Abstract
Mutationally activated BRAFV600E (BRAFVE) is detected in ∼6% of human malignancies and promotes sustained MEK1/2–ERK1/2 pathway activation. We have designed BRafCA mice to express normal BRaf prior to Cre-mediated recombination after which BRafVE is expressed at physiological levels. BRafCA mice infected with an Adenovirus expressing Cre recombinase developed benign lung tumors that only rarely progressed to adenocarcinoma. Moreover, BRafVE-induced lung tumors were prevented by pharmacological inhibition of MEK1/2. BRafVE expression initially induced proliferation that was followed by growth arrest bearing certain hallmarks of senescence. Consistent with Ink4a/Arf and TP53 tumor suppressor function, BRafVE expression combined with mutation of either locus led to cancer progression.
Keywords: BRaf, mouse model, lung tumors, senescence
The RAS-activated RAF–MEK–ERK mitogen-activated protein (MAP) kinase signaling pathway is directly implicated in the development of a wide variety of human malignancies. Although mutational activation of RAS in human cancer was first demonstrated in 1982, somatic activating mutations in BRAF were only detected in 2002 (Davies et al. 2002). Mutationally activated BRAF is detected in melanoma (70%), colorectal (15%), papillary thyroid (40%), ovarian (30%), and non-small-cell lung cancers (NSCLCs)(3%) (Davies et al. 2002; Zebisch and Troppmair 2006). Of the ∼40 BRAF mutations identified to date, a T1799A transversion encoding constitutively active BRAF-V600E (BRAFVE hereafter) accounts for ∼90% of all BRAF mutations (Wan et al. 2004). Mutationally activated BRAFVE promotes the sustained activation of the ERK1/2 MAP kinases, pleiotropic regulators of the aberrant physiology of the cancer cell.
Lung cancer is the most prevalent cancer in the industrialized world and was responsible for ∼165,000 deaths in the United States in 2005. Adenocarcinomas represent approximately one-half of all NSCLC subtypes. Despite its prevalence and characteristically high mortality rates, the cellular and genetic origins of the disease remain obscure. Mutational activation of KRAS or BRAF has been detected in ∼25% of NSCLCs, and, while the majority of these harbor KRAS mutations, activating mutations in BRAF account for ∼3% of all NSCLCs (Brose et al. 2002; Davies et al. 2002; Naoki et al. 2002). In addition, somatic mutations in the genes encoding the EGF receptor or ERBB2, both of which activate RAS signaling, are detected in ∼13% of lung adenocarcinomas (Shigematsu and Gazdar 2006). Finally, ERK1/2 activation is associated with disease aggression, suggesting a more general role for this pathway in NSCLC pathogenesis (Vicent et al. 2004).
Here we describe mice carrying a genetically modified allele of BRaf, BRafCA, which expresses normal BRaf prior to Cre-mediated recombination after which BRafVE is expressed. BRafVE expression in the lung elicited the growth of benign neoplastic adenomas, reminiscent of the effects of KRasG12D expression (Jackson et al. 2001). Tumor formation was potently inhibited by pharmacological MEK1/2 inhibition. Evidence suggested that BRafVE-induced lung tumors arose as a consequence of an initial burst of cell proliferation followed by an indolent phase, characterized by decreased proliferation accompanied by expression of senescence markers. Indeed, rarely did BRafVE-induced adenomas display spontaneous progression to adenocarcinoma unless mice were deliberately engineered to lack the TP53 or Ink4a/Arf tumor suppressor genes (TSGs).
Results and Discussion
Generation of BRafCA mice by homologous recombination in embryonic stem (ES) cells
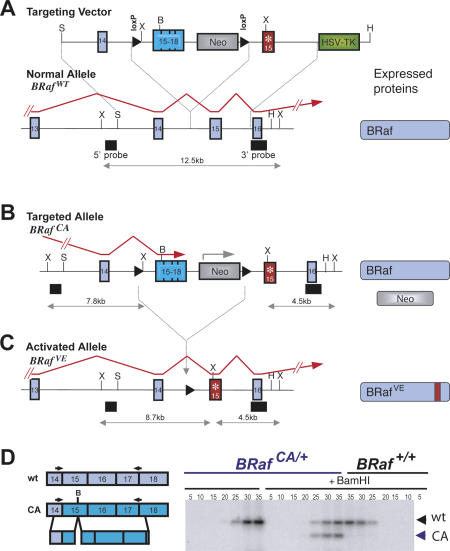
The development of new mouse models of human cancer places great emphasis on temporal and spatial control of oncogene expression, with particular attention paid to the levels of oncogene expression. This is especially important for BRAF since relatively small differences in RAF activity can promote quiescence, proliferation, or cell cycle arrest/senescence (Woods et al. 1997; Zhu et al. 1998). We sought to develop a mouse to accurately model the role of BRafVE in cancer initiation, progression, and therapy with an emphasis on temporal and spatial control in tissues of interest. Using a suitably designed targeting vector (Fig. 1A), we used homologous recombination in ES cells to generate mice carrying a Cre-activated allele of BRaf (BRafCA) (Fig. 1B). BRafCA is designed to express normal BRaf prior to Cre-mediated recombination at which time BRafVE expression is initiated at physiological expression levels and subject to normal patterns of alternate splicing and differential exon usage (Fig. 1C). By design, this model recapitulates the situation in human cells, where one copy of normal BRAF is converted by somatic mutation to BRAFVE. To discriminate the expression of the various BRaf mRNAs, silent restriction enzyme polymorphisms were incorporated into the exon 15 sequences of BRafCA (BamHI, B) and BRafVE (XbaI, X) (Fig. 1A–C).
Figure 1.
Schematic representation and generation of Cre-activated BRaf allele (BRafCA). (A) The genomic BRaf locus (BRafwt), the targeting vector used, and the proteins expressed from the locus are depicted. Red arrows depict fully processed BRaf mRNAs. External 5′ and 3′ probe locations and the size of XbaI restriction enzyme fragments are indicated. The modified exon 15 encoding V600E is indicated (*). (B) The targeted BRafCA allele and following Cre-mediated recombination, BRafVE (C). (D) Semiquantitative RT–PCR analysis of targeted and wild-type ES cell-derived mRNAs using a radiolabeled reverse primer. PCR products amplified for the indicated numbers of cycles were treated with BamHI where indicated to distinguish BRaf transcripts. Expression of the BRafCA allele was 98% that of endogenous BRaf mRNA.
Multiple correctly targeted ES cell clones were identified by Southern blotting (Supplementary Fig. 1A) and by RT–PCR analysis of BRaf and BRafCA mRNA expression. The latter analysis confirmed that BRaf and BRafCA mRNAs were expressed at the same level (Fig. 1D), whereas BRafVE mRNA was not detected (data not shown). Two independent clones gave rise to chimeric mice that transmitted the BRafCA allele through the germline. Breeding of BRafCA/+ mice to mice heterozygous for the CA allele, or a null allele, generated mice with all possible genotypes at normal Mendelian frequencies, demonstrating that the BRafCA allele functions analogously to normal BRaf prior to Cre-mediated recombination (Supplemental Material).
BRafVE expression in the lung elicits tumor formation
To test the utility of BRafCA mice, we initiated expression of BRafVE in the lungs by intranasal instillation of an adenovirus expressing Cre recombinase (Ad-Cre) (Fasbender et al. 1998). We chose this strategy for four reasons: (1) Mutationally activated BRAF has been described in human NSCLC (Zebisch and Troppmair 2006). (2) The use of Ad-Cre provides temporal control over BRafVE expression and therefore permits prospective evaluation of BRafVE effects. (3) Ad-Cre titration allows control of the number of cells in which BRafVE expression is initiated. (4) Others have previously described the effects of Raf-1- or Ad-Cre-induced KRasG12D expression in the lungs (Kerkhoff et al. 2000; Jackson et al. 2001; Tuveson et al. 2004).
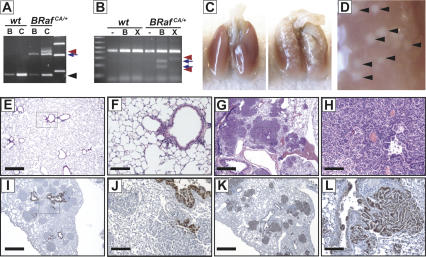
Six- to eight-week-old wild-type or BRafCA/+ mice were administered with 5 × 107 PFU (plaque-forming units) of either Ad-Cre or Ad-βGal. Mice were monitored for 7–8 wk, at which time all of the BRafCA mice administered with Ad-Cre were euthanized due to evidence of labored breathing and general wasting. BRafCA genomic rearrangement (Fig. 2A) and BRafVE mRNA expression (Fig. 2B) were Cre-dependent and entirely restricted to the lung, as we detected no abnormalities or induction of BRafCA somatic recombination in any other tissue analyzed. Lungs of BRafVE-expressing mice displayed evidence of the formation of multiple lung tumors with an adenomatous morphology (Fig. 2C–H). Such lesions were not detected in BRafCA mice even up to 6 mo after administration of a high dose of Ad-βGal or in wild-type mice infected with Ad-Cre. The effects of BRafVE expression in the lung were highly penetrant in that all BRafCA mice (n = 31) treated with Ad-Cre (107 PFU) developed lung tumors over the course of nine experiments. Moreover, induction of BRafVE expression in the lung using a ubiquitously expressed, conditionally active CreER transgene (Guerra et al. 2003) elicited the formation of multiple lung tumors of the same type.
Figure 2.
Expression of BRafVE in lung induces adenomas. (A) PCR detection of BRafCA rearrangement from lungs of BRaf+/+ (wt) or BRafCA/+ mice infected with Ad-βGal (B) or Ad-Cre (C). The black arrowhead, blue arrow, and red arrowhead highlight the wild-type, the unrearranged BRafCA, and the BRafVE alleles, respectively. (B) Detection of BRafVE mRNA in BRafCA mice following Ad-Cre infection. Total lung RNA from BRaf+/+ (wt) or BRafCA/+ mice isolated 8 wk after Ad-Cre infection (5 × 107 PFU) was subjected to RT–PCR analysis as in Figure 1D. BRafCA and BRafVE transcripts are distinguished through the use of restriction polymorphisms (BamHI, B; XbaI, X) with the diagnostic digestion products indicated by blue arrows and red arrowheads, respectively. (C–L) BRafCA/+ mice were infected with 5 × 107 PFU (C–H) or 106 PFU (I–L) of Ad-Cre intranasally and were analyzed 7 wk following infection. (C) Macroscopic lesions are present on the surface of BRafCA/+ (right and magnified in D) but not wild-type (left) lungs. (E–H) Hematoxylin and eosin staining of histological sections of wild-type (E,F) or BRafCA/+ (G,H). Note papillary adenomas in higher power magnification (H) and wide-scale involvement of lungs (G). (I,J) Adenomas stain negative for CCA (I,J) and positive for SP-C (K,L). Bars: F,H,J,L, 100 μm; E,G,I,K, 500 μm.
The cellular origins of human lung adenocarcinoma and adenomas are unclear. NSCLCs frequently express markers of Clara cells or alveolar type II pneumocytes (ATII). Indeed, it has been suggested that they arise from a common bronchio-alveolar stem cell (BASC) (Kim et al. 2005). To assess the properties of BRafVE-induced tumors, immunohistochemical analyses to detect Clara Cell antigen (CCA/CC10) and Surfactant Protein-C (SP-C), a surface marker of ATII pneumocytes, were performed. Staining for CCA detected cells lining the bronchioles, but the BRafVE-induced tumors were largely CCA negative (Fig. 2I,J). In contrast, the majority of cells within BRafVE-induced tumors expressed SP-C, suggesting that they have properties of ATII pneumocytes (Fig. 2K,L). Furthermore, analysis of the earliest BRafVE-induced lesions (2 wk after Ad-Cre) revealed them to express SP-C.
Requirement for BRAFVE–MEK–ERK activity for lung tumor formation
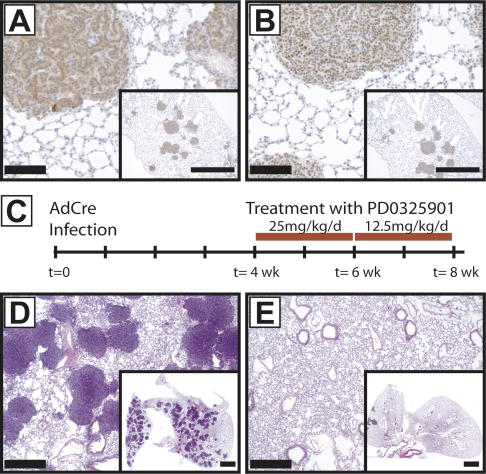
Clear genetic and biochemical evidence indicates that the MEK–ERK pathway plays an essential role in cancer cell proliferation, yet recent studies have suggested that alternate RAF-regulated pathways may exist (Chen et al. 2001). To address the effects of BRafVE on canonical ERK signaling, we assessed phosphorylation of downstream effector proteins MEK1/2, ERK1/2, and the ERK-dependent phosphorylation of the Ets1/2 transcription factors. Indeed, using serial tissue sections, positive staining for all three of these surrogate markers of BRafVE activity overlapped in all tested cases, demonstrating ERK MAP kinase activation (Fig. 3A,B; Supplementary Fig. 2A–C). It is reported that KRasG12D-induced lung tumors do not routinely display elevated pERK1/2 but display stress-activated MAP kinase (SAPK/JNK) activation (Lee et al. 2002). Hence, to address the importance of MEK1/2 in BRafVE-induced tumor formation, we used PD0325901, a highly specific and selective MEK1/2 pharmacological inhibitor (a generous gift of Pfizer, Inc.). PD0325901 displays a nanomolar IC50 in cells and, unlike most protein kinase inhibitors, acts noncompetitively with its substrates (Sebolt-Leopold and Herrera 2004). Tumors were initiated in BRafCA/+ mice by administration of 5 × 106 PFU of Ad-Cre. Four weeks later, at a time when hyperplastic lung epithelium exists (Supplementary Fig. 2D), PD0325901, or the appropriate vehicle control, was administered daily by oral gavage for an additional 28 d (Fig. 3C). As expected, mice receiving the vehicle control displayed numerous lung tumors (Fig. 3D), whereas mice receiving PD0325901 were largely tumor free (Fig. 3E). These data indicate that the ability of BRafVE to elicit lung tumors is dependent on MEK1/2–ERK1/2 signaling. However, this does not eliminate the possibility that additional BRafVE-regulated pathways may contribute to tumor maintenance.
Figure 3.
MEK1/2 dependency of BRafVE-induced adenoma formation. (A,B) Adenomas stain positive for active MEK (A) and active ERK (B). (C) Time line of PD0325901 treatment. BRafCA mice infected with Ad-Cre (5 × 106 PFU) were grouped randomly into one of two arms: One arm received PD0325901, and the other received vehicle control. Drug treatment was initiated 4 wk after initial infection of mice and was sustained at the indicated concentrations for 28 d. Eight weeks post-infection, all mice receiving vehicle develop multiple adenomas (D), whereas no mouse treated with PD0325901 developed adenomas (E). (Insets) Lower magnification. Bars: A,B,D,E, 100 μm; A,B, insets, 1 mm; E,F, insets, 2 mm.
BRafVE-induced cell cycle arrest as a barrier to lung adenocarcinoma progression
Induction of BRafVE expression by high dose Ad-Cre induced the formation of sufficiently large numbers of tumors that such mice rapidly succumbed to breathing difficulties and general wasting. In subsequent experiments, we used lower Ad-Cre doses to elicit smaller numbers of tumors to determine if such lesions would display progression given sufficient time for additional events to occur. In addition, mice were euthanized at different times following Ad-Cre administration, and the histological appearance of the tumors was assessed. Lesions were characterized according to consensus criteria established by a panel of lung cancer biologists (Nikitin et al. 2004).
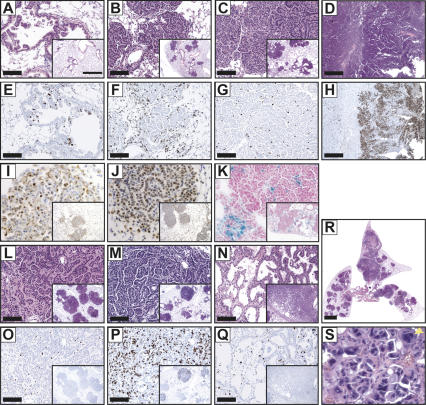
At early times after induced BRafVE expression, we detected evidence of epithelial hyperplasia (classified as 1.1.1.1) arising within the terminal bronchioles and within the central lung parenchyma. This hyperplastic epithelium displayed papillary excresences; however, at early time points (2–4 wk), there was no nodule formation (Fig. 4A). Papillary adenomas (1.2.1.2.2) were detected 6–8 wk after BRafVE expression and appeared to increase in number and size. These lesions were bronchiolocentric, but did not appear to involve the terminal bronchioles. Rather, the lesions appeared to arise in alveolar ducts and expand outward and around bronchioles (Fig. 4B). At later times (∼14 wk) after BRafVE induction, we detected changes in the papillary adenomas such that they appeared to undergo alterations at their periphery, changing to a more solid architectural appearance (Fig. 4C). The cells also displayed additional morphologic alterations where the cytoplasm became more abundant and eosinophilic. This is consistent with the appearance of mixed papillary and solid adenomas (1.2.1.2.3). It is striking that the BRafVE-induced lung adenomas formed rapidly (6–8 wk), but did not appear to grow beyond ∼900 μm in diameter. Moreover, we rarely detected spontaneous progression to adenocarcinoma in these mice. To date, only two out of 57 BRafCA mice possessing adenomas displayed focal adenocarcinomas at the time of necropsy (Fig. 4D). The adenocarcinoma cells in these two mice displayed nuclear enlargement, hyperchromasia, and contour irregularities. The cells were arranged in solid sheets, histologically consistent with adenocarcinoma (1.2.3.2). Thus, expression of BRafVE appears sufficient to rapidly induce benign lung tumors that only rarely progress to adenocarcinoma, suggesting that additional events are required for cancer progression.
Figure 4.
Sustained BRafVE expression induces a proliferative arrest with hallmarks of oncogene-induced senescence. (A–D) Lung histology was analyzed from BRafCA mice at 4 wk (A), 6 wk (B), 14 wk (C), and 41 wk (D) post-infection with 107 PFU (A–C) or 105 PFU (D) of Ad-Cre. Note that the hyperplastic tissue in A was observed as early as 14 d post-infection with high doses of Ad-Cre. (E–H) Ki67 immunochemical staining of lung sections at 4 wk (E), 6 wk (F), 8 wk (G), and 41 wk (H) post-infection using serial section of those depicted in A–D. (I–K) BRafVE-induced adenomas express p19ARF (I) and Dec1 (J) and are negative for SA-βGal (K). (L–S) BRafCA mice (L,O) and those homozygous for conditional alleles of TP53 (M,P,R,S) or Ink4a/Arf (N,Q) were analyzed at 7 wk (R,S), 9 wk (L,M,O,P) or 14 wk (N,Q) post-infection with Ad-Cre. (O–Q) Ki67 immunochemical staining of serial lung sections from L–N. (R,S) Low-powered (R) and high-powered (S) views of BRafCA;TP53lox/lox lungs demonstrate the extent of lesions and disordered morphology, enlarged size (yellow arrow), and heterchromatic staining of nuclei. Bars, A–C,E–G,L–Q, 100 μm; D,H, 500 μm; insets, 1 mm; R, 2 mm.
By a similar strategy, others assessed the effects of BRafVE expression in the hematopoietic system. In this case, BRafVE elicits hematopoietic dysplasia with characteristics of histiocytic sarcoma (Mercer et al. 2005). In this model, BRafVE expression is uniformly lethal due to bone marrow failure, but mice do not die due to acute myeloid leukemia, suggesting that BRafVE induces a premalignant myelodysplastic state. It is interesting to note that the consequences of induced BRafVE lung expression bear both interesting similarities and differences in comparison to those reported for KRasG12D (Jackson et al. 2001). Ad-Cre-induced expression of KRasG12D in the lungs of KRasLSL mice leads initially to development of atypical adenomatous hyperplasia (AAH, ∼2 wk) that progresses to adenoma (∼6 wk). Interestingly, the major difference appears to be that expression of KRasG12D in the lung leads to more rapid and consistent progression to adenocarcinoma than that elicited by BRafVE. These data suggest that either additional KRas effector pathways are required for cancer progression or that BRafVE-induced adenomas are more highly constrained from progression.
Oncogene-induced cell cycle arrest with features of senescence (OIS) is reported to be a cellular defense mechanism restraining the proliferation of normal cells in response to inappropriate activation of RAS or its signaling effectors (Collado et al. 2005; Michaloglou et al. 2005). Recently, it was reported that KRasG12D-induced lung tumors are arrested in the cell cycle and express OIS markers (Collado et al. 2005). To determine if BRafVE-induced lung tumors display evidence of OIS, tissue sections from lungs harvested at different times after BRafVE expression were stained for markers of cell proliferation (phospho-histone H3, Ki67, or BrdU incorporation) and for OIS markers (SA-βGal, p19ARF, Dec1). In hyperplastic lung tissues and in tumors induced at early times after BRafVE expression, a high percentage of cells stain positive for Ki67 (Fig. 4E,F), phospho-histone H3, and BrdU incorporation. However, in tumors observed at later times, the percentage of Ki67-positive cells is dramatically decreased even though the BRafVE–MEK–ERK signaling pathway remains active in these cells (Figs. 3A,B, 4G). Further analysis of adenomatous lesions revealed that they were positive for expression of p19ARF and Dec1 (Fig. 4I,J). p19ARF expression was detected in ∼15% of tumor cells with staining localized to nucleoli, a pattern consistent with previous data (Weber et al. 1999). While these markers of OIS were readily detected, these adenomas were largely negative for SA-βGal activity (Fig. 4K). In the BRafVE-induced adenocarcinomas found in the two mice displaying tumor progression, however, a very high rate of Ki67 staining was observed (Fig. 4H). These data are consistent with the hypothesis that sustained activation of BRafVE promotes an initial period of cell proliferation followed by cell cycle arrest constraining further tumor progression.
BRafVE cooperates with TSG loss to promote adenocarcinoma formation
If BRafVE-induced senescence is a barrier to lung carcinogenesis, then BRafVE expression combined with loss of TSG expression should lead to cancer progression. To this end, we crossed BRafCA mice to obtain mice homozygous for floxed alleles of either TP53 or Ink4a/Arf. These mice express normal levels of the various TSGs until the modified alleles undergo Cre-mediated deletion of critical exons. Concomitant loss of TP53 led to increased proliferation at ∼8 wk post-infection with Ad-Cre when compared with BRafCA/+; TP53+/+ mice (Fig. 4L,M,O,P). Histological analyses of lungs from BRafCA/+; TP53lox/lox mice revealed rapid cancer progression (Fig. 4L,M,R) and showed nodules composed of central papillary structures with solid peripheral areas (similar to older BRafCA/+ mice) consistent with formation of mixed adenomas (1.2.1.2.3). Several small airways also displayed papillary hyperplasia, which was never observed in BRafCA/+ mice. In each case analyzed (n = 5), mice lacking TP53 function displayed an increased proliferative index (Fig. 4P; Supplementary Fig. 3), and cells displayed altered nuclear morphology (Fig. 4S). Collections of cells with enlarged nuclei stained positive for Ki67, indicating ongoing proliferation (Supplementary Fig. 3A–D). Significantly, at ∼8 wk post-infection, all BRaf+/+ mice homozygous for the TP53lox allele had a normal lung phenotype (data not shown). Upon euthanasia, 2/5 BRafCA/+; TP53lox/lox mice displayed prominent adenocarcinomas (Fig. 4R) composed of glandular structures lined by highly atypical cells with nuclear enlargement, hyperchromasia, and contour irregularities and displayed prominent nucleoli (Fig. 4S). The adenocarcinomas possessed large areas of necrosis and showed evidence of vascular and lymphatic invasion. This is significant, as we have not detected adenocarcinoma in any BRafCA mice prior to 40 wk of age (n = 55).
Similarly, BRafVE expression in an Ink4a/Arflox/lox background led to scattered papillary adenomas with evidence of subpleural nodules harboring cords of atypical cells trapped within regions of mesenchymal proliferation. 2/4 BRafCA/+; Ink4a/Arflox/lox mice contained multiple lung tumors with a classical bronchioloalveolar component and, when tumor cells were present, growing along alveolar septa without architectural destruction. This pattern is peculiar and is not categorized in the recent classification scheme but displays phenotypic similarities to human bronchioloalveolar carcinoma. Ki67 staining demonstrates that these lesions display increased proliferation relative to BRafVE-induced adenomas (Fig. 4Q).
The data presented here provide support for the hypothesis that oncogenic BRAF can provoke cell cycle arrest accompanied by induction of some, but not all, markers of OIS (Zhu et al. 1998; Collado et al. 2005). That KRasG12D (Jackson et al. 2005) or BRafVE can cooperate with loss of TSGs believed to mediate OIS is further, albeit circumstantial, evidence that OIS may serve as a bona fide tumor-suppressive mechanism in vivo. However, there remains at least one conundrum in this hypothesis. Adenomas that arise from expression of KRasG12D or BRafVE were initiated by oncogene activation in a single cell that subsequently underwent ∼15–20 population doublings to give rise to an adenoma. Clearly then, the initial response to KRasG12D or BRafVE expression is not cell cycle arrest but a dramatic induction of cell proliferation. However, at some time temporally distinct from oncogene activation, mechanisms that promote cell cycle arrest/senescence are engaged. This interpretation is consistent with the observation that BRAFVE-induced melanocytic nevi are clonal, having undergone multiple rounds of proliferation (Pollock et al. 2003), but eventually undergo senescence (Michaloglou et al. 2005). One possibility is that these biologically distinct outcomes reflect differences in the level of oncogene expression/activation at different stages of tumor progression. Alternatively, secondary signals may cooperate with KRasG12D or BRafVE to initiate and/or maintain the OIS program. These observations contrast with those in cultured cells, where activation of RAF robustly induces p16INK4A and irreversible cell cycle arrest/senescence within 12–24 h and in the absence of any initial cell proliferation (Zhu et al. 1998). While in vitro culture conditions may cooperate with RAF in the induction of senescence, the nature of the secondary trigger for OIS in vivo remains a key unanswered question.
Materials and methods
Generation of a conditionally active BRaf allele
DNA encompassing mouse BRaf exons 14–16 was modified such that exon 16 was replaced with a HSV-thymidine kinase cassette. A LoxP-flanked cassette containing the 3′ 382 base pairs (bp) of intron 14, the human BRAF cDNA containing exons 15–18, the mouse BRaf polyadenylation sequences, and a PGK-neo cassette was inserted into intron 14 upstream of a modified exon 15 that encodes BRafV600E and a silent XbaI restriction site polymorphism. The details of construction, Southern blotting, and RNA analysis are available as Supplemental Material.
Mice used in this study
Mice carrying conditional alleles of TP53 or Ink4A/Arf were genotyped as described in the Supplemental Material. BRafCA mouse genotypes were determined by standard PCR of tail DNA with primer pair AD (AD FwdA1, 5′-TGAGTATTTTTGTGGCAACTGC-3′; and AD RevB1, 5′-CTCTGCTGGGAAAGCGGC-3′) to produce diagnostic PCR products of 185 bp, 308 bp, and 335 bp for BRaf, BRafCA, and the BRafVE alleles, respectively.
Adenoviral Cre delivery
Ad-Cre and Ad-βGal were purified and titred by standard means (Viraquest), and 105 to 5 × 107 PFU were administered to the nasal septum of 6- to 8-wk-old mice by intranasal instillation as a calcium phosphate precipitate (Fasbender et al. 1998).
Immunohistochemistry
Animals were euthanized at the specified times or upon display of visible signs of disease. Lung tissues were prepared through standard techniques. Immunostaining was performed as described in the Supplemental Material using antibodies to CC10 (1:800); SP-C (Santa Cruz Biochemicals; 1:200); pMEK (Cell Signaling; 1:50), pERK (Cell Signaling; 1:100), and Ki67 (NeoMarkers; 1:200) with secondary antibodies and detection kits from DakoCytomation.
Acknowledgments
We thank Allan Balmain, Anton Berns, David Carretto, Ron DePinho, Byron Hann, Takashi Hirano, Nigel Killeen, Akiko Kobiyashi, Tyler Jacks, Leisa Johnson, Jennifer LeCouter, Anil Mukherjee, Daniel Murphy, Judith Sebolt-Leopold, and the UCSF transgenic, preclinical therapeutics, and mouse pathology core facilities for mice, materials, reagents, technical assistance, and advice. We thank Mike Fried and members of the Stokoe, Balmain, and McMahon laboratories for discussion. We acknowledge financial support from the General Motors Cancer Research Scholars Program (D.D.), Mouse Models of Human Cancer Consortium CA084244, and U.C. Discovery Grant (M.M.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1516407
References
- Brose M.S., Volpe P., Feldman M., Kumar M., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Volpe P., Feldman M., Kumar M., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Feldman M., Kumar M., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Kumar M., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Rishi I., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Gerrero R., Einhorn E., Herlyn M., Minna J., Nicholson A., Einhorn E., Herlyn M., Minna J., Nicholson A., Herlyn M., Minna J., Nicholson A., Minna J., Nicholson A., Nicholson A., et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Chen J., Fujii K., Zhang L., Roberts T., Fu H., Fujii K., Zhang L., Roberts T., Fu H., Zhang L., Roberts T., Fu H., Roberts T., Fu H., Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK–ERK independent mechanism. Proc. Natl. Acad. Sci. 2001;98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Benguria A., Zaballos A., Flores J.M., Barbacid M., Zaballos A., Flores J.M., Barbacid M., Flores J.M., Barbacid M., Barbacid M., et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., Teague J., Woffendin H., Garnett M.J., Bottomley W., Woffendin H., Garnett M.J., Bottomley W., Garnett M.J., Bottomley W., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Fasbender A., Lee J.H., Walters R.W., Moninger T.O., Zabner J., Welsh M.J., Lee J.H., Walters R.W., Moninger T.O., Zabner J., Welsh M.J., Walters R.W., Moninger T.O., Zabner J., Welsh M.J., Moninger T.O., Zabner J., Welsh M.J., Zabner J., Welsh M.J., Welsh M.J. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J. Clin. Invest. 1998;102:184–193. doi: 10.1172/JCI2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Mijimolle N., Dhawahir A., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M., Mijimolle N., Dhawahir A., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M., Dhawahir A., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M., Barradas M., Serrano M., Campuzano V., Barbacid M., Serrano M., Campuzano V., Barbacid M., Campuzano V., Barbacid M., Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A., Crowley D., Montoya R., Jacks T., Tuveson D.A., Montoya R., Jacks T., Tuveson D.A., Jacks T., Tuveson D.A., Tuveson D.A. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Olive K.P., Tuveson D.A., Bronson R., Crowley D., Brown M., Jacks T., Olive K.P., Tuveson D.A., Bronson R., Crowley D., Brown M., Jacks T., Tuveson D.A., Bronson R., Crowley D., Brown M., Jacks T., Bronson R., Crowley D., Brown M., Jacks T., Crowley D., Brown M., Jacks T., Brown M., Jacks T., Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E., Fedorov L.M., Siefken R., Walter A.O., Papadopoulos T., Rapp U.R., Fedorov L.M., Siefken R., Walter A.O., Papadopoulos T., Rapp U.R., Siefken R., Walter A.O., Papadopoulos T., Rapp U.R., Walter A.O., Papadopoulos T., Rapp U.R., Papadopoulos T., Rapp U.R., Rapp U.R. Lung-targeted expression of the c-Raf-1 kinase in transgenic mice exposes a novel oncogenic character of the wild-type protein. Cell Growth Differ. 2000;11:185–190. [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T., Vogel S., Crowley D., Bronson R.T., Jacks T., Crowley D., Bronson R.T., Jacks T., Bronson R.T., Jacks T., Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Suh Y.A., Lee J.I., Hassan K.A., Mao L., Force T., Gilbert B.E., Jacks T., Kurie J.M., Suh Y.A., Lee J.I., Hassan K.A., Mao L., Force T., Gilbert B.E., Jacks T., Kurie J.M., Lee J.I., Hassan K.A., Mao L., Force T., Gilbert B.E., Jacks T., Kurie J.M., Hassan K.A., Mao L., Force T., Gilbert B.E., Jacks T., Kurie J.M., Mao L., Force T., Gilbert B.E., Jacks T., Kurie J.M., Force T., Gilbert B.E., Jacks T., Kurie J.M., Gilbert B.E., Jacks T., Kurie J.M., Jacks T., Kurie J.M., Kurie J.M. Inhibition of oncogenic K-ras signaling by aerosolized gene delivery in a mouse model of human lung cancer. Clin. Cancer Res. 2002;8:2970–2975. [PubMed] [Google Scholar]
- Mercer K., Giblett S., Green S., Lloyd D., DaRocha Dias S., Plumb M., Marais R., Pritchard C., Giblett S., Green S., Lloyd D., DaRocha Dias S., Plumb M., Marais R., Pritchard C., Green S., Lloyd D., DaRocha Dias S., Plumb M., Marais R., Pritchard C., Lloyd D., DaRocha Dias S., Plumb M., Marais R., Pritchard C., DaRocha Dias S., Plumb M., Marais R., Pritchard C., Plumb M., Marais R., Pritchard C., Marais R., Pritchard C., Pritchard C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S., Shay J.W., Mooi W.J., Peeper D.S., Mooi W.J., Peeper D.S., Peeper D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Naoki K., Chen T.H., Richards W.G., Sugarbaker D.J., Meyerson M., Chen T.H., Richards W.G., Sugarbaker D.J., Meyerson M., Richards W.G., Sugarbaker D.J., Meyerson M., Sugarbaker D.J., Meyerson M., Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- Nikitin A.Y., Alcaraz A., Anver M.R., Bronson R.T., Cardiff R.D., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Alcaraz A., Anver M.R., Bronson R.T., Cardiff R.D., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Anver M.R., Bronson R.T., Cardiff R.D., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Bronson R.T., Cardiff R.D., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Cardiff R.D., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C., Gabrielson E.W., Gunning W.T., Haines D.C., Gunning W.T., Haines D.C., Haines D.C., et al. Classification of proliferative pulmonary lesions of the mouse: Recommendations of the Mouse Models of Human Cancers Consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- Pollock P.M., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Moses T.Y., Hostetter G., Wagner U., Kakareka J., Hostetter G., Wagner U., Kakareka J., Wagner U., Kakareka J., Kakareka J., et al. High frequency of BRAF mutations in nevi. Nat. Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Sebolt-Leopold J.S., Herrera R., Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- Shigematsu H., Gazdar A.F., Gazdar A.F. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int. J. Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- Tuveson D.A., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., Mercer K.L., Grochow R., Hock H., Crowley D., Grochow R., Hock H., Crowley D., Hock H., Crowley D., Crowley D., et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Vicent S., Lopez-Picazo J.M., Toledo G., Lozano M.D., Torre W., Garcia-Corchon C., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Lopez-Picazo J.M., Toledo G., Lozano M.D., Torre W., Garcia-Corchon C., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Toledo G., Lozano M.D., Torre W., Garcia-Corchon C., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Lozano M.D., Torre W., Garcia-Corchon C., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Torre W., Garcia-Corchon C., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Garcia-Corchon C., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Quero C., Soria J.C., Martin-Algarra S., Manzano R.G., Soria J.C., Martin-Algarra S., Manzano R.G., Martin-Algarra S., Manzano R.G., Manzano R.G., et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br. J. Cancer. 2004;90:1047–1052. doi: 10.1038/sj.bjc.6601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P.T., Garnett M.J., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., Garnett M.J., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., Jones C.M., Marshall C.J., Springer C.J., Barford D., Marshall C.J., Springer C.J., Barford D., Springer C.J., Barford D., Barford D., et al. Mechanism of activation of the RAF–ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Weber J.D., Taylor L.J., Roussel M.F., Sherr C.J., Bar-Sagi D., Taylor L.J., Roussel M.F., Sherr C.J., Bar-Sagi D., Roussel M.F., Sherr C.J., Bar-Sagi D., Sherr C.J., Bar-Sagi D., Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Woods D., Parry D., Cherwinski H., Bosch E., Lees E., McMahon M., Parry D., Cherwinski H., Bosch E., Lees E., McMahon M., Cherwinski H., Bosch E., Lees E., McMahon M., Bosch E., Lees E., McMahon M., Lees E., McMahon M., McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebisch A., Troppmair J., Troppmair J. Back to the roots: The remarkable RAF oncogene story. Cell. Mol. Life Sci. 2006;63:1314–1330. doi: 10.1007/s00018-006-6005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Woods D., McMahon M., Bishop J.M., Woods D., McMahon M., Bishop J.M., McMahon M., Bishop J.M., Bishop J.M. Senescence of human fibroblasts induced by oncogenic Raf. Genes & Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]