SUMMARY
Histone lysine methylation has been linked to the recruitment of mammalian DNA repair factor 53BP1 and putative fission yeast homolog Crb2 to DNA double-strand breaks (DSBs), but how histone recognition is achieved has not been established. Here we demonstrate that this link occurs through direct binding of 53BP1 and Crb2 to histone H4. Using X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, we show that, despite low amino acid sequence conservation, both 53BP1 and Crb2 contain tandem tudor domains that interact with histone H4 specifically dimethylated at Lys20 (H4-K20me2). The structure of 53BP1/H4-K20me2 complex uncovers a unique five-residue 53BP1 binding cage, remarkably conserved in the structure of Crb2, that best accommodates a dimethyllysine but excludes a trimethyllysine, thus explaining the methylation state-specific recognition of H4-K20. This study reveals an evolutionarily conserved molecular mechanism of targeting DNA repair proteins to DSBs by direct recognition of H4-K20me2.
INTRODUCTION
Although histone lysine methylation has a well-documented regulatory role in DNA transcription (Martin and Zhang, 2005; Strahl and Allis, 2000), its involvement in other DNA functions is now beginning to emerge. Two recent studies have uncovered a link between histone lysine methylation and the DNA damage checkpoint and double-strand break (DSB) repair proteins 53BP1 in mammals and fission yeast putative homolog Crb2, where the relocalization of 53BP1 and Crb2 to DNA DSBs necessitates histone methylation (Huyen et al., 2004; Sanders et al., 2004). The mechanism of this dependency on histone methylation is not yet understood (Vidanes et al., 2005). It is unclear whether 53BP1 and Crb2 directly interact with methylated histones. There is also uncertainty in the identity of the histone partner, as biochemistry and cell biology studies seem to correlate 53BP1 to methylated Lys79 of histone H3 (H3-K79) (Huyen et al., 2004) while genetic data from fission yeast compellingly connect Crb2 to methylated Lys20 of histone H4 (H4-K20) (Nakamura et al., 2005; Sanders et al., 2004). Furthermore, it is not known whether the histone methylation state (mono-, di- or trimethylated) is relevant to the specificity and affinity of the interaction.
53BP1 contains canonical tandem tudor domains (Charier et al., 2004) that are putative methylated histone-binding modules (Maurer-Stroh et al., 2003). Thus, the current thinking is that 53BP1 is recruited to chromatin regions flanking DNA DSBs via interaction of its tudor domains with methylated H3-K79 and that Crb2 may also have a tandem of tudor domains that directly recognize methylated H4-K20. However, in the absence of quantitative binding studies and three-dimensional (3D) structural information on canonical tandem tudor domain complexes, one cannot draw any hypothesis on the mechanism of such interactions, if they do exist.
Here, we investigate the molecular mechanism linking methylated histones to 53BP1 and Crb2. We demonstrate a direct interaction between 53BP1 tandem tudor domains and histone H4 specifically dimethylated at Lys20 (H4-K20me2) and show that dimethylated H4-K20, and not H3-K79, contributes to the relocation of 53BP1 to sites of DNA DSBs. The 3D structures and dynamics of free and H4-K20me2-bound 53BP1 tudor domains reveal a unique five-residue cage in the first tudor domain that becomes ordered upon interaction. This binding pocket best accommodates a dimethyllysine but blocks a trimethyllysine, explaining the methylation state-specific recognition of histone H4. By 3D structure determination, we show that despite low amino acid sequence similarity, Crb2 is structurally related to 53BP1 in having two tudor domains and a conserved dimethyllysine-binding pocket, and that, like 53BP1, it directly binds H4-K20me2.
RESULTS AND DISCUSSION
53BP1 Selectively Recognizes Histone H4 Dimethylated at Lys20 via Its Tandem Tudor Domains
To investigate the mechanism of methylated histone recognition in DNA DSB repair and to test the possibility that 53BP1 might bind both histone H3 and histone H4, we examined by isothermal titration calorimetry (ITC) the interaction of human 53BP1 tandem tudor domains (residues 1484–1603) with a series of H4-K20 (residues 12–25) and H3-K79 (residues 74–83) peptides carrying various lysine methylation states (Figure 1). ITC measurements revealed that 53BP1 binds to histone H4-K20 with a stoichiometry of one H4 peptide for two tudor domains. Interestingly, 53BP1 is highly selective for the dimethyllysine-containing H4 peptide (H4-K20me2) with a dissociation constant (KD) of 19.7 μM (Figure 1A). In contrast, the affinity of 53BP1 for non- and trimethylated H4 peptides is low (KD, ~1.0 mM). Under the same conditions, the KD of 53BP1 for a monomethylated histone H4 peptide is 52.9 μM. Because there is no precedent for the selective recognition of dimethyllysine over trimethyllysine, we verified the integrity of the trimethylated H4-K20 peptide by titrating it to the hybrid tudor domains of JMJD2A, a protein known to nonselectively bind methylated histones H3-K4 and H4, including trimethylated H4-K20 (Kim et al., 2006). A tight interaction was measured (KD, 0.7 μM), ruling out any defect in the trimethylated H4-K20 peptide (Figure 1A).
Figure 1. Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 Tandem Tudor Domains.
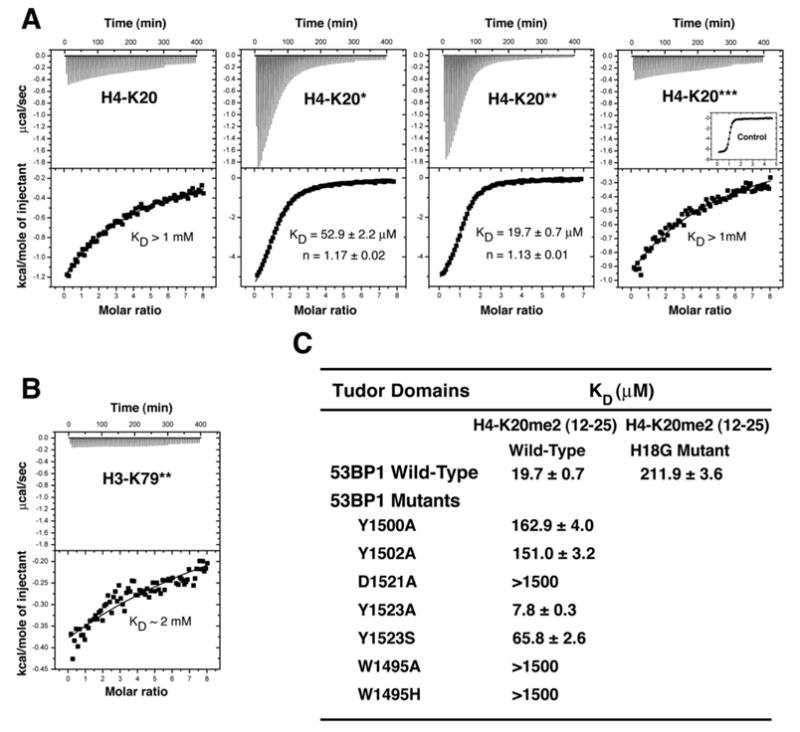
(A) Representative ITC results for the titration of 53BP1 tandem tudor domains with, from left to right panels, nonmethylated, monomethylated, dimethylated, and trimethylated H4-K20 (residues 12–25). The number of stars (★) indicates the number of methyl groups attached to Nζ of Lys20. Shown for each experiment are the integrated heat measurements from raw titration data as well as curve fitting with a standard one-site model. Inset in the last panel is a control titration of JMJD2A hybrid tudor domains with trimethylated H4-K20.
(B) ITC titration of 53BP1 with dimethylated H3-K79 (residues 74–83).
(C) Effects of point mutations in 53BP1 tandem tudor domains on the interaction with H4-K20me2. For each interaction, the dissociation constant (KD) derived by fitting a standard one-binding site model (Wiseman et al., 1989) is reported with the associated error determined by nonlinear least squares analysis.
Using ITC, we also detected an interaction, albeit with very low affinity (KD, ~2.0 mM), between 53BP1 and histone H3 specifically dimethylated at lysine 79 (H3-K79me2) (Figure 1B). This interaction is three orders of magnitude weaker than what was previously estimated (Huyen et al., 2004). No binding could be measured for the non-, mono-, and trimethylated H3-K79 peptides (data not shown). In a recent study using protein domain microarrays, no interaction was detected between 53BP1 and methylated H3-K79 peptides (Kim et al., 2006).
The region of histone H3 containing methylated lysine 79 is buried and in close proximity to DNA in nucleosomes (Luger et al., 1997). Upon this histone H3 segment becoming exposed, the tudor domains of 53BP1 may contact not only H3-K79me2 but also DNA, resulting in higher affinity. In fact, a number of studies have shown nucleic acid binding to a region of 53BP1 that encompasses the tandem tudor domains (Charier et al., 2004; Iwabuchi et al., 2003; Pryde et al., 2005). Thus, even if the binding of 53BP1 to H3-K79me2 is extremely weak in vitro, such interaction might still have some biological significance. Histone H4-K20 is likewise buried and in close vicinity to DNA in stacked nucleosomes (Luger et al., 1997). The two orders of magnitude higher affinity measured for the interaction of 53BP1 with H4-K20me2 compared to H3-K79me2 (Figures 1A and 1B) then suggests that if both H3-K79me2 and H4-K20me2 function in DSB repair, the latter may be the primary target of 53BP1.
Structural Basis for the Recognition of Histone H4 Dimethylated at Lys20 by 53BP1 Tandem Tudor Domains
To understand the selective recognition of H4-K20me2 by 53BP1, we determined the 3D structures of 53BP1 tandem tudor domains in the absence and presence of a histone H4 peptide (residues 16–25) at 1.25 Å and 1.70 Å resolution, respectively (Figure 2A). This ten-residue H4-K20me2 peptide binds as tightly as the 14 amino acid peptide used in previous experiments (Figure 1A). The structures were solved by molecular replacement with the 2.8 Å resolution structure of free 53BP1 (Huyen et al., 2004). The crystallographic statistics are given in the Supplemental Data available with this article online (Table S1).
Figure 2. 3D Structures and Interaction of Human 53BP1 Tandem Tudor Domains.
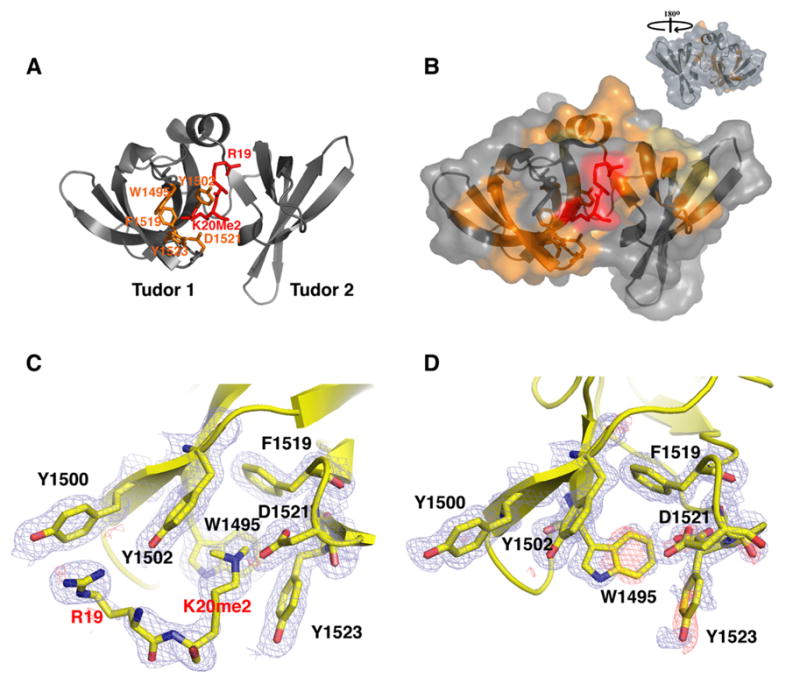
(A) Ribbon representation of the tandem tudor domains of 53BP1 in complex with dimethylated H4-K20 peptide (red stick representation). The side chains of Trp1495, Tyr1502, Phe1519, Tyr1523, and Asp1521, forming the dimethyllysine-binding cage, are shown in orange. The dimethyllysine is labeled K20me2.
(B) Molecular surface representation of 53BP1 tandem tudor domains in complex with dimethylated H4-K20. 53BP1 residues with NMR weighted average chemical shift differences >0.04 ppm between free and H4-K20me2-bound 53BP1 forms are in orange. 53BP1 residues for which resonances disappeared upon peptide binding are in yellow. The H4-K20me2 peptide is in red. Inset is the view after a 180° rotation along the vertical axis.
(C and D) Close-up view of the peptide-binding site in 53BP1/H4-K20me2 complex (C) and in free 53BP1 (D). The Fo – Fc and 2Fo – Fc electron density maps are overlaid for both structures (contoured at 1 σ level, blue and red mesh). Note the negative densities (red) for Trp1495 and Tyr1523 in the free protein, indicative of conformational disorder (D). Two alternative conformations are shown for the side chain of Asp1521 (D).
The structures of free and H4-K20me2-bound 53BP1 explain the high selectivity of 53BP1 for dimethyllysine 20 of histone H4 and allow the accurate description of an induced fit mechanism that accompanies this interaction. Clearly seen in the electron density map of the complex are histone H4 residues Arg19 and dimethylated Lys20 (Figure 2C). The guanidinium group of Arg19 makes a cation-π interaction with the phenyl ring of Tyr1500 in 53BP1 while the dimethyllysine is caged by four aromatic residues (Trp1495, Tyr1502, Phe1519, and Tyr1523) and Asp1521, all from the first tudor domain of 53BP1 and arranged approximately orthogonally. These aromatic amino acids participate in van der Waals and cation-π interactions with the dimethyllysine ammonium group. The favorable coulombic interaction in the form of a salt bridge between the carboxylate group of Asp1521 and the dimethylammonium ion of H4-K20me2 also likely contributes to the affinity and high specificity of 53BP1 for H4-K20me2. This is supported by semiempirical and ab initio molecular orbital calculations that predict that the ion-pair interaction involving a carboxylate group is significantly stronger with a dimethylammonium than with a trimethylammonium group (Mavri and Vogel, 1994). A direct hydrogen bond between the carboxylate group of Asp1521 and the amino proton of H4-K20me2, as attested by the optimal 2.77 Å Nζ-O distance and ~125° θ bond angle, further stabilizes the complex. This is another likely key determinant of 53BP1 specificity for a dimethyllysine- over a trimethyllysine-containing H4 peptide, since a trimethyllysine does not have an amino proton and thus cannot form a hydrogen bond. The very low affinity of 53BP1 for trimethylated H4-K20 could also be partly explained by steric hindrance, since in the absence of any side-chain reorientation in 53BP1 a trimethyllysine would be too bulky to fit into the five-residue enclosure. While non- and monomethylated H4-K20 can participate in energetically favorable hydrogen bond and salt bridge interactions with Asp1521, they have less optimal van der Waals and cation-π interactions with the four aromatic rings in the binding cage of 53BP1, likely explaining their lower affinities compared to H4-K20me2.
The dimethyllysine-binding pocket of 53BP1 is distinct from any other known methyllysine recognition motif in protein domains such as the chromodomain of HP1 (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002), the double chromodomains of CHD1 (Flanagan et al., 2005), the hybrid tudor domains of JMJD2A (Huang et al., 2006), and the PHD finger of NURF and ING2 (Li et al., 2006; Pena et al., 2006), which all bind tightest to trimethylated peptides from histone H3 and also bind other lysine methylation states with relatively high affinities. It is remarkable that the tudor domains of JMJD2A adopt a fold topology radically different from that of 53BP1 despite significant amino acid sequence similarity, and hence are classified as hybrid tudor domains in opposition to the canonical tudor domains of 53BP1 (Huang et al., 2006). JMJD2A uses the second hybrid tudor domain for trimethyllysine recognition of histone H3-K4 (Huang et al., 2006), whereas it is the first tudor domain of 53BP1 that binds the dimethyllysine of H4-K20. The lower selectivity for the different methylation states and preference for trimethyllysine-containing sequences by HP1, CHD1, JMJD2A, NURF, and ING2 may be explained by a more open binding cavity involving fewer residues in direct contact with the methylated lysine side chain: two to four (all aromatic) versus five in 53BP1. Importantly, only in 53BP1 is there a direct hydrogen bond and salt bridge formed between a carboxylate group (of Asp1521) and the dimethylammonium group.
His18 is another residue of H4-K20me2 crucial for its interaction with 53BP1, as attested by the ~11-fold decrease in affinity detected after His18 was mutated to a glycine (Figure 1C). His18 was not modeled in the 3D structure, as no obvious electron density was observed for its side chain, presumably because of the dynamics of interaction as explained in the next subsection. However, from the structure of the 53BP1/H4-K20me2 complex, His18 is expected to lie in a shallow pocket in 53BP1 defined by the side chains of Tyr1502, Leu1547, Glu1549, and Met1584. Noticeably, these last three residues are part of the second tudor domain, indicating that the two tudor domains function in concert to recognize H4-K20me2.
We note that, unlike H4-K20me2, the methylated sites K9 and K27 of histone H3 are preceded by an arginine but lack a histidine before the arginine. This may explain the low affinities of H3-K9me2 (residues 1–15) and H3-K27me2 (residues 19–33) peptides for 53BP1 tandem tudor domains: i.e., KD, 385.0 ± 34.0 μM and 434.8 ± 14.4 μM, respectively, as determined by ITC.
Mutations, Stability, and Dynamics of 53BP1/H4-K20me2 Complex
The 53BP1 residues in the dimethyllysine-binding cage were selectively mutated to evaluate their contribution to the stability of the complex with H4-K20me2 (Figure 1C). Trp1495, Tyr1502, Tyr1523, and Asp1521 were considered good targets for mutation. They are only involved in binding H4-K20me2, are solvent accessible, and do not contribute to the protein fold as verified by gel filtration chromatography of all mutants and from the 1H-15N heteronuclear single-quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectra of the W1495A and Y1523A 53BP1 mutants (data not shown). Phe1519 participates not only in binding H4-K20me2 but also in the tertiary structure of 53BP1 and was not mutated.
A striking feature of Trp1495 and Tyr1523 side chains is that their electron densities become visible only in the complex, indicating that they become ordered upon peptide binding (Figures 2C and 2D). In the 1.25 Å resolution crystal structure of free 53BP1, two alternative conformations can be modeled for the side chain of Asp1521 (Figure 2D), another indication of structural flexibility. Mutation of Trp1495 to an alanine or histidine abrogates 53BP1 binding to H4-K20me2 peptide, underscoring the importance of Trp1495 for interaction (Figure 1C). Serine substitution of Tyr1523 shows a significantly higher KD (65.8 μM) whereas the Y1523A mutant has a decreased KD (7.8 μM) relative to the wild-type protein (KD, 19.7 μM), suggesting that the hydrophobicity of residue 1523 is key to the high affinity of 53BP1 for H4-K20me2 (Figure 1C). The Y1502A 53BP1 mutant results in a significantly weaker interaction with H4-K20me2, with a binding affinity reduced by a factor of ~8. The D1521A mutant abolishes 53BP1 interaction with H4-K20me2 (Figure 1C), hence demonstrating the importance of the hydrogen bond and ion-pair interaction that Asp1521 forms with the dimethyllysine (Figure 2C). Tyr1500 is not part of the dimethyllysine-binding pocket of 53BP1 but contacts Arg19 of histone H4 (Figure 2C). Replacement of Tyr1500 by an alanine greatly weakens 53BP1 binding to H4-K20me2 with an affinity one-eighth that of the wild-type protein (Figure 1C).
The fact that only two amino acids of histone H4, Arg19 and dimethylated Lys20, can be modeled in the electron density map of the complex indicates that some of the other H4 residues may be involved in conformational exchange. To test this possibility, we compared the 1H-15N HSQC spectra of 53BP1 tandem tudor domains with and without H4-K20me2 (Figures 2B and 3A). While the majority of changes in backbone amide chemical shifts of 53BP1 are localized in the first tudor domain (24 residues affected) where, as seen from the 3D structure, Arg19 and dimethylated Lys20 interact, other changes occur in the second tudor domain (12 residues affected), specifically at the interface with the first domain where His18 of histone H4 is expected to be positioned (Figure 2B). A subset of the signals corresponding to this interfacial region of the second tudor domain are missing in the 1H-15N HSQC spectrum of H4-K20me2-bound 53BP1 due to severe exchange broadening. This indicates slow conformational exchange on the microsecond to millisecond time scale of the 53BP1/H4-K20me2 complex (Figures 2B and 3A).
Figure 3. 53BP1 Uses the Same Binding Site for H4-K20me2 and H3-K79me2.
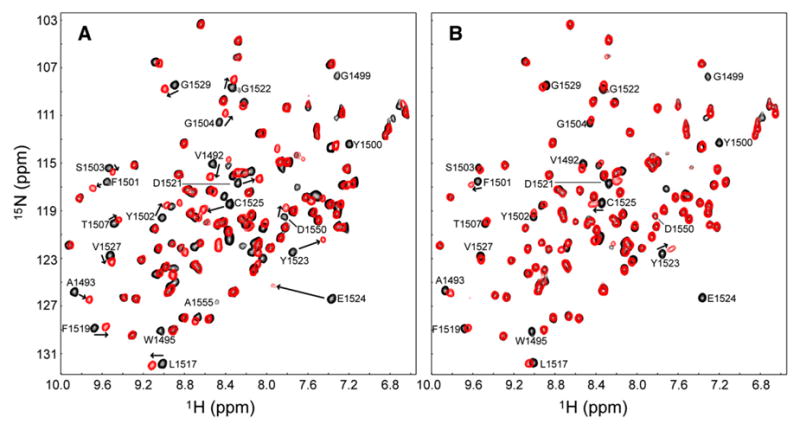
(A and B) (A) 1H-15N HSQC spectra of H4-K20me2-bound (red) versus free (black) 53BP1 and (B) H3-K79me2-bound (red) versus free (black) 53BP1. H4-K20me2 (residues 16–25) and H3-K79me2 (residues 74–83) peptides were nonlabeled, while 53BP1 was 15N labeled. Peptides were added to the protein at peptide:protein ratios of 1.2:1 in (A) and 10:1 in (B). Representative 53BP1 residues perturbed (shifted or disappeared peaks) by addition of H4 and H3 peptides are labeled.
The conformational flexibility of 53BP1 observed here may explain why 53BP1 is able to bind both H4-K20me2 (strongly) and H3-K79me2 (weakly), two peptides that do not share any sequence similarity with the exception of the dimethyllysine. Indeed, although the very low affinity of 53BP1 for H3-K79me2 precluded crystallographic studies of the complex, by using NMR spectroscopy we were able to establish that H4-K20me2 and H3-K79me2 peptides share the same binding site on 53BP1. We titrated nonlabeled H3-K79me2 (residues 74–83) into 15N-labeled 53BP1 tandem tudor domains and monitored the changes in the 1H-15N HSQC spectrum of 53BP1 (Figure 3B). The 53BP1 residues exhibiting chemical shift changes upon addition of H3-K79me2 are the same as those perturbed by H4-K20me2 (residues 16–25), although more strongly with the latter, demonstrating that the two peptides bind the same region of 53BP1 (Figure 3). While the exchange between the H4-K20me2-bound and free states of 53BP1 is intermediate to fast on the NMR chemical shift time scale, this exchange is fast for free and H3-K79me2-bound 53BP1, consistent with the affinity of H3-K79me2 for 53BP1 being lower than that of H4-K20me2: micromolar range KD for H4-K20me2 versus milli-molar range for H3-K79me2, in agreement with the ITC data (Figures 1A and 1B).
The Interaction between 53BP1 and H4-K20me2, and Not H3-K79me2, Is Necessary for the Accumulation of 53BP1 to DNA DSBs
In a first attempt to evaluate the relative contributions of H3-K79me2 and H4-K20me2 to the relocation of 53BP1 to DNA DSBs, we used short hairpin RNA (shRNA) to inactivate the enzyme Dot1 responsible for H3-K79 methylation (Feng et al., 2002). Stable Dot1 knockdown HeLa and A549 cell lines were generated. Details of the procedure are given in the Supplemental Data. Downregulation of Dot1 expression in either HeLa or A549 cells had virtually no effect on the relocalization of 53BP1 to ionizing radiation (IR)-induced DNA DSBs (Figure S1A), suggesting that H3-K79me2 does not play an important role in this process. We verified that Dot1 was effectively downregulated (Figures S1B and S1C), that nonradiated cells had a normal appearance, and that activation of ATM kinase and phosphorylation of histone H2AX, which signal the presence of DNA DSBs, were normal in irradiated cells (data not shown).
To further evaluate the contribution of H3-K79me2 to the 53BP1-mediated DNA damage response, we set out to generate Dot1-deficient mice or cells. Although lack of a functional Dot1 gene resulted in late embryonic lethality, we were able to culture viable Dot1-deficient mouse embryonic fibroblasts (MEFs). Western blot analysis showed that methylation of H3-K79 was abolished in Dot1 null (Dot1−/−) cells (Figure 4C), a confirmation that Dot1 is the enzyme required for H3-K79 methylation in vivo. In these Dot1 null cells, recruitment of 53BP1 to sites of DNA DSBs still proceeded normally as revealed by immunofluorescence microscopy (Figures 4A and 4B). IR-induced foci formation of 53BP1 in Dot1−/− cells was indistinguishable from that observed in wild-type cells (Figures 4A and 4B). Activation of ATM kinase and phosphorylation of histone H2AX appeared normal in irradiated cells lacking Dot1 (Figure 4B). We also verified that no detectable change occurred in the level of histone H4-K20 dimethylation in Dot1−/− compared to wild-type cells. These data therefore demonstrate that an interaction between 53BP1 and methylated H3-K79 is not a requirement for the localization of 53BP1 to DNA DSBs and strongly suggest that such interaction does not participate in the initiation of the DNA damage response.
Figure 4. Lack of Histone H3 Methylation at Lys79 in Dot1-Deficient Cells Does Not Affect 53BP1 Localization to the Sites of DNA DSBs.
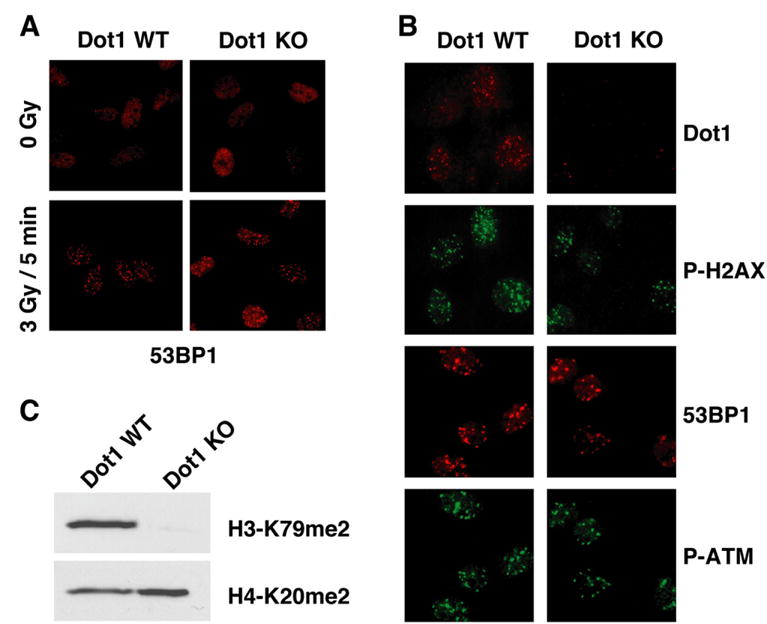
(A) Wild-type Dot1 (Dot1 WT) and Dot1 null (Dot1 KO) primary MEFs were mock treated or irradiated with 3 Gy of IR and analyzed 5 min later by immunostaining using anti-53BP1 (53BP1) antibody.
(B) Wild-type Dot1 (Dot1 WT) and Dot1 null (Dot1 KO) primary MEFs were irradiated with 1 Gy of IR and analyzed 1 hr later by immunostaining using anti-Dot1 (Dot1), anti-phosphorylated-H2AX (Ser139) (P-H2AX), anti-53BP1 (53BP1), and anti-phosphorylated-ATM kinase (Ser1981) (P-ATM) antibodies.
(C) Methylation of Lys79 of histone H3 in Dot1 WT and Dot1 KO MEFs was analyzed by anti-dimethylated H3-K79 (H3-K79me2) immunoblotting. Anti-dimethylated H4-K20 (H4-K20me2) immunoblotting was used as loading control.
Next, we downregulated H4-K20 methylation by short interfering RNA (siRNA) knockdown of the PR-Set7/Set8 histone methyltransferase. PR-Set7/Set8 catalyzes the monomethylation of H4-K20 (Couture et al., 2005; Fang et al., 2002; Nishioka et al., 2002; Xiao et al., 2005), a prerequisite for the subsequent H4-K20 di- and trimethylation (Karachentsev et al., 2005). HeLa cells triple transfected with PR-Set7/Set8 siRNA showed severe impairment in 53BP1 foci formation upon treatment with IR (Figure S2A). Western blot analysis confirmed that PR-Set7/Set8 down-regulation was associated with reduced methylation of H4-K20 (Figure S2B). Although these data suggest a key role for H4-K20 in vivo, we could not rule out other contributions to lack of 53BP1 foci since some of the PR-Set7/Set8 siRNA-transfected cells appeared to be arrested in S phase. Future genetic studies are needed to confirm these observations.
These findings lend support to a preponderant role of dimethylated histone H4-K20 in targeting 53BP1 to DSBs. Our results parallel the recent discovery in fission yeast that methylation of H4-K20 is required for efficient IR-induced foci formation of the 53BP1 homolog Crb2 (Sanders et al., 2004; Du et al., 2006). Loss of the methyltransferase Set9 that methylates H4-K20 in fission yeast results in increased DNA damage sensitivity (Sanders et al., 2004).
Key Residues for the Interaction of 53BP1 with Dimethylated H4-K20 In Vitro Are Essential for 53BP1 Binding to Chromatin In Vivo
With several lines of evidence presenting H4-K20me2 as the binding partner of 53BP1 in DNA DSB repair, we assessed the contribution of 53BP1 residues to its ability to bind H4-K20me2 and be recruited to DSBs in vivo. Having knowledge of the 3D structure and dynamics of 53BP1/H4-K20me2 complex, we selectively mutated key residues in the dimethyllysine-binding cage to probe their role in the interaction of full-length 53BP1 with chromatin in the cell (Figure 5). Substitution of Trp1495 with a valine compromised the targeting of 53BP1 to DNA DSBs, even more so when replaced with an alanine (Figure 5), which is consistent with the total loss of interaction between W1495A 53BP1 mutant and H4-K20me2 as monitored in vitro by ITC. In contrast, the W1495F substitution does not have any effect on the localization of 53BP1 to DNA DSBs. This is expected since this mutation mimics the H4-K20me2-binding site of Crb2 as explained in the next subsection. Y1523A substitution does not affect 53BP1 localization to DNA DSBs, in complete agreement with ITC results showing that this mutant has a slightly higher affinity than wild-type 53BP1 for H4-K20me2 (Figure 1C).
Figure 5. 53BP1 Localization to Sites of DNA DSBs Is Mediated by the Interaction between 53BP1 Tandem Tudor Domains and Dimethylated Lys20 of Histone H4.
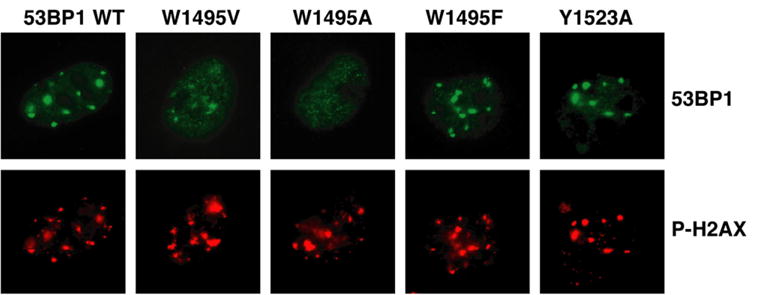
53BP1 relocalization (53BP1) in irradiated 53BP1 null MEFs transiently transfected with 53BP1 wild-type (WT) or 53BP1 mutated in the H4-K20me2-binding cage. Sites of DNA DSBs were marked by costaining using anti-phosphorylated-H2AX (Ser139) (P-H2AX) antibody.
Previously, other surface residue mutants of human 53BP1 were tested in vivo in an effort to identify amino acids important for the localization of 53BP1 to DNA DSBs (Huyen et al., 2004). In retrospect, the effects of these mutations are well explained by the 3D structure of 53BP1/H4-K20me2 complex. Mutating Tyr1552 and Phe1553 to alanines was found to have no influence on the recruitment of 53BP1 to DSBs. There is no change in chemical shift for these amino acids in the 1H-15N HSQC spectrum of 53BP1 upon titration with H4-K20me2 (Figure 3A). The structure clearly shows that these two residues are distant from the binding interface of 53BP1 for histone H4. Y1502Q and Y1502L mutations abrogated the ability of 53BP1 to relocate to DNA damage sites. Y1502 is one of the residues in the dimethyllysine-binding cage. Mutating it to an alanine significantly reduces the affinity of 53BP1 for H4-K20me2, as we showed by ITC (Figure 1C). The same rationale explains why a corresponding point mutation (Y1487L) in murine 53BP1 abolished the ability of 53BP1 to relocalize to laser-induced DSB tracks (Bekker-Jensen et al., 2005). The mutants D1521R and D1521A completely prevented 53BP1 relocation to DSBs. From the structure, Asp1521 is located in the binding pocket and forms a hydrogen bond and salt bridge with the dimethyllysine. We showed by ITC that the D1521A 53BP1 mutant does not interact with H4-K20me2 in vitro (Figure 1C). The mutational analyses presented above illustrate the importance of 53BP1 interaction with dimethylated H4-K20 for the binding of 53BP1 to chromatin in vivo.
Crystallographic and NMR Studies Reveal that Crb2 Contains Tandem Tudor Domains with a Conserved Dimethylated H4-K20-Binding Site
Our finding that 53BP1 specifically recognizes dimethylated histone H4-K20 in vitro and relocates to DSBs via this interaction correlates well with results from genetic experiments in fission yeast linking the accumulation of Crb2 at DNA damage sites to histone H4-K20 methylation (Nakamura et al., 2005; Sanders et al., 2004). Since 53BP1 directly binds H4-K20me2 as we have shown, then by analogy the genetic link between Crb2 and histone H4-K20 may also involve direct binding of Crb2 to methylated H4-K20. In fact, a segment of Crb2 has limited sequence similarity to the first tudor domain of 53BP1 (Figure 6E), suggesting that, like 53BP1, Crb2 may contain a tandem of tudor domains. To examine such possibility, multiple Crb2 DNA constructs encompassing the region of similarity were generated and the corresponding 15N-labeled proteins were screened by 1H-15N HSQC spectroscopy to identify folded domains with well-defined boundaries.
Figure 6. Crb2 Contains Tandem Tudor Domains and Interacts with Dimethylated Lys20 of Histone H4 Like 53BP1.
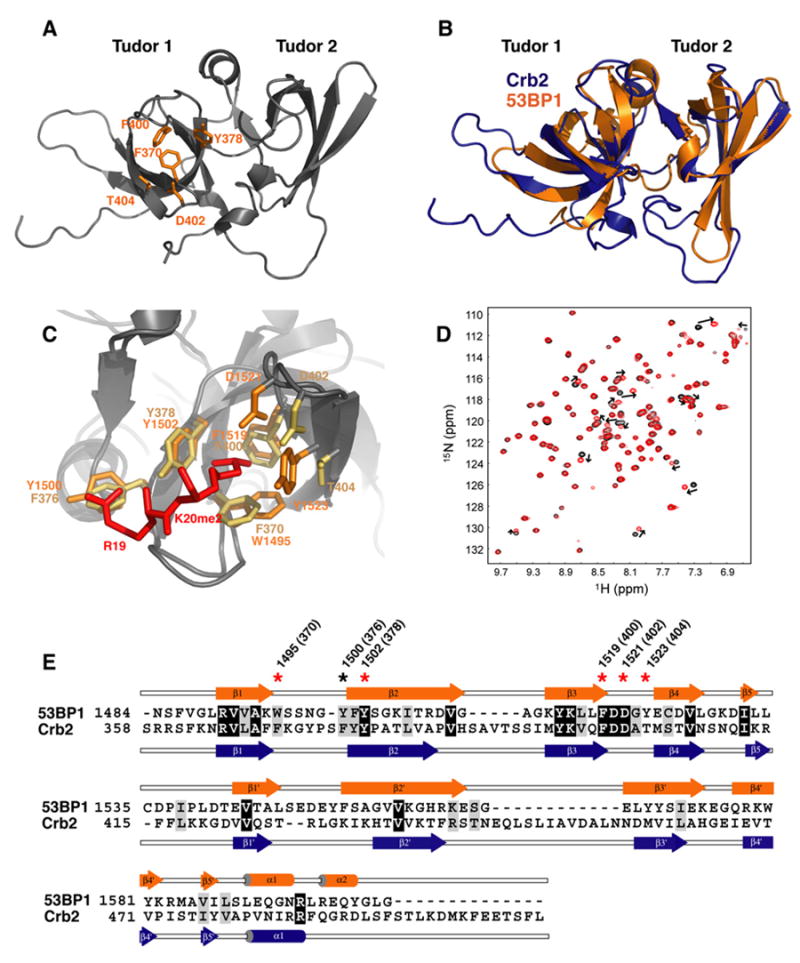
(A) Ribbon representation of the Crb2 structure. The side chains of Phe370, Tyr378, Phe400, Thr404, and Asp402, forming the dimethyllysine-binding cage, are shown in orange.
(B) Superposition of the 3D structures of 53BP1 (orange) and Crb2 (blue) in ribbon representation.
(C) Magnified view of the superimposed dimethyllysine- and Arg19-binding pocket of Crb2 (brown) and 53BP1 (orange) showing key interacting residues: Phe370, Phe376, Tyr378, Phe400, Thr404, and Asp402 of Crb2; Trp1495, Tyr1500, Tyr1502, Phe1519, Tyr1523, and Asp1521 of 53BP1; and Arg19 and Lys20 (K20me2) of histone H4 peptide (red).
(D) 1H-15N HSQC spectrum of 15N-labeled Crb2, free (black) and in complex with H4-K20me2 (red), showing chemical shift perturbation of a number of residues.
(E) Alignment of 53BP1 and Crb2 amino acids based on their 3D structures. Red and black stars indicate 53BP1 residues interacting with K20me2 and Arg19 of H4-K20me2, respectively. In parentheses are corresponding Crb2 residues.
We found the segment 358–507 of Crb2 suitable for X-ray crystallography analysis. The 3D structure was determined to a resolution of 2.4 Å by single-wavelength anomalous diffraction (SAD) using a selenomethionine-enriched protein (Figure 6). The statistics are provided in Table S2, and a sample of Crb2 electron density map is shown in Figure S3. The structure of Crb2 reveals the presence of a second tudor domain (Figure 6A). A structural alignment with 53BP1 in Figure 6B further shows that the relative orientation of the tudor domains of Crb2 is remarkably similar to what is seen in 53BP1. The minimized root-mean-square distance between 59 Cα atoms from the secondary structure elements of both proteins is only 0.91 Å. Based on amino acid sequence alignment alone, the presence and extent of the second tudor domain of Crb2 could not have been correctly predicted (Figure 6E). As seen in Figures 6B and 6E, both tudor domains of Crb2 are significantly bigger than those of 53BP1, with the additional amino acids located in the loop regions between β strands 3 to 4 and 3′ to 4′. An extra C-terminal segment devoid of any regular secondary structure element is also present in Crb2.
Importantly, on the basis of the structural alignment a dimethyllysine-binding pocket similar to that in 53BP1 is readily predicted in Crb2 (Figure 6C). Tyr1502, Phe1519, and Asp1521 participating in the binding pocket of 53BP1 have corresponding identical amino acids (Tyr378, Phe400, and Asp402) in Crb2. Tyr1523, another residue of 53BP1 that contacts H4-K20me2, has a counterpart in Crb2 (Thr404) that preserves the hydrophobicity deemed important at the Tyr1523 position (see above). A significant difference is the replacement of 53BP1 Trp1495 with a phenylalanine (Phe370) in Crb2. However, we showed that a W1495F mutation in 53BP1 does not impede 53BP1 nuclear localization (Figure 5). The aromatic residue Tyr1500 in 53BP1, engaged in cation-π interaction with histone H4 Arg19, is also conserved in Crb2 (Phe376).
With these similarities between Crb2 and 53BP1, we then verified by NMR spectroscopy the direct binding of Crb2 to dimethylated H4-K20. Indeed, titration of 15N-labeled Crb2 with a nonlabeled H4-K20me2 peptide led to the selective perturbation of 17 amide proton/nitrogen resonances in the 1H-15N HSQC spectrum of Crb2 (Figure 6D). The exchange between free and H4-K20me2-bound Crb2 is fast on the time scale of NMR chemical shifts, indicating a low affinity with a KD value in the milli-molar range. It is likely that the effective affinity is higher inside the cell if one considers, as we suggested for 53BP1, that Crb2 not only interacts with histone H4 but also with DNA in the context of nucleosomes. In the process of purifying Crb2, we noticed some affinity for nucleic acids (data not shown). The crystal structure of Crb2 reveals the presence of nine phosphate ions in all three Crb2 molecules in the asymmetric unit. It is not uncommon for negatively charged ions to adhere to nucleic acid-binding surfaces in proteins (Ghosh et al., 2004). By also taking into account the multimeric functional state of Crb2 (Du et al., 2004), one can reasonably expect additional contribution to affinity inside the cell.
The evolutionarily conserved mode of methylated H4-K20 recognition from fission yeast to higher eukaryotes described in this study strongly suggests that Crb2 is a functional homolog of 53BP1. The methylation state of H4-K20 important for the relocalization of Crb2 to DNA DSBs has not been examined in vivo in fission yeast, but our work suggests that the target of Crb2, just like for 53BP1, is H4-K20me2. It is likely that genetic studies in fission yeast will uncover the precise role of this methylation state-specific recognition of histone H4-K20 by Crb2.
Conclusions
Our study explores the molecular mechanism linking histone lysine methylation to the accumulation of mammalian DNA repair factors 53BP1 and putative fission yeast homolog Crb2 to DNA DSBs. Contrary to what was previously thought, we show that the mechanism by which these factors are recruited to chromatin is evolutionarily conserved as 53BP1 and Crb2 have the same methylated histone target in vivo: histone H4-K20. Furthermore, we demonstrate that their recruitment to DNA damage sites is through direct interaction with methylated H4-K20. This is achieved by their similar 3D structures despite low amino acid sequence similarity. We show that both 53BP1 and Crb2 contain tandem tudor domains that form a specific complex with dimethylated histone H4-K20. The preferential interaction with H4-K20me2 is explained by the presence of a conserved five-residue binding pocket in their first tudor domain that best fits a dimethylammonium but blocks a trimethylammonium group. Our discovery of a methylation state-specific recognition of histone H4-K20 uncovers an added level of complexity in the DNA DSB repair process. In mammals, dimethylated histone H4-K20, identified in this study as the binding partner of 53BP1, is generally present together with monomethylated H4-K20 in euchromatic regions of chromosomes, which are often under active transcription (Karachentsev et al., 2005; Rice et al., 2002). On the other hand, trimethylated histone H4-K20, which is not a target of 53BP1, is usually associated with transcriptionally inactive pericentric heterochromatin (Schotta et al., 2004; Sims et al., 2006). Thus, DNA DSB repair may proceed through different mechanisms depending on the structure of chromatin (i.e., euchromatin versus heterochromatin).
Although H4-K20me2 is clearly a docking site for 53BP1, we did not detect any significant change in the levels of H4-K20 mono-, di-, or trimethylation after DNA damage by irradiation (data not shown). Similarly, while the methylation state of H4-K20 was not specifically addressed in fission yeast, bulk levels of H4-K20 methylation are not altered after DNA damage (Sanders et al., 2004). What then links the recognition of H4-K20me2 by 53BP1 and Crb2 to DSBs? A possible explanation is that H4-K20me2, which is buried in the context of stacked nucleosomes, becomes exposed and accessible to interaction after DSB (Sanders et al., 2004). However, this seems unlikely with regards to recent measurements showing that nucleosomes are highly dynamic structures allowing proteins rapid access, even to buried regions of chromatin (Li et al., 2005). We favor the idea that there is a DSB-dependent signal that in conjunction with the recognition of H4-K20me2 by 53BP1 and Crb2 would trigger their relocalization to DSBs. Phosphorylated histone H2AX (γH2AX), a well-characterized signal for the recruitment of DNA damage response proteins, is necessary for the efficient accumulation of 53BP1 at sites of DSBs (Stucki et al., 2005; van Attikum and Gasser, 2005; Ward et al., 2003). While MDC1 is the primary target of γH2AX (Stucki et al., 2005), it was also shown that 53BP1 can interact with γH2AX (Ward et al., 2003). It may be that simultaneous recognition of γH2AX and H4-K20me2 through two different domains of 53BP1 is needed to provide an affinity high enough to relocate 53BP1 to DNA damage sites. This may partly explain the ordered assembly of repair proteins at DNA DSBs (Bekker-Jensen et al., 2005) where MDC1 is first recruited through high-affinity binding to γH2AX and promotes ATM-dependent amplification of the γH2AX signal at DSB sites (Lou et al., 2006), which would then facilitate the relocalization of 53BP1 through interaction with both γH2AX and H4-K20me2 near DSB-flanking chromatin. This hypothesis of dual interaction by 53BP1 is consistent with recent genetic studies in fission yeast indicating that H4-K20 methylation and H2A phosphorylation function in the same pathway to regulate Crb2 recruitment to DNA DSBs (Du et al., 2006). Alternatively, another possible targeting mechanism may be through phosphorylation of histone H4. It has been known for a long time that there are histidine kinases in mammals that phosphorylate His18 and His79 of histone H4 (Besant and Attwood, 2005; Fujitaki et al., 1981; Smith et al., 1973; Steeg et al., 2003). Interestingly, His18 is located at the binding interface of the 53BP1/H4-K20me2 complex, in a region that experiences conformational exchange as detected by NMR spectroscopy (Figure 2B). It is tempting to speculate that H4-H18 phosphorylation, in conjunction with H4-K20me2, could stabilize the interaction of 53BP1 with nucleosomes and function as yet another signal involved in DSB repair. The instability of phosphohistidine has so far precluded extensive studies in vitro and in vivo. New methods will need to be developed to test this hypothesis.
EXPERIMENTAL PROCEDURES
Preparation of Proteins and Peptides
The preparation and purification of wild-type and mutant 53BP1, Crb2, and JMJD2A domains, as well as isotope enrichment of 53BP1 and Crb2 with 15N and 15N/13C and labeling of Crb2 with selenomethionine, follow previously published procedures (Botuyan et al., 2004) and are described in detail in the Supplemental Data. Nonmethylated and methylated H3-K9, H3-K27, H3-K79, and H4-K20 peptides were prepared and purified at the Mayo Clinic peptide synthesis facility and checked by mass spectrometry.
X-Ray Crystallography
Crystallization and cryoprotection conditions of 53BP1, 53BP1/H4-K20me2, and Crb2 are given in the Supplemental Data.
All diffraction patterns were indexed, integrated, and scaled with HKL2000 (Otwinowski and Minor, 1997). Initial phases for both the native and complex crystals of 53BP1 were obtained via molecular replacement by using the coordinates of the 2.8 Å resolution structure of the tandem tudor domains of 53BP1 (PDB ID 1XNI) as a search model with the program MOLREP (version 9.2) (Vagin and Teplyakov, 1997). For Crb2, initial phases were determined using SAD at the selenium wavelength. The initial models were completed with manual building in COOT (Emsley and Cowtan, 2004) and refined with Refmac5 (Murshudov et al., 1997) using reflections ranging from 50–1.25 Å (for free 53BP1), 50–1.70 Å (for 53BP1/H4-K20me2), and 50–2.4 Å (for Crb2) (Tables S1 and S2). Molecular representations were generated using PyMol (Delano, 2002).
NMR Spectroscopy
All NMR data were collected at 25°C in 50 mM perdeuterated Tris/HCl (pH 7.0), 100 mM NaCl on Bruker Avance spectrometers operating at 500 MHz and 600 MHz with a cryoprobe. 1H-15N HSQC spectra were acquired, initially on ~0.8 mM 15N- or 15N/13C-labeled 53BP1 protein and subsequently after each addition of small amounts of concentrated nonlabeled dimethylated H4-K20 peptide (residues 16–25), until an ~1:1.2 protein:peptide complex was attained. Changes in backbone and side-chain amide chemical shifts of 53BP1 were determined for each peak from overlaying the titration spectra collected. Assignment of the peaks was achieved by performing standard triple resonance experiments (Ferentz and Wagner, 2000; Mer et al., 2000) collected for the 15N/13C-labeled 53BP1, both free and bound to a nonlabeled dimethylated H4-K20 peptide. NMR titrations involving 15N-labeled 53BP1 and Crb2 with nonlabeled H3-K79me2 (residues 74–83) and H4-K20me2 (residues 16–25), respectively, were also done. All data were processed with NMRPipe/NMRDraw (Delaglio et al., 1995) and analyzed with NMRView (Johnson and Blevins, 1994) software.
Isothermal Titration Calorimetry
All ITC measurements were recorded at 10°C with a VP-ITC titration calorimeter (MicroCal, Northampton, MA). All protein and peptide stock samples were in the target buffer, i.e., 50 mM Tris/HCl (pH 7.5) and 20 mM NaCl, and then diluted in the same buffer to achieve the desired concentrations: 40–100 μM protein and 3 to 4 mM peptide. A 298 μl syringe was used to deliver the peptide, typically as 79 injections (1 × 3 μl followed by 59 × 4 μl then 19 × 3 μl) at 5 min intervals into the calorimetric cell containing 1.41 ml of protein solution. Control experiments were performed under identical conditions to determine the heat signals that arise from injecting buffer into the protein solution and the peptide into the buffer. The initial data point was routinely deleted. Curve fitting was done by Lavenberg-Marquardt nonlinear regression using Origin 7.0 with a standard one-site model provided by MicroCal (Turnbull and Daranas, 2003; Wiseman et al., 1989).
Generation of Dot1 Genetic Knockout
An embryonic stem (ES) cell line from BayGenomics (RRR032) was used to generate Dot1+/− mice, using a standard protocol (Lou et al., 2006). An ES clone containing an insertion mutation at the mDot1 locus was injected into C57BL/6 blastocysts to obtain Dot1 heterozygous mice. Dot1−/− MEFs were isolated from Dot1+/− intercrosses at embryonic day 12.5. Immunofluorescence and western blotting procedures are explained in the Supplemental Data.
Supplemental Data
Supplemental Data include three figures, two tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/127/7/1361/DC1/.
Supplementary Material
Acknowledgments
We acknowledge use of the X12C beamline of Brookhaven National Laboratory’s National Synchrotron Light Source (NSLS) and 19ID beamline of Argonne National Laboratory’s Advance Photon Source (APS). We thank S. Sclafani at NSLS and R. Alkire and Y. Kim at APS for their assistance with X-ray data collection, D. McCormick for advice on peptide syntheses, E. Wasielewski for assistance with figures, N. Juranic and S. Macura for assistance with NMR, and Y. Nominé for help with the calorimetry experiments. Use of Argonne National Laboratory Structural Biology Center beamlines at APS was supported by the U.S. Department of Energy under contract number W-31-109-ENG-38. This work was partially supported by an NIH grant (CA100109) and a DOD breast cancer Era of Hope Scholar Award (to J.C.). G.M. acknowledges partial support from the March of Dimes Basil O’Connor Starter Scholar Research Award and the Human Frontier Science Program Organization.
Footnotes
Accession Numbers
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 2G3R and 2IG0 for free and H4-K20me2-bound 53BP1, respectively, and 2FHD for Crb2.
References
- Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besant PG, Attwood PV. Mammalian histidine kinases. Biochim Biophys Acta. 2005;1754:281–290. doi: 10.1016/j.bbapap.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Nominé Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charier G, Couprie J, Alpha-Bazin B, Meyer V, Qué méneur E, Guérois R, Callebaut I, Gilquin B, Zinn-Justin S. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure. 2004;12:1551–1562. doi: 10.1016/j.str.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PyMol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Du LL, Moser BA, Russell P. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J Biol Chem. 2004;279:38409–38414. doi: 10.1074/jbc.M403326200. [DOI] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Fujitaki JM, Fung G, Oh EY, Smith RA. Characterization of chemical and enzymatic acid-labile phosphorylation of histone H4 using phosphorus-31 nuclear magnetic resonance. Biochemistry. 1981;20:3658–3664. doi: 10.1021/bi00515a055. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Uthaiah R, Howard J, Herrmann C, Wolf E. Crystal structure of IIGP1: a paradigm for interferon-inducible p47 resistance GTPases. Mol Cell. 2004;15:727–739. doi: 10.1016/j.molcel.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Basu BP, Kysela B, Kurihara T, Shibata M, Guan D, Cao Y, Hamada T, Imamura K, Jeggo PA, et al. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J Biol Chem. 2003;278:36487–36495. doi: 10.1074/jbc.M304066200. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView: a computer program for visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Mavri J, Vogel HJ. Ion pair formation involving methylated lysine side chains: a theoretical study. Proteins. 1994;18:381–389. doi: 10.1002/prot.340180408. [DOI] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Moser BA, Du LL, Russell P. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol Cell Biol. 2005;25:10721–10730. doi: 10.1128/MCB.25.24.10721-10730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, Melton DW, Jullien D, Adachi Y. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- Smith DL, Bruegger BB, Halpern RM, Smith RA. New histone kinases in nuclei of rat tissues. Nature. 1973;246:103–104. doi: 10.1038/246103a0. [DOI] [PubMed] [Google Scholar]
- Steeg PS, Palmieri D, Ouatas T, Salerno M. Histidine kinases and histidine phosphorylated proteins in mammalian cell biology, signal transduction and cancer. Cancer Lett. 2003;190:1–12. doi: 10.1016/s0304-3835(02)00499-8. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Duaa M, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, Wilson JR. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


