Abstract
Because migratory cells in all animals share common properties, we hypothesized that genetic networks involved in cell migration may be conserved between nematodes and vertebrates. To explore this, we performed comparative genomic analysis to identify vertebrate orthologs of genes required for hermaphrodite-specific neuron (HSN) migration in Caenoryhabditis elegans, and then examined their expression and function in the vertebrate neural crest. The results demonstrate high conservation of regulatory components involved in long-range migrations across diverse species. Although the neural crest is a vertebrate innovation, the results suggest that its migratory properties evolved by utilizing programs already present in the common vertebrate–invertebrate ancestor.
Keywords: Neural crest, cell migration, C. elegans HSN, comparative genomic analysis
During animal development, selected cell populations undergo complex and stereotypic migrations that are critical for proper pattern formation of the embryo. Cell migration is important for both embryonic and adult tissues, since its abnormal regulation causes developmental defects and can result in metastasis. However, the molecular mechanisms controlling cell migration remain poorly understood, particularly in vertebrates.
The neural crest is a stem cell-like multipotent cell population unique to vertebrates. Neural crest progenitors migrate from their site of origin in the central nervous system to numerous tissues in the periphery (Fig. 1A); for example, cranial neural crest cells leave the neural tube and migrate in defined streams to surround the eye, the frontonasal process, and the branchial arches. They subsequently differentiate into neurons, glia, pigment cells, and much of the craniofacial skeleton (Knecht and Bronner-Fraser 2002; Crane and Trainor 2006). The neural crest is one of the most motile of cell types in vertebrate embryos, and thus a potential model for understanding the molecular basis of migratory processes. However, due to the difficulty of visualizing motile cells in living vertebrate embryos, the problem of neural crest migration has not been particularly amenable to genetic dissection, and only a limited number of genes that affect aspects of neural crest migration have been identified (Halloran and Berndt 2003).
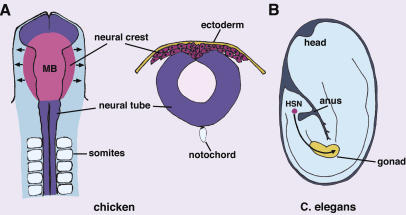
Figure 1.
Schematic diagram of long-range cell migration of vertebrate neural crest and C.elegans hermaphrodite-specific neurons. (A) Cranial neural crest cells emigrate from the neural tube and migrate to different sites in the periphery of the embryo (stage 9 chick embryo). The diagram on the right shows a transverse section through the midbrain level with neural crest cells emigrating out of dorsal neural tube and migrating into the head mesenchyme. (MB) Midbrain. (B) HSN neurons migrate from the tail to gonadal primodium in the C. elegans embryo.
The feasibility of genetic screens in invertebrates, such as Caenorhabditis elegans and Drosophila melanogaster, has allowed the isolation of mutants that disrupt various aspects of cell migration (Garriga and Stern 1994; Montell 1999). For example, C. elegans hermaphrodite-specific neurons (HSN) undergo long-range migrations from their location at birth in the tail to positions flanking the gonadal primordium in the midbody of the animal (Fig. 1B). Forward genetic screening has identified 15 genes required for HSN migration, including transcription factors, signaling, and adhesion molecules (Garriga and Stern 1994).
In the present study, we take a comparative genomic analysis, which combines the information of invertebrate genetics and complete genome sequences from animal models. We address the question of whether the genetic networks involved in long-range cell migrations are conserved across the large evolutionary distance between C. elegans HSN neurons and vertebrate neural crest cells. Our study demonstrates that the neural crest, a uniquely vertebrate cell type, shares common regulatory elements for cell migration programs utilized in the nematode, suggesting that the role of these genes was present in the common ancestor of vertebrates and invertebrates. We offer this comparative genomic analysis as a powerful tool for identifying regulatory genes, including transcription factors, governing vertebrate-specific processes.
Results and Discussion
Genomic screening of vertebrate orthologs of HSN migration genes
Given that morphological and biochemical aspects of cell migratory behavior appear to be largely conserved across species, we posited that the genetic mechanisms controlling long-range cell migration may also be conserved between evolutionarily distant animals. To test this idea, we asked whether genes involved in the migration of C. elegans HSN neurons were also utilized for a similar purpose in the neural crest, thus offering a bioinformatics approach for identifying candidate cell migration genes in vertebrates. To this end, we picked 15 genes that are essential for HSN migration and identified their putative vertebrate orthologs. We have tentatively defined the vertebrate ortholog as the gene having the highest identity score in the tBLAST analysis against all available vertebrate genome sequences. We then used the putative vertebrate orthologs to identify chicken or Xenopus laevis genes and their available EST sequences (Table 1; Supplementary Table S1).
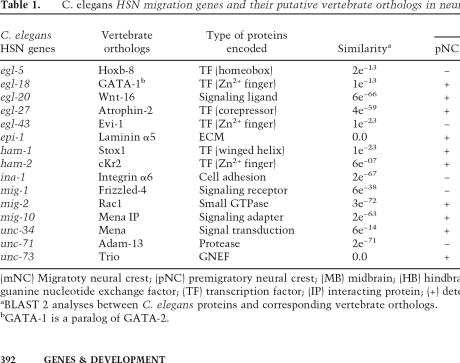
Table 1.
C. elegans HSN migration genes and their putative vertebrate orthologs in neural crest migration
(mNC) Migratoty neural crest; (pNC) premigratory neural crest; (MB) midbrain; (HB) hindbrain; (ECM) extracellular matrix; (GNEF) guanine nucleotide exchange factor; (TF) transcription factor; (IP) interacting protein; (+) detected; (−) not detected.
aBLAST 2 analyses between C. elegans proteins and corresponding vertebrate orthologs.
bGATA-1 is a paralog of GATA-2.
These 15 genes included six transcription factors, (egl-5, egl-18, egl-27, egl-43, ham-1, and ham-2) (Chisholm 1991; Garriga et al. 1993; Garriga and Stern 1994; Guenther and Garriga 1996; Baum et al. 1999; Herman et al. 1999), three signaling ligands or receptors (egl-20, ina-1, and mig-1) (Harris et al. 1996; Baum and Garriga 1997; Maloof et al. 1999), one extracellular matrix molecule (epi-1) (Forrester and Garriga 1997), four intracellular signaling molecules (mig-2, mig-10, unc-34, and unc-73) (Forrester and Garriga 1997; Manser et al. 1997; Zipkin et al. 1997; Steven et al. 1998), and one protease (unc-71) (Huang et al. 2003). We obtained all 15 chicken orthologs from the complete chick genome sequence and five X. laevis orthologs from available partial genome sequence and EST databases (Table 1; Supplementary Table S1).
Reciprocal BLAST analysis to the C. elegans genome matched nine vertebrate orthologs as having the highest identity scores to their corresponding C. elegans genes (Supplementary Table S1); the other six putative orthologs were matched to their corresponding best match by virtue of the KOGs (eukaryotic orthologous groups), to which the C. elegans gene belongs (Tatusov et al. 2003). We further analyzed these six genes by OrthoMCL, a method that constructs ortholog groups using a Markov Cluster algorithm (Li et al. 2003). This program assigned mig-2 to an ortholog group OG1_290 together with other C. elegans paralogs and RAC1 of other species, suggesting that mig-2 may be one of the “recent” paralogs or co-orthologs, which are equally related to orthologs in other species. The ortholog groups for egl-5 (OG1_29675), egl-18 (OG1_64731), ham-2 (OG1_39625), and mig-1 (OG1_39065) contained only nematode genes, and the one for unc-71 (OG1_17783) contained yeast, nematode, and fly genes, suggesting that they are related as “ancient” paralogs, perhaps accounting for why the reciprocal BLAST from their corresponding putative vertebrate orthologs identified their paralogs as best matches in C. elegans genome.
Expression of HSN migration genes in neural crest
We first examined the expression patterns of these transcripts in chicken embryos, focusing on initial stages of neural crest migration in the head. The EST sequences or partial sequences obtained by PCR, based on genomic sequences, were used to prepare RNA probes for subsequent in situ hybridization. Gene expression in neural crest cells was examined as previously described (Kee and Bronner-Fraser 2001, 2005). At early stages of nervous system formation, most of the chicken orthologs were expressed in premigratory and migrating neural crest cells in the midbrain as shown in whole-mount and sections of in situ hybridized embryos (Fig. 2A). The chicken orthologs of egl-20, egl-27, epi-1, ham-1, ham-2, mig-2, mig-10, unc-34, and unc-73 were expressed in premigratory neural crest cells in the dorsal neural tube. Sections through these embryos revealed that most genes were also expressed on newly emigrated neural crest cells on the dorsal neural tube as they embark on their migratory routes. The exception was the ham-1 ortholog, which was down-regulated as neural crest cells emigrated out of the neural tube (Fig. 2A).
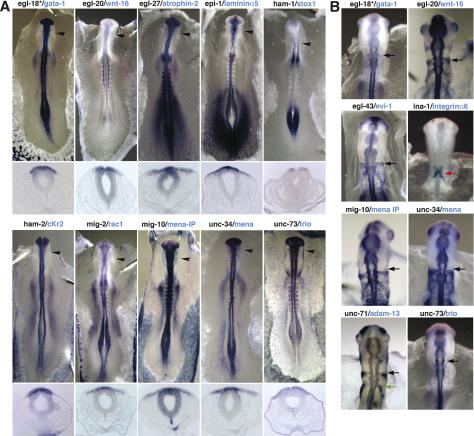
Figure 2.
C. elegans genes essential for HSN migration are conserved in migrating cranial neural crest cells. (A) Vertebrate orthologs of the indicated C. elegans HSN migration genes are expressed in migrating neural crest cells of chicken at the level of midbrain (arrowhead). (Top) Embryos were processed by whole-mount in situ hybridization using chick probes to the orthologs of C. elegans genes. (Bottom) Sections through each embryo at the level of midbrain (arrowhead in top panel) show expression in emigrating and migrating neural crest cells. (B) Migrating neural crest cells in the chick hindbrain express vertebrate orthologs of the indicated C. elegans genes. Gene names are in alphabetical order by the convention of each C. elegans name together with that of the vertebrate ortholog. Each arrow indicates neural crest stream migrating from rhombomere 4 (r4) (black), r5 (red), or r6 (green).
The putative vertebrate ortholog of egl-18 was identified as the transcription factor GATA-2 by tBLAST search, but was not detected in neural crest cells (data not shown). The vertebrate GATA transcription factor family has been recently duplicated (Lowry and Atchley 2000), suggesting that a paralog of GATA-2 might have assumed the activity of egl-18 in migrating neural crest cells. To test this possibility, we examined the expression patterns of six GATA genes that were most recently duplicated as two groups of GATA1, GATA2, and GATA3, and GATA4, GATA5, and GATA6, and found that only GATA-1 was expressed in the premigratory and migrating neural crest cells (Fig. 2A,B). This result suggests GATA transcription factor function has been evolutionarily conserved in migrating neural crest cells and indicates that co-orthologs by gene duplication should be considered in this kind of comparative genomic analysis.
Of the 15 genes, we failed to see neural crest expression only in the putative orthologs of two genes, egl-5 (Hoxb-8 ortholog) and mig-1 (Frizzled-4 ortholog). However, there is good evidence that family members related to Hoxb-8 are expressed in migrating neural crest cells (Prince and Lumsden 1994; Saldivar et al. 1996).
Several of the vertebrate orthologs were expressed in interesting patterns in subsets of neural crest cells. Particularly striking is the ina-1 ortholog, integrin α6, which was specifically expressed in neural crest cells emanating from rhombomere 5 (r5) and joining the r4 and r6 migration streams in the hindbrain (Fig. 2B). The egl-43 and unc-71 orthologs were expressed in the r4 and r6 streams of hindbrain, but not in the midbrain streams (Fig. 2B). Orthologs of egl-18, egl-20, mig-10, unc-34, and unc-73 were also expressed in the r4 and/or r6 neural crest streams in the hindbrain (Fig. 2B) as well as in the midbrain migratory streams (Fig. 2A). Many of the genes appeared to be expressed at relatively low levels, suggesting that they may be transiently expressed or tightly regulated in migrating neural crest cells.
We next examined the distribution patterns of HSN orthologs in X. laevis, an organism highly amenable to functional analysis, to determine whether these expression profiles were conserved in other vertebrates. As described above, we identified five X. laevis orthologs, but were unable to obtain others due to the absence of complete genome sequence. The orthologs of egl-43, ina-1, mig-2, and unc-71 were clearly expressed in the cranial neural crest cells that migrate in prominent streams from the neural tube toward the branchial arches in frogs (Fig. 3A).
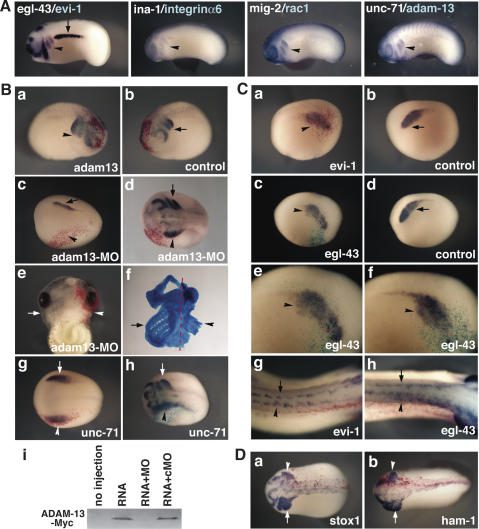
Figure 3.
Functional analysis of the migration genes in Xenopus neural crest development. (A) Xenopus orthologs of HSN migration genes are expressed in migrating cranial neural crest cells (arrowheads), as shown by in situ hybridization of Xenopus orthologs corresponding to the indicated C. elegans genes. Arrow points to the pronephric duct, which also expresses the egl-43 ortholog, evi-1. (B) unc-71 and its ortholog adam-13 in neural crest development. Lateral view of the embryo injected with adam-13 (Red-Gal) has more migrating neural crest cells (panel a; arrowhead) than the uninjected control side (panel b; arrow). (Panel c) Dorsal view of stage14 embryo with anterior at left shows that slug expression is dramatically decreased on the side injected with MO against adam-13 (arrowhead, Red-Gal) compared with the uninjected control side (arrow). (Panel d) Dorsal view of a stage 22 embryo (with anterior to the left) shows that neural crest migration is delayed or inhibited on the side injected with MO (Red-Gal). (Panel e) Head cartilage formation is defective on the side injected with MO (arrowhead, Red-Gal) compared with the uninjected side, as shown in ventral view of stage 45 embryo. (Panel f) Cartilage staining of this embryo clearly shows a profound defect in branchial arch formation on the injected side (arrowhead). Dotted red line indicates midline. (Panel g) Ectopic expression of unc-71 (arrowhead, Red-Gal) does not affect neural crest induction as assayed by slug expression. (Panel h) However, neural crest migration is defective on the side injected with unc-71 (arrowhead, turquoise, X-Gal), compared with the uninjected side (arrow). (Panel i) Immunoblot analysis shows ADAM-13 protein expression is inhibited by adam-13 MO (MO) but not by control MO (cMO). (RNA) adam-13 RNA injected into frog embryos. (C) egl-43 and its vertebrate ortholog evi-1 in neural crest development. Lateral view of a stage 14 embryo shows that neural crest cells are more dispersed and loosely associated on the side on which evi-1 is overexpressed (panel a; arrowhead, Red-Gal staining), while the uninjected control side retains a compact premigratory neural crest (panel b; arrow), as shown by in situ hybridization with slug. Ectopic expression of egl-43 causes dramatic effects including premature migration of neural crest cells and increased cell dispersal (arrowhead) on the injected side (panel c; turquoise, X-Gal staining), compared with the control side (panel d). (Panels e,f) Higher-magnification images clearly show streams of neural crest cells coming out of the premigratory neural crest domain where egl-43 is expressed (arrowhead). Dorsal view of stage 26 embryo (anterior at left) shows that ectopic expression of either evi-1 (panel g) or egl-43 (panel h) causes muscle precursors, another motile cell type, to migrate prematurely on the injected side (arrowhead), compared with the uninjected control side (arrow), as shown by in situ hybridization with twist. (D) ham-1 and its ortholog stox1 in neural crest development. Dorsal views of stages 21–22 embryos show that the migrating neural crest cell population is decreased and migration is delayed or inhibited at the side injected with stox1 (panel a) or ham-1 (panel b) (arrowhead), compared with the control side (arrow).
Functional analysis of HSN migration genes
The conserved expression of these genes in migrating cells in both vertebrates and nematodes suggests that their function in aspects of cell migration might also be conserved, since genetic analysis has revealed functional roles for the genes in HSN migration in C. elegans. Although little is known about the role of these genes in vertebrates, two of the identified genes have ascribed function in the neural crest. First, the adam-13 (unc-71 ortholog) was previously shown to be expressed by Xenopus neural crest cells, and ectopic expression of the protease-defective forms caused abnormalities in neural crest migration (Alfandari et al. 2001). Second, the laminin α5 (epi-1 ortholog) knockout mice have defects in neural crest migration (Coles et al. 2006). To further test this possibility, we used the vertebrate and/or nematode genes for gain- and loss-of- function studies in Xenopus embryos to ask whether these genes play a role in neural crest migration. C. elegans or vertebrate full-length genes were used for gain-of-function and morpholino antisense oligonucleotides (MO) for loss-of-function experiments; the nucleotides were injected into one blastomere of the four- or eight-cell stage embryos to target the presumptive neural crest region. Subsequent effects were ascertained by examining the early expression of neural crest markers, slug, twist, or sox10 in premigratory or migrating neural crest by in situ hybridization.
Overexpression of Xenopus ADAM-13 (unc-71 ortholog) resulted in expansion of the cranial neural crest population, in agreement with previous studies (Fig. 3B, panels a,b; n = 46, 48%; Alfandari et al. 2001). To examine whether loss of ADAM-13 function would lead to a reciprocal phenotype, we designed a MO to block translational initiation of endogenous ADAM-13. Injection of the antisense MO into one blastomere of the four- or eight-cell stage embryo resulted in defects in early neural crest development (Fig. 3B, panels c,d; n = 78, 63%). Rather than migrating in distinct streams in the cranial region, neural crest cells in morphant embryos exhibited delayed induction and migration or failed to leave the dorsal portion of the neural tube. At later tadpole stages, severe loss of branchial arch tissues, which are derived from cranial neural crest cells, was noted on the ADAM-13-depleted side (Fig. 3B; panels e,f, n = 47, 49%). The specificity of the ADAM13-MO was tested and confirmed for inhibiting protein expression of ADAM-13-Myc in vivo in frog embryos, while control MO had no effect (Fig. 3B, panel i). We next examined the effect of overexpressing the C. elegans unc-71 gene in Xenopus. Overexpression of unc-71 had a similar effect as the loss-of-function phenotype, causing defective neural crest migration on the injected side of the embryo, even though early neural crest induction was unaffected (Fig. 3B, panels g,h; n = 42, 67%). The opposite phenotype may be explained by the fact that protease-defective ADAM-13 disrupts neural crest migration (Alfandari et al. 2001), while UNC-71 lacks protease activity (Huang et al. 2003).
Overexpression of the human evi-1 proto-oncogene (egl-43 ortholog) promoted dispersal of premigratory cranial neural crest cells or decreased the cell population on the injected side compared with the uninjected side of the same embryo (Fig. 3C, panels a,b; n = 75, 68%). Injection of C. elegans egl-43 transcripts caused a similar effect, producing clear streams of prematurely migrating neural crest cells, with the nematode gene being more efficacious than the human gene (Fig. 3C, panels c–f; n = 65, 69%). At later stages of development, embryos were stained with twist, a basic helix–loop–helix (bHLH) transcription factor that serves as a molecular marker for both cranial neural crest cells and muscle precursors in the trunk. The muscle precursors, another migratory cell population, also appeared to prematurely initiate migration on the side of the embryo injected with either human evi-1 or nematode egl-43 (Fig. 3C, panels g,h), suggesting that these genes may be involved in cell migration in other cell types as well.
Expression of stox1, the ham-1 ortholog, in premigratory neural crest was transient at stage 8 in chicken embryo (Fig. 2A) and down-regulated as neural crest cells emigrated from neural tube. Interestingly, overexpression of stox1a (Fig. 3D, panel a; n = 54, 61%) or ham-1 (Fig. 3D, panel b; n = 45, 47%) caused a decrease in the migrating neural crest population and sometimes inhibited normal migration on the injected side compared with the control side. Overexpression of ina-1 or unc-34 did not affect neural crest development, but embryos injected with mig-2 developed abnormally at early embryonic stages, suggesting it may have pleiotropic effects (data not shown).
Evolutionary conservation of regulatory genes
This study shows that the genes necessary for HSN migration in C. elegans are also expressed by migrating vertebrate neural crest cells, demonstrating a high degree of conservation of regulatory components involved in long-range cell migrations between evolutionarily far-distant animals. Thus, many key components of the regulatory networks underlying cell motility and migration must have been fixed early in animal evolution. Although the neural crest is unique to vertebrates, our study suggests that its migration might have evolved by utilizing a genetic program underlying cell migration programs in cells already present in the common ancestor of vertebrates and invertebrates. In addition, specific subsets of neural crest cells give rise to diverse tissues and, therefore, may require different combinations of regulatory proteins to control specific migration paths and destinations. We speculate that the genes governing migration of other types of cells in nematodes and insects may also be conserved in different subsets of migrating neural crest cells.
Evolutionary conservation of individual genes involved in development or cell motility has been shown based on analyses of a single gene at a time. In this study, we have screened a number of genes by combining the rich information from C. elegans genetics with complete genome sequences of different model organisms, and demonstrate that long-range cell migrations of C. elegans HSN and vertebrate neural crest share common genetic networks. Interestingly, many of the genes identified in this screen were not previously reported to function in neural crest migration. Thus, comparative genomics, combined with invertebrate genetics, can be used as a powerful tool to uncover genes regulating vertebrate-specific processes for which complete genome sequences are available.
Materials and methods
Bioinformatic identification of vertebrate orthologs of C. elegans HSN migration genes
To identify the putative chicken (Gallus gallus) and frog (X. laevis) orthologs of the 15 C. elegans genes essential for the migration of HSN neurons, full-length cDNA sequences of the 15 C. elegans genes were downloaded from WormBase (http://www.wormbase.org), and their translated cDNA sequences were used as queries to tBLAST search against all vertebrate genome sequences. Vertebrate orthologs were tentatively defined as the genes having the highest identity scores in this tBLAST analysis. Next, the vertebrate sequences were used as queries to BLAST search against the chicken genome, for which complete sequence information is available, and against the frog genome database, which contains partial genome and EST sequences of X. laevis and Xenopus tropicalis. Putative chicken orthologs were defined as the genes having the highest identity scores in the BLAST search against chicken genome sequence. We were able to identify the orthologs corresponding to all 15 C. elegans HSN genes from the completely sequenced genome. Only X. laevis genes that were almost identical to the translated vertebrate protein sequences, which were defined as having either an E value of 0.0 or at least 90% sequence identity, were considered as putative orthologs because of the incomplete genome sequence. For this reason, we were able to identify only five orthologs from the X. laevis genome. Finally, the ortholog sequences were used as queries to search EST sequences available in the databases of http://www.openbiosystems.com and http://www.chick.umist.ac.uk. Information about the putative orthologs and their reciprocal BLAST analysis are summarized in Supplementary Table S1.
Embryo manipulation and in situ hybridization
Chicken and frog (X. laevis) embryo manipulation and in situ hybridization were performed as described previously (Kee and Bronner-Fraser 2001, 2005). The EST plasmids were used as templates in the synthesis of RNA probes for in situ hybridization to determine their expression in the neuralcrest cells. Capped RNA was injected into embryos with n-β-galactosidase RNA as tracer, and the injected region was visualized as red color with Red-Gal staining or as turquoise color with X-gal staining.
Materials and Methods on DNA Constructs and RNA Synthesis, Morpholino Experiments and Western Blot Analysis, and Cartilage Staining are described in the Supplemental Material.
Acknowledgments
We thank Samuel Ki for technical assistance and colleagues in the M.B.-F. laboratory for technical advice and discussion. We especially thank Drs. Titus Brown, Scott Fraser, Mihoko Kato, Daniel Meulemans, Ellen Rothenberg, Tatjana Sauka-Spengler, Erich Schwarz, and J.K. Sky Yu for helpful discussions on the manuscript, and Drs. Dominique Alfandari, Tamara Allison, Rik Derynck, Todd Evans, Cees Oudejans, and Richard Harland for sharing the constructs. This work was supported by NIH grants NS36585 and NS051051 (to M.B.-F.); by the Howard Hughes Medical Institute, of which P.W.S. is an Investigator; and by NHGRI Genome Scholar Development and Faculty Transition Award (K22) (to B.J.H.).
Footnotes
Supplemental material is available at http://www.genesdev.org
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1509307
References
- Alfandari D., Cousin H., Gaultier A., Smith K., White J.M., Darribere T., DeSimone D.W., Cousin H., Gaultier A., Smith K., White J.M., Darribere T., DeSimone D.W., Gaultier A., Smith K., White J.M., Darribere T., DeSimone D.W., Smith K., White J.M., Darribere T., DeSimone D.W., White J.M., Darribere T., DeSimone D.W., Darribere T., DeSimone D.W., DeSimone D.W. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Baum P.D., Garriga G., Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Baum P.D., Guenther C., Frank C.A., Pham B.V., Garriga G., Guenther C., Frank C.A., Pham B.V., Garriga G., Frank C.A., Pham B.V., Garriga G., Pham B.V., Garriga G., Garriga G. The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron. Genes & Dev. 1999;13:472–483. doi: 10.1101/gad.13.4.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- Coles E.G., Gammill L.S., Miner J.H., Bronner-Fraser M., Gammill L.S., Miner J.H., Bronner-Fraser M., Miner J.H., Bronner-Fraser M., Bronner-Fraser M. Abnormalities in neural crest cell migration in laminin α5 mutant mice. Dev. Biol. 2006;289:218–228. doi: 10.1016/j.ydbio.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Crane J.F., Trainor P.A., Trainor P.A. Neural crest stem and progenitor cells. Annu. Rev. Cell Dev. Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- Forrester W.C., Garriga G., Garriga G. Genes necessary for C. elegans cell and growth cone migrations. Development. 1997;124:1831–1843. doi: 10.1242/dev.124.9.1831. [DOI] [PubMed] [Google Scholar]
- Garriga G., Stern M.J., Stern M.J. Hams and Egls: Genetic analysis of cell migration in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 1994;4:575–580. doi: 10.1016/0959-437x(94)90075-e. [DOI] [PubMed] [Google Scholar]
- Garriga G., Guenther C., Horvitz H.R., Guenther C., Horvitz H.R., Horvitz H.R. Migrations of the Caenorhabditis elegans HSNs are regulated by egl-43, a gene encoding two zinc finger proteins. Genes & Dev. 1993;7:2097–2109. doi: 10.1101/gad.7.11.2097. [DOI] [PubMed] [Google Scholar]
- Guenther C., Garriga G., Garriga G. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development. 1996;122:3509–3518. doi: 10.1242/dev.122.11.3509. [DOI] [PubMed] [Google Scholar]
- Halloran M.C., Berndt J.D., Berndt J.D. Current progress in neural crest cell motility and migration and future prospects for the zebrafish model system. Dev. Dyn. 2003;228:497–513. doi: 10.1002/dvdy.10374. [DOI] [PubMed] [Google Scholar]
- Harris J., Honigberg L., Robinson N., Kenyon C., Honigberg L., Robinson N., Kenyon C., Robinson N., Kenyon C., Kenyon C. Neuronal cell migration in C. elegans: Regulation of Hox gene expression and cell position. Development. 1996;122:3117–3131. doi: 10.1242/dev.122.10.3117. [DOI] [PubMed] [Google Scholar]
- Herman M.A., Ch’ng Q., Hettenbach S.M., Ratliff T.M., Kenyon C., Herman R.K., Ch’ng Q., Hettenbach S.M., Ratliff T.M., Kenyon C., Herman R.K., Hettenbach S.M., Ratliff T.M., Kenyon C., Herman R.K., Ratliff T.M., Kenyon C., Herman R.K., Kenyon C., Herman R.K., Herman R.K. EGL-27 is similar to a metastasis-associated factor and controls cell polarity and cell migration in C. elegans. Development. 1999;126:1055–1064. doi: 10.1242/dev.126.5.1055. [DOI] [PubMed] [Google Scholar]
- Huang X., Huang P., Robinson M.K., Stern M.J., Jin Y., Huang P., Robinson M.K., Stern M.J., Jin Y., Robinson M.K., Stern M.J., Jin Y., Stern M.J., Jin Y., Jin Y. UNC-71, a disintegrin and metalloprotease (ADAM) protein, regulates motor axon guidance and sex myoblast migration in C. elegans. Development. 2003;130:3147–3161. doi: 10.1242/dev.00518. [DOI] [PubMed] [Google Scholar]
- Kee Y., Bronner-Fraser M., Bronner-Fraser M. Temporally and spatially restricted expression of the helix–loop–helix transcriptional regulator Id1 during avian embryogenesis. Mech. Dev. 2001;109:331–335. doi: 10.1016/s0925-4773(01)00574-3. [DOI] [PubMed] [Google Scholar]
- Kee Y., Bronner-Fraser M., Bronner-Fraser M. To proliferate or to die: Role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes & Dev. 2005;19:744–755. doi: 10.1101/gad.1257405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht A.K., Bronner-Fraser M., Bronner-Fraser M. Induction of the neural crest: A multigene process. Nat. Rev. Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J., Jr., Roos D.S., Stoeckert C.J., Jr., Roos D.S., Roos D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry J.A., Atchley W.R., Atchley W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- Maloof J.N., Whangbo J., Harris J.M., Jongeward G.D., Kenyon C., Whangbo J., Harris J.M., Jongeward G.D., Kenyon C., Harris J.M., Jongeward G.D., Kenyon C., Jongeward G.D., Kenyon C., Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Manser J., Roonprapunt C., Margolis B., Roonprapunt C., Margolis B., Margolis B. C. elegans cell migration gene mig-10 shares similarities with a family of SH2 domain proteins and acts cell nonautonomously in excretory canal development. Dev. Biol. 1997;184:150–164. doi: 10.1006/dbio.1997.8516. [DOI] [PubMed] [Google Scholar]
- Montell D.J. The genetics of cell migration in Drosophila melanogaster and Caenorhabditis elegans development. Development. 1999;126:3035–3046. doi: 10.1242/dev.126.14.3035. [DOI] [PubMed] [Google Scholar]
- Prince V., Lumsden A., Lumsden A. Hoxa-2 expression in normal and transposed rhombomeres: Independent regulation in the neural tube and neural crest. Development. 1994;120:911–923. doi: 10.1242/dev.120.4.911. [DOI] [PubMed] [Google Scholar]
- Saldivar J.R., Krull C.E., Krumlauf R., Ariza-McNaughton L., Bronner-Fraser M., Krull C.E., Krumlauf R., Ariza-McNaughton L., Bronner-Fraser M., Krumlauf R., Ariza-McNaughton L., Bronner-Fraser M., Ariza-McNaughton L., Bronner-Fraser M., Bronner-Fraser M. Rhombomere of origin determines autonomous versus environmentally regulated expression of Hoxa-3 in the avian embryo. Development. 1996;122:895–904. doi: 10.1242/dev.122.3.895. [DOI] [PubMed] [Google Scholar]
- Steven R., Kubiseski T.J., Zheng H., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J., Kubiseski T.J., Zheng H., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J., Zheng H., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J., Kulkarni S., Mancillas J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J., Mancillas J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J., Ruiz Morales A., Hogue C.W., Pawson T., Culotti J., Hogue C.W., Pawson T., Culotti J., Pawson T., Culotti J., Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Mekhedov S.L., Nikolskaya A.N., Nikolskaya A.N., et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkin I.D., Kindt R.M., Kenyon C.J., Kindt R.M., Kenyon C.J., Kenyon C.J. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell. 1997;90:883–894. doi: 10.1016/s0092-8674(00)80353-0. [DOI] [PubMed] [Google Scholar]