fraction 5) while the ratio of mature RBCS in the Rubisco holoenzyme (represented by mature RBCS in fraction 13, see below) to the supercomplex gradually increased. These data suggested that the lighter peak might represent an earlier step, and the Rubisco holoenzyme a later step, than the Tic/Toc supercomplex in the import process.
After chasing for 20 min, most mature RBCS were in the center of the gradient (Figure 2a, bottom panel, chase 20 min) where they coincided with the endogenous large subunit of Rubisco (RBCL, immunoblot of RBCL below the 0-min chase sample in Figure 2a). The BN-PAGE analysis of these fractions in the center of the gradient also revealed the presence of newly imported [35S]RBCS in the Rubisco holoenzyme (Figure 3a, fractions 11 and 13). These data indicated that the imported RBCS had been incorporated into the Rubisco holoenzyme and the chase was successful. The presence of Rubisco holoenzyme in the membrane fractions has always been observed and is most likely a result of contamination due to the abundance of the Rubisco holoenzyme (Werner-Washburne et al., 1983).
Figure 3. Blue-native PAGE analyses of fractions from the two precursor peaks.
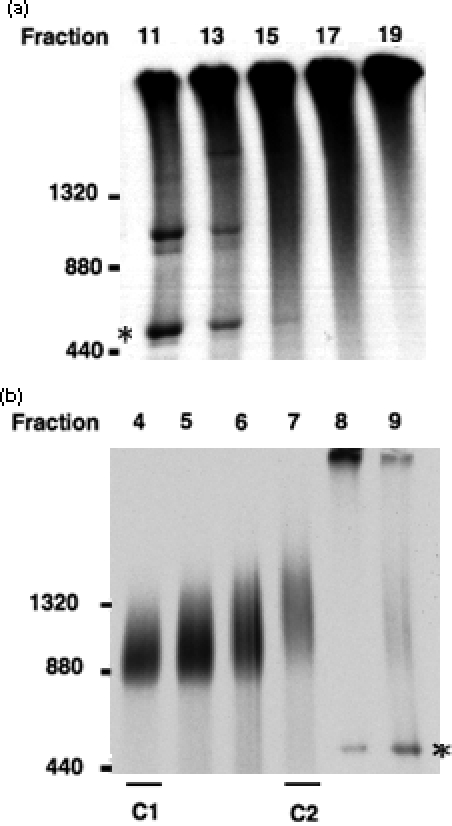
Fractions 11–19 (a) or fractions 4–9 (b) of the 20-min import samples as shown in Figure 2(a) were analyzed by BN-PAGE. Molecular masses (in kDa) are labeled according to the migration positions of ferritin monomer (440), dimer (880), and trimer (1320). Asterisks indicate the position of Rubisco holoenzyme.
Processing of transit peptides most likely occurs while precursors are still associated with the supercomplex
To identify the number and size of complexes in the two precursor peaks in the sucrose density gradient, peak fractions from the 20-min-import samples (Figure 2a, top panel) were analyzed by BN-PAGE. When fractions 17 and 19 containing the heavier peak were analyzed, prRBCS-containing complexes were too large to enter the gel (Figure 3a). For a comparison, fractions 11 and 13 that contained a small amount of Rubisco holoenzyme were also analyzed. Rubisco holoenzyme at a position slightly larger than ferritin monomer (approximately 440 kDa, Figure 3a, asterisk) as previously observed (Poetsch et al., 2000; Rexroth et al., 2003) was indeed present. The large size of the complexes present in the heavier peak made it difficult to analyze their number or composition. From previous sucrose density gradient (Nielsen et al., 1997) and cross-linking (Akita et al., 1997) studies, it is most likely that complexes in these fractions contain all the Tic and Toc components. However, other methods are required to analyze their sizes, number, and composition.
It is interesting to note that during the chase, mature RBCS first appeared and concentrated at the same fractions as the supercomplex (Figure 2a, chase 0 and 10 min, underlined) then shifted to the center of the gradient where the Rubisco holoenzyme was (chase 20 min). The BN-PAGE analyses indicated that mature RBCS in the supercomplex fractions was not in Rubisco holoenzyme but mixed in the smear of supercomplex (Figure 3a, compare fractions 11 and 13 with fractions 15–19). These data suggested that processing of the transit peptide most likely occurred while prRBCS was still associated with the supercomplex.
The lighter peak contains at least two complexes
When fractions 4–7 around the lighter peak were analyzed by BN-PAGE, at least two complexes were revealed (Figure 3b). The [35S]prRBCS in fraction 4 was associated with a complex slightly larger than ferritin dimer of 880 kDa. The [35S]prRBCS in fraction 7 was associated with a complex at the position of ferritin trimer (1320 kDa). Fractions 5 and 6 contained both complexes in different proportions. Hereafter the 880-kDa complex is referred to as C1 and the 1320-kDa complex as C2 (Figure 3b).
The compositions of C1 and C2 were analyzed by antibody-shift BN-PAGE assays. If C1 contains a certain translocon component, incubation of sucrose-density gradient fractions containing C1 with specific antibody against this translocon component, followed by addition of Protein-A-conjugated Sepharose, should precipitate C1 from the soluble phase or make C1 form aggregates too large to enter the gel resulting in the accumulation of aggregates in the wells. As shown in Figure 4(a), C1 was removed by antibodies against Toc75 and Toc 34 (lanes 2 and 7) but not by an antibody against Tic110 (lane 4) or the monoclonal antibody SPA-820 (lane 9), which was used to identify the hsp70 homolog associated with importing precursors (Schnell et al., 1994). These results indicated that C1 contained the two Toc components but not Tic110 or any hsp70 homolog recognized by SPA-820. An anti-Toc159 antibody (α-Toc159-1; Figure 4a, lane 6 and Figure 4b, lane 2) removed only part of C1 and shifted the remaining C1 to a slower-migrating position. Increasing the amount of antibody added had no effect (data not shown). Furthermore, this anti-Toc159 antibody caused complete removal of the C2 complex (see below), suggesting that the incomplete removal of C1 was not due to insufficient efficiency of the antibody. This antibody was raised against the C-terminal domain (the ‘M’ domain; Bauer et al., 2000) of Arabidopsis Toc159. To further confirm that the incomplete shift was not due to this particular antibody, another anti-Toc159 antibody made against both the G and M domains of Arabidopsis Toc159 was tested. This second anti-Toc159 antibody gave similar shifting results (Figure 4b, lane 3). Therefore there might be at least two forms of the C1 complex: one form which contained Toc159 and the other form which did not contain Toc159 or contained Toc159 in a conformation not accessible to the antibodies.
Figure 4. Antibody-shift BN-PAGE analyses of the composition of the C1 and C2 complexes.
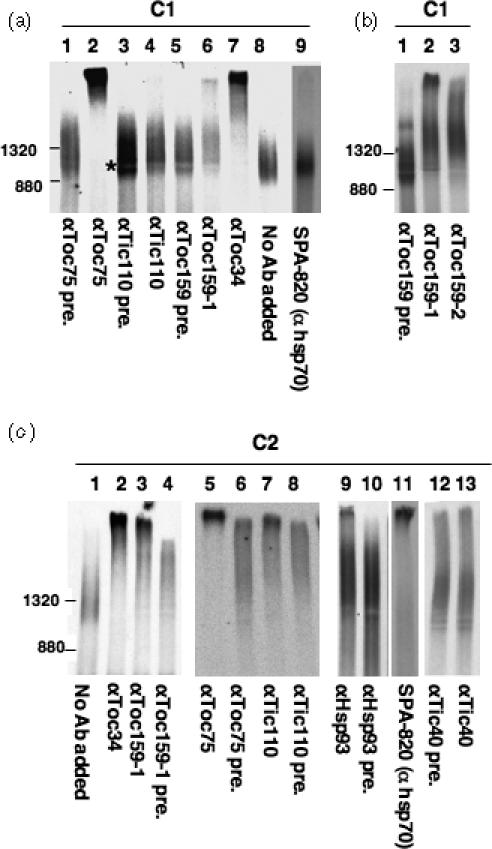
(a) The sucrose-density gradient fraction containing C1 was incubated with various antibodies, as labeled at the bottom, to remove possible components in C1 (pre., pre-immune serum of the respective antibody). The sharp band between 880 and 1320 kDa was formed due to the presence of a major serum protein at the position marked by the asterisk.
(b) Same as in (a) and the experiment was repeated with a second anti-Toc159 serum (αToc159-2).
(c) The sucrose-density gradient fraction containing C2 was incubated with various antibodies, as labeled at the bottom, to remove possible components in C2.
The composition of C2 was also analyzed by BN-PAGE antibody-shift assays. For unknown reasons, all pre-immune sera we tested caused a similar smearing of C2 in BN-PAGE (Figure 4c, lanes 4, 6, 8, 10 and 12). The smearing might be caused by some rabbit serum proteins reacting with some chloroplast proteins in the sucrose-density fraction containing C2. Nonetheless, antibodies against Toc75 and Toc34 still caused complete removal of C2 (lanes 2 and 5). In contrast to the situations with C1, SPA-820 and antibodies against Toc159 and Tic110 caused complete removal of C2 (lanes 3, 7 and 11). Antibodies against the Tic40 C-terminal domain caused no additional shifting or aggregation compared to its pre-immune serum (lanes 12 and 13). Two other anti-Tic40 antibodies, made against the N-terminal half and the entire stromal domain, respectively, gave the same results (data not shown). These data suggested that Tic40 was not present in C2. Since Tic40 has been shown to be associated with Toc75, Tic110 and importing precursors (Chou et al., 2003), it is likely that Tic40 was assembled into the active translocon complex later in the import process. An anti-Hsp93 antibody removed part of C2 and caused some aggregation in the well (lane 9), suggesting that at least part of C2 contained Hsp93. Therefore C2 contained the three major Toc components, Tic110, and an hsp70 homolog but not Tic40. At least some C2 might also contain Hsp93.
Precursors to PORB are associated with the same C1 and C2 complexes during import
To confirm that C1 and C2 are general import intermediates, a different precursor was tested. We performed the same import, sucrose-density fractionation and BN-PAGE analyses with in vitro-translated [35S]-labeled precursors to Arabidopsis protochlorophyllide oxidoreductase B (prPORB; At4g27440). Membrane-bound prPORB also formed a two-peak distribution pattern in the sucrose density gradient (Figure 5a). The BN-PAGE analyses of fractions from the lighter peak also revealed the presence of two complexes at similar migration positions to C1 and C2 (Figure 5b). Fractions 5–9 contained both prPORB and mature PORB (Figure 5a). It was likely that imported mature PORB was assembled into its functional complex which had a similar sedimentation coefficient to C1 and C2. Therefore it was unclear whether the C1 and C2 complex bands in BN-PAGE (Figure 5b) were generated by prPORB or mature PORB. However, fraction 3 contained mostly mature PORB (Figure 5a) and when fraction 3 was analyzed by BN-PAGE, complexes associated with mature PORB could not be detected on the gel (Figure 5b), possibly due to their small sizes. Therefore it is most likely that the C1 and C2 bands on BN-PAGE in fractions 5 and 8 were formed by prPORB, not mature PORB. Antibody-shift assays again revealed that C1 contained Toc75 and Toc34 but not Tic110 (Figure 5c, lanes 2, 4 and 8). Both antibodies against Toc159 removed part of C1 and shifted the remaining C1 to a slower migrating position (Figure 5c, lanes 6 and 7). C2 also contained the three Toc components (Figure 5d, lane 2 and data not shown) and Tic110 (lane 4) but not Tic40 (data not shown). Similarly, only part of prPORB-C2 was shifted by the anti-Hsp93 antibodies (lane 6).
Figure 5. Translocon complexes associated with importing prPORB.
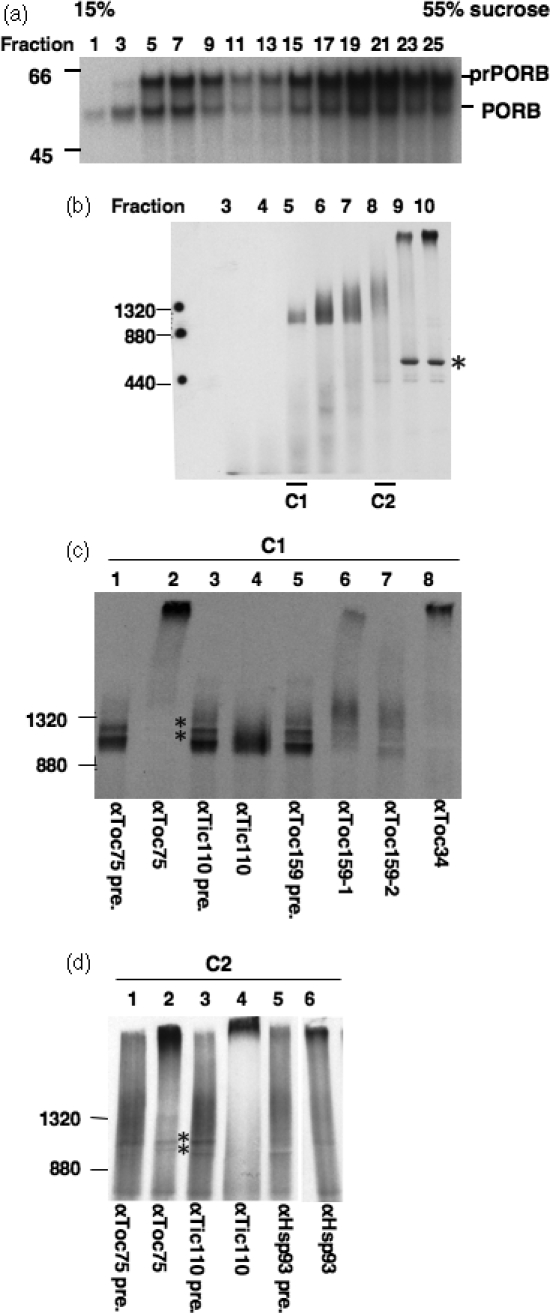
(a) [35S]prPORB was incubated with chloroplasts under import conditions for 20 min. Total membranes from lysed chloroplasts were analyzed by sucrose density gradient as in Figure 2(a).
(b) Fractions 3–10 from (a) were analyzed by BN-PAGE. The asterisk indicates the position of Rubisco holoenzyme.
(c and d) Antibody-shift BN-PAGE analyses of the composition of the C1 and C2 complexes, respectively. Sucrose-density gradient fractions containing C1 or C2 were incubated with various antibodies, as labeled at the bottom, to remove possible components (pre., pre-immune serum of the respective antibody). The sharp bands between 880 and 1320 kDa were formed due to the presence of major serum proteins at the positions marked by asterisks. Anti-Tic110 antibodies were affinity purified and therefore did not contain the serum proteins.
Discussion
We have identified at least two translocon complexes, C1 and C2, which were associated with precursors during active import. Subsequent chase kinetics (Figure 2b) suggested that at least C1 represented an earlier step than the Tic/Toc supercomplex. C1 contained the three major Toc proteins but did not contain any of the Tic protein tested. Precursor engagement with the Toc complex alone had only previously been shown when further translocation was blocked by low-ATP conditions (Akita et al., 1997; Perry and Keegstra, 1994; Schnell et al., 1994). Here we demonstrated that an independent Toc complex is indeed one of the first steps in the active import process. The size of C2 was very close to C1, and C2 contained several additional components which function after initial precursor binding. Furthermore, unlike the supercomplex, which was associated with precursors and processed mature proteins, C2 was only associated with precursors. Therefore it is plausible to speculate that C2 might be an assembly intermediate between C1 and the supercomplex. However, the ratio of C2 to the supercomplex remained constant during the chase (Figure 2b). We cannot exclude the possibility that C2 represented a fixed proportion of precursors dissociated from the supercomplex. Furthermore, to identify components in C1 and C2, we used the antibody-shift assay, which is subjected to variability in antibody efficiency and in the native gel systems. The presence and conformation in the complex of components for which no clean removal of the complex was observed, like Toc159 in C1 and Hsp93 in C2, may need to be confirmed by other techniques.
Based on our data, we propose a working model for the assembly of translocon components during active import (Figure 6). Precursors first associate with the complex C1 composed of the Toc components. The incomplete shift of C1 by anti-Toc159 antibodies suggests that Toc159 may be present in multiple conformations or assembly states. Tic110, Hsp93, and hsp70 are probably among the first components to assemble with the Toc components to form C2. Due to the lack of effective antibodies and the low abundance of Tic20 and Tic22, we were unable to investigate whether they were also present in C2. Other translocon components like Tic40, which is probably among the last to be recruited, then assemble to form the Tic/Toc supercomplex. Processing of transit peptides most likely occurs at the supercomplex.
Figure 6. Model of translocon complex assembly states during precursor import into chloroplasts.
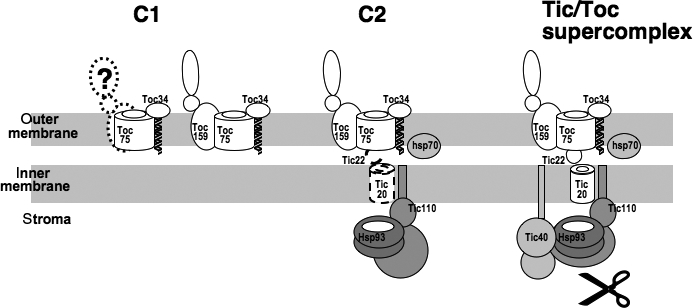
During import, precursors first associate with the C1 complex composed of the Toc components. A population of C1 may contain Toc159 in a different conformation. Another distinct complex, C2, composed of the Toc components, Tic110, Hsp93, and hsp70 exists. However, whether C2 is an intermediate step between C1 and the Tic/Toc supercomplex is not clear. The presence of Tic22 and Tic20 in C2 has also not yet been demonstrated. Other additional translocon components, like Tic40, then assemble to form the Tic/Toc supercomplex. The scissor represents the stromal transit-peptide processing peptidase and its position indicates that processing most likely occurs at the supercomplex.
Due to the large increase in size from C1/C2 to the Tic/Toc supercomplex, it is possible that more copies of existing components or complexes are further assembled together. When a C-terminally truncated form of atTic110 was co-expressed with endogenous full-length atTic110 in Arabidopsis, the truncated form was only detected to be associated with the endogenous Tic110 when assembled in the supercomplex (Inaba et al., 2005). This observation suggests that Tic110 may form multiplemers when assembled into the supercomplex.
Kikuchi et al. (2006) have recently reported the existence of a stable Toc complex of 800–1000 kDa when chloroplasts were directly solubilized and analyzed by BN-PAGE. This Toc complex is most likely the same complex as the C1 complex we have identified. Kikuchi et al. (2006) have further reported that when chloroplasts were subjected to mild proteolysis, or proteolysis plus harsher detergent solubilization, a smaller Toc complex and a partial Toc complex with only Toc159 and Toc75 were observed. Therefore the two smaller complexes observed by Akita et al. (1997) are most likely protease degradation products of the intact Toc complex. In addition, it has been shown that an isolated Toc core complex was about 500 kDa when analyzed by size exclusion chromatography (Schleiff et al., 2003b). The different molecular mass estimations could be due to the different methods used or partial degradation of the isolated complexes. Taken together, these results suggest that the chloroplast outer membrane contained a stable pre-assembled Toc complex that is around 800–1000 kDa on BN-PAGE. We show here that interaction with this Toc complex is one of the earliest steps during active import into chloroplasts.
Hsp70 homologs have been suggested to function in chloroplast protein import in at least three different suborganellar locations: the cytosolic side of the outer membrane (Kourtz and Ko, 1997; May and Soll, 2000; Waegemann et al., 1990), the intermembrane-space side of the outer membrane (Marshall et al., 1990; Schnell et al., 1994) and the stroma (Ivey et al., 2000). However, none of the hsp70 studied has been shown to assemble into complexes with other translocon components. We have identified the presence of an Hsp70 homolog in C2. This protein is most likely the same protein as the integral outer-membrane hsp70 homolog associated with importing precursors (Schnell et al., 1994) since they are identified by the same monoclonal antibody. Our demonstration of its presence in the C2 complex further supports its involvement in chloroplast protein import. Furthermore, recently a J-domain-containing protein, Toc12, has been shown to bind to this integral outer-membrane hsp70 (Becker et al., 2004). It is possible that Toc12 may also be present in C2.
Only a portion of the C1 complex was removed by two anti-Toc159 antibodies. C1 ran as a compact band on BN-PAGE, suggesting that it was composed of a single complex. It is possible that C1 contained Toc159 in different conformations or assembly states. At least two functions have been proposed for Toc159, and both require conformational changes in Toc159. Toc159 has been proposed to function as the soluble receptor in the cytosol that brings chloroplast precursors to the translocon complex in the chloroplast envelope (Hiltbrunner et al., 2001; Smith et al., 2002). Therefore Toc159 is expected to convert between the soluble form in the cytosol and the Toc complex-associated form inserted in the outer membrane. Toc159 has also been proposed to function as the propelling motor that actively pushes precursors into the Toc75 channel (Schleiff et al., 2003a). In this case Toc159 has been proposed to change between an exposed conformation and a conformation partially buried in the Toc75 channel. In both cases, multiple forms of Toc complexes would be expected: some Toc complexes have their Toc159 readily exposed and some have Toc159 protected by other Toc components. It is possible that only those with readily exposed Toc159 could be removed by the anti-Toc159 antibodies in the BN-PAGE antibody shift assays. However, we cannot exclude the possibility that C1 represents two distinct complexes of similar sizes and surface charges. One complex contains Toc159/Toc75/Toc34 and the other contains only Toc75/Toc34 in different copy numbers or with other as yet unidentified components.
Similarly only a portion of the C2 complex was removed by the anti-Hsp93 antibody. However, C2 had a much more smeared migration pattern in BN-PAGE and might indeed represent multiple complexes with only some containing Hsp93. It is also possible that some Hsp93 failed to react with the cross-linker due to the reduced environment of the chloroplast stroma and dissociated from the C2 complex during sample preparation.
Experimental procedures
Protein import into isolated chloroplasts and sucrose density gradient centrifugation
Isolation of chloroplasts from 12-day-old pea (Pisum sativum cv. Little Marvel) seedlings and synthesis of [35S]methionine-labeled precursors were performed as described (Perry et al., 1991). Import was performed in a total volume of 1.5–2 ml which contained chloroplasts at 0.5 mg chlorophyll ml−1 30–50% (v/v), [35S]-labeled precursor proteins and 3 mm ATP in import buffer (50 mm HEPES-KOH pH 8.0 and 300 mm sorbitol). Reactions were incubated at 20°C in the light for the amount of time indicated in each figure and terminated by diluting with cold import buffer and sedimenting the intact chloroplasts. For chase incubation, chloroplasts from a 20-min import sample were isolated and resuspended in cold import buffer containing 3 mm ATP, and further incubated at 20°C for the amount of time indicated. For sucrose density centrifugation, re-isolated chloroplasts from indicated time points were treated with 0.5 mm DSP as described (Akita et al., 1997). Briefly, chloroplasts were resuspended in import buffer to a concentration of 1 mg chlorophyll ml−1. Then DSP, dissolved in DMSO, was added to the chloroplast suspension to a final concentration of 0.5 mm. The cross-linking reaction was carried out in the dark for 15 min on ice. An equal volume of 50 mm glycine in import buffer was then added to quench the reaction. After DSP cross-linking, chloroplasts were re-isolated and lysed by resuspending in a hypotonic buffer (25 mmHEPES-KOH pH 8.0, 4 mm MgCl2). Total membranes were collected by pelleting the lysate at 125 000 g for 45 min. Membranes were solubilized by 2%n-decyl-β-d-maltopyranoside in resuspension buffer [20% (w/v) glycerol, 25 mm BisTris-HCl, pH 7.0] as described (Cline and Mori, 2001) and the lysate was clarified by centrifuging at 100 000 g for 5 min. The supernatant was layered over a 15–55% linear sucrose (in 25 mm BisTris-HCl, pH 7.0) density gradient and sedimented at 150 000g for 18 h in a SW41 rotor. Fractions were removed and the sedimentation patterns were analyzed by SDS-PAGE and fluorography. For immunoblotting, secondary antibodies were linked to alkaline phosphatase. Quantifications of gel images were performed on Fuji LAS-1000plus pictrography 3000 (Fuji, Tokyo, Japan).
BN-PAGE and antibody-shift analyses
Blue-native PAGE was carried out as described (Schägger et al., 1994) with the following modifications. Samples from sucrose density gradients were combined with 1/10 volume of 5% Serva blue G prepared in 100 mm BisTris-HCl, pH 7.0, 0.5 m 6-amino-n-caproic acid, and 30% sucrose, and applied to 0.75-mm-thick 5–13.5% acrylamide gradient gels in a Hoefer Mighty Small vertical electrophoresis unit connected to a cooling circulator (Amersham GE Healthcare, Piscataway, NJ, USA). Electrophoresis was performed for 15–20 h at 50–100 V and 2–4°C. For antibody-shift assays, 16 μl of an antiserum was added to 35 μl sample from the sucrose density gradient and incubated for 30 min on ice. Twenty-four microliters of Protein A-Sepharose (Pierce, Rockford, IL, USA) was then added and further incubated for 30 min. After centrifuging at 1000 g for 5 min, the supernatant was analyzed by BN-PAGE. Antibodies against Toc75, Toc34, Toc159 (antiToc159-1), and Tic110 were produced as described (Tu et al., 2004). Antibodies against Tic40 were produced by subcloning the coding region of Arabidopsis Tic40 residues 385–447 into pGEX5X-1 (Amersham, Piscataway, NJ, USA), overexpressing the recombinant protein with an N-terminal GST fusion in Escherichia coli and injecting the purified recombinant protein into rabbits. Antibodies against Hsp93 were produced by subcloning the coding region for the mature region of pea Hsp93 into pET28a (Invitrogen, San Diego, CA, USA), overexpressing the recombinant protein with an N-terminal His6 tag in E. coli and injecting the purified recombinant protein into rabbits. Monoclonal antibody SPA-820 against Hsp70 was purchased from Stressgen (Victoria, Canada). Some pre-immune sera were not available but all pre-immune sera tested gave the same results (data not shown). Anti-Tic110 serum used in Figures 4(c) and 5 was purified by affinity purification with atTic11093−966 (Inaba et al., 2003). Materials used in this study will be available upon request.
Acknowledgments
We thank Lih-Jen Chen for her excellent technical assistance. We thank Dr Danny Schnell for the anti-Toc159-2 antibody and the construct for atTic11093−966, Dr Shingo Kikuchi and Dr Masato Nakai for advice on BN-PAGE and Ming-Yan Kuo and Dr Ming-Lun Chou for the anti-Tic40 antibodies. This work was supported by grants from the National Science Council (NSC 93-2321-B001-011, H-mL) and Academia Sinica (AS92-IMB2, H-mL).
References
- Akita M. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Becker T. Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts Mol. Biol. Cell. 2004;15:5130–5144. doi: 10.1091/mbc.E04-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Chou M-L. Tic40, a membrane-anchored co-chaperone homologue in the chloroplast protein translocon. EMBO J. 2003;22:2970–2980. doi: 10.1093/emboj/cdg281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 2001;154:719–729. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D. An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 2004;38:93–106. doi: 10.1111/j.1365-313X.2004.02024.x. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A. Targeting of an abundant cytosolic form of the protein import receptor atToc159 to the outer chloroplast membrane. J. Cell Biol. 2001;154:309–316. doi: 10.1083/jcb.200104022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah SC. Reconstitution of a chloroplast protein import channel. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T. atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 2003;278:38617–38627. doi: 10.1074/jbc.M306367200. [DOI] [PubMed] [Google Scholar]
- Inaba T. atTic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 2005;17:1482–1496. doi: 10.1105/tpc.105.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D. Two novel proteins in the mitochondrial outer membrane mediate b-barrel protein assembly. J. Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey RA., III Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol. 2000;122:1289–1299. doi: 10.1104/pp.122.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DT. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 1998;273:16583–16588. doi: 10.1074/jbc.273.26.16583. [DOI] [PubMed] [Google Scholar]
- Kessler F. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 2006;7:248–257. doi: 10.1111/j.1600-0854.2005.00382.x. [DOI] [PubMed] [Google Scholar]
- Kessler F. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Kikuchi S. Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol. 2006;47:363–371. doi: 10.1093/pcp/pcj002. [DOI] [PubMed] [Google Scholar]
- Kourtz L. The early stage of chloroplast protein import involve Com70. J. Biol. Chem. 1997;272:2808–2813. doi: 10.1074/jbc.272.5.2808. [DOI] [PubMed] [Google Scholar]
- Kubis S, et al. Functional Specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl Acad. Sci. USA. 1990;87:374–378. doi: 10.1073/pnas.87.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–63. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 1992;267:433–439. [PubMed] [Google Scholar]
- Olsen LJ. ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 1989;246:6724–6729. [PubMed] [Google Scholar]
- Perry SE. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE. In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol. 1991;34:327–344. doi: 10.1016/s0091-679x(08)61688-x. [DOI] [PubMed] [Google Scholar]
- Poetsch A. Dye removal, catalytic activity and 2D crystallization of chloroplast H+-ATP synthase purified by blue native electrophoresis. Biochim. Biophys. Acta. 2000;1466:339–349. doi: 10.1016/s0005-2736(00)00191-7. [DOI] [PubMed] [Google Scholar]
- Reumann S. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc. Natl Acad. Sci. USA. 1999;96:784–789. doi: 10.1073/pnas.96.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroth S. Thyalkoid membrane at altered metabolic state: challenging the forgotten realms of the proteome. Electrophoresis. 2003;24:2814–2823. doi: 10.1002/elps.200305543. [DOI] [PubMed] [Google Scholar]
- Schägger H. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schleiff E. A GTP-driven motor moves proteins across the outer envelope of chloroplasts. Proc. Natl Acad. Sci. USA. 2003a;100:4604–4609. doi: 10.1073/pnas.0730860100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E. Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 2003b;160:541–551. doi: 10.1083/jcb.200210060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Smith MD. The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 2002;159:833–843. doi: 10.1083/jcb.200208017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 2004;165:323–334. doi: 10.1083/jcb.200311074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J. Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- Sveshnikova N. TOC34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl Acad. Sci. USA. 2000;97:4973–4978. doi: 10.1073/pnas.080491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S-L. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell. 2004;16:2078–2088. doi: 10.1105/tpc.104.023952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K. Translocation of proteins into isolated chloroplasts requires cytosolic factors to obtain import competence. FEBS Lett. 1990;261:89–92. [Google Scholar]
- Werner-Washburne M. Analysis of pea chloroplast inner and outer envelope membrane proteins by two dimensional gel electrophoresis and their comparison with stromal proteins. Plant Physiol. 1983;73:569–575. doi: 10.1104/pp.73.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]


