Abstract
Plants must perceive and rapidly respond to changes in ambient temperature for their successful reproduction. Here we demonstrate that Arabidopsis SHORT VEGETATIVE PHASE (SVP) plays an important role in the response of plants to ambient temperature changes. The loss of SVP function elicited insensitivity to ambient temperature changes. SVP mediates the temperature-dependent functions of FCA and FVE within the thermosensory pathway. SVP controls flowering time by negatively regulating the expression of a floral integrator, FLOWERING LOCUS T (FT), via direct binding to the CArG motifs in the FT sequence. We propose that this is one of the molecular mechanisms that modulate flowering time under fluctuating temperature conditions.
Keywords: Ambient temperature, SVP, floral repressor, FT, the thermosensory pathway
Plants are sessile organisms and are, consequently, exposed to a wide variety of environmental stresses, both abiotic and biotic, exerted by their surroundings. The most common of these is temperature. Within the range of temperatures tolerable to plants, the response to low temperature, particularly near-freezing temperature, is well understood. Plants have evolved a number of adaptive mechanisms to meet the challenge of low temperature. In Arabidopsis, flowering is accelerated by prolonged exposure to cold, a process called vernalization. The epigenetic silencing of the FLOWERING LOCUS C (FLC) (Michaels and Amasino 1999; Sheldon et al. 1999) is central to the vernalization process (Sung and Amasino 2005), and this silencing has been attributed to the activities of the VERNALIZATION1 (VRN1), VERNALIZATION2 (VRN2), and VERNALIZATION INSENSITIVE3 (VIN3) genes (Gendall et al. 2001; Levy et al. 2002; Sung and Amasino 2004). Cold acclimation is another well-characterized response to low temperature (Guy 1990). Plants become tolerant to freezing temperatures by being previously exposed to short periods of low but nonfreezing temperatures. Analyses of mutant plants have identified C-Repeat-binding factor (CBF)-dependent and CBF-independent signaling pathways in cold acclimation (Sharma et al. 2005), suggesting that plants use distinct mechanisms to respond to low temperature.
There is increasing concern about the potential impact of global temperature changes, which significantly affect ambient temperature, on plant development. Several lines of evidence suggest that the recently observed alterations in the flowering times of many plant species and the increase in plant respiration rates are closely associated with these changes in ambient temperature (Fitter and Fitter 2002; Atkin and Tjoelker 2003). Although a great deal of progress has been made in our understanding of the regulation of plant development by low temperature, less is currently known about the molecular mechanisms underlying the responses of plants to changes in ambient temperature (Coupland and Prat Monguio 2005; Samach and Wigge 2005). Here, we show that the SHORT VEGETATIVE PHASE (SVP) gene mediates ambient temperature signaling in Arabidopsis and that the SVP-mediated control of FLOWERING LOCUS T (FT) expression is one of the molecular mechanisms evolved by plants to modulate the timing of the developmental transition to flowering phase in response to changes in the ambient temperature.
Results and Discussion
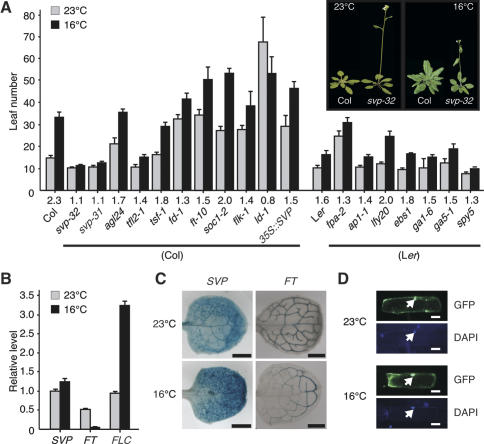
As a first step to determining the mechanism underlying the perception and transduction of ambient temperature signaling in plants, we assessed mutants in known flowering time genes for their insensitivity to changes in ambient growth temperature. Of the flowering time mutants tested, one with a lesion in svp was indeed insensitive to such changes. The flowering of the majority of these flowering time mutants was noticeably delayed at 16°C, with flowering time ratios (16°C/23°C) ranging from 1.1 to 2.0 (Fig. 1A), the exception being ld-1. However, svp-31 and svp-32 mutants, the T-DNA alleles of SVP (Supplementary Fig. 1; Hartmann et al. 2000), manifested almost identical flowering times at 23°C and 16°C (Fig. 1A). svp mutants were early flowering, especially at 16°C, suggesting that a reduction in SVP activity significantly decreased plant response to lower temperature and that the loss of SVP activity would result in the loss of the effects of low temperature. In contrast, SVP overexpressor plants were late flowering, especially at 23°C, suggesting that overexpression of SVP can mimic the effect of low temperature. Reduced FT expression was likely responsible for this late flowering phenotype of 35S∷SVP plants (Supplementary Fig. 2). The weak temperature response seen in 35S∷SVP plants can be explained by the differential expression of FT, such that FT expression at 23°C was higher than that at 16°C in 35S∷SVP plants. SVP probably performs a nonredundant role in ambient temperature sensing, as loss of the function of AGL24 (Yu et al. 2002), the closest homolog of SVP, did not induce temperature insensitivity.
Figure 1.
Role of SVP in the temperature-dependent control of flowering in Arabidopsis. (A) Flowering time of a group of flowering time mutants at 23°C and 16°C under long-day conditions. The numbers listed above the genotypes denote the ratios of flowering time at 16°C and 23°C (16°C/23°C). Error bars indicate the standard deviation. The inset shows wild-type Columbia (Col) plants and svp-32 plants grown at 23°C and 16°C. (B) Effects of low temperature on SVP expression in wild-type plants (Col). SVP, FLC, and FT expression levels were measured by real-time PCR in the leaf of 10-d-old seedlings grown at the indicated temperatures. The average of three technical replicates is shown. FLC and FT were used as control genes, the expressions of which are thermo-regulated (Blázquez et al. 2003). Tubulin was used as an internal control. (C) Histochemical analysis of 10-d-old seedlings of SVP∷GUS and FT∷GUS plants grown at 23°C and 16°C. Bars, 500 μm. (D) Nuclear localization of SVP-GFP fusion protein in onion epidermal cells incubated at 16°C and 23°C. The nucleus is indicated by an arrow. DAPI (4′-6-Diamidino-2-phenylindole) was used for nuclear staining. Bars, 10 μm.
Characterization of the pattern of SVP expression at different temperatures in wild-type plants by real-time PCR analyses revealed that SVP expression slightly increased in the leaf at 16°C (Fig. 1B). In contrast, FT expression was strongly repressed in the leaf at l6°C. Histochemical β-glucuronidase (GUS) analysis detected both SVP and FT expression throughout the expanded leaves, but SVP expression was up-regulated and FT expression was down-regulated (Fig. 1C). Since this up-regulation of SVP may not be significant in itself in explaining this dramatic down-regulation of FT expression, it is possible that the post-transcriptional regulation of SVP or altered protein–protein interaction of SVP at the lower temperature may also be responsible for the reduction in FT transcription. Considering that SVP acts as a floral repressor (Hartmann et al. 2000), these data suggested that additional flower-inhibitory factors exist in the leaf at lower temperature. Subcellular localization analysis showed that the SVP-green fluorescence protein (GFP) fusion protein localized in the nucleus at both 16°C and 23°C (Fig. 1D). Taken together, these results indicate that SVP expression is weakly temperature dependent, similar to the thermosensory genes of other species (Johansson et al. 2002).
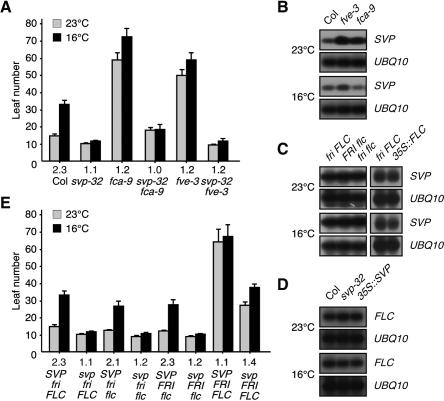
Ambient temperature is perceived via a genetic pathway (thermosensory pathway) that requires both FCA and FVE in Arabidopsis (Blázquez et al. 2003). An analysis of the genetic interaction of svp mutants with fca and fve mutants was conducted to ascertain whether or not SVP operates within the same genetic pathway as FCA and FVE. The late flowering phenotypes observed in the fca-9 and fve-3 mutants under long-day conditions were largely masked by the loss of SVP function (Fig. 2A), demonstrating that svp is epistatic to the fca and fve mutants. In addition, the temperature insensitivity induced by the fca and fve mutations persisted even in the absence of SVP function, which suggests that SVP functions downstream from FCA and FVE within the thermosensory pathway and that SVP mediates temperature signaling. Consistent with this view, SVP expression was elevated in the fca and fve mutants (Fig. 2B), but not in other autonomous pathway mutants (Supplementary Fig. 3), and the flk and fpa mutants were temperature sensitive (Fig. 1A). SVP expression, however, was regulated neither by vernalization nor by CONSTANS (CO) (Samach et al. 2000), a central regulator of the long-day pathway. The observation that both the svp-32 mutants and wild-type plants responded similarly to gibberellin (GA) treatment or to differing light conditions (Supplementary Fig. 4) supports the premise that SVP functions primarily within the thermosensory pathway.
Figure 2.
Genetic interaction of SVP with FCA, FVE, and FLC. (A) Flowering time of the svp-32 fca-9, and svp-32 fve-3 double mutants at 23°C and 16°C under long-day conditions. The numbers listed below denote the ratios of flowering time (16°C/23°C). (B) Effects of fca and fve mutations on SVP expression in 10-d-old seedlings. (C) SVP expression in 10-d-old seedlings of loss- and gain-of-function alleles of FLC. The FRI locus originates from the Sf2 ecotype, and flc is flc-3 (Michaels and Amasino 1999). 35S∷FLC plants were used rather than an allele harboring functional FRI and FLC. (D) FLC expression in 10-d-old seedlings of the loss- and gain-of-function mutants of SVP. (E) Flowering time of mutants harboring various combinations of svp, fri, and flc mutations at 23°C and 16°C. SVP fri FLC and svp fri FLC indicate wild-type Columbia plants and svp-32 plants, respectively. The numbers listed below denote the ratios of flowering time (16°C/23°C).
The genetic interaction of svp mutants with flc mutants was assessed in an attempt to determine whether or not SVP interacts with FLC, since FLC is an important regulator that mediates vernalization effects in the autonomous pathway (Michaels and Amasino 1999), and both FLC and SVP function downstream from FCA and FVE (Fig. 2A,B). The results indicate that SVP acts independently of FLC at the transcriptional level within the thermosensory pathway: SVP expression was unchanged in the presence of functional alleles of FRI or FLC (Fig. 2C) and FLC expression remained unaffected by increases or reductions in SVP activity (Fig. 2D). Based on these results, we propose that SVP is very likely a thus-far unidentified repressor that mediates the temperature-dependent role of FCA and FVE (Blázquez et al. 2003). SVP appears to function, at least in part, downstream from FLC by modulating flowering time in response to ambient temperatures. The flowering of fri flc, FRI flc, and FRI FLC mutants was accelerated by the svp-32 mutation (Fig. 2E). Conversely, the temperature responsiveness exhibited by the fri flc and FRI flc mutants disappeared in the absence of SVP function, thereby suggesting that SVP exerts its effects principally within the thermosensory pathway. Interestingly, at flowering, FRI FLC plants had a similar number of leaves at 23°C and 16°C (64 vs. 67 leaves), which was also found in fca and fve mutants in which FLC levels were elevated even in the absence of functional FRI. A possible explanation of this flowering time phenotype of FRI FLC plants is that the floral repressive activity of FLC may be highly elevated in FRI FLC mutants and, consequently, further floral repression at 16°C may be masked. Consistent with this premise, temperature responses were restored in fve flc and fca flc double mutants to a level similar to that shown by flc single mutants (Blázquez et al. 2003; Balasubramanian et al. 2006). Of particular interest is that the severe late flowering of FRI FLC plants was largely suppressed by the svp-32 mutation, suggesting that FLC requires SVP to inhibit flowering. Considering that the MADS-box proteins are known to interact physically in a protein complex (Riechmann and Meyerowitz 1997), a possible scenario to explain this suppression by the svp-32 mutation is that SVP and FLC proteins may interact in a complex during temperature signaling. This proposal is supported by recent findings that FLC is a component of a multimeric protein complex in vivo and that SVP interacts with several MADS-box proteins (de Folter et al. 2005; Helliwell et al. 2006).
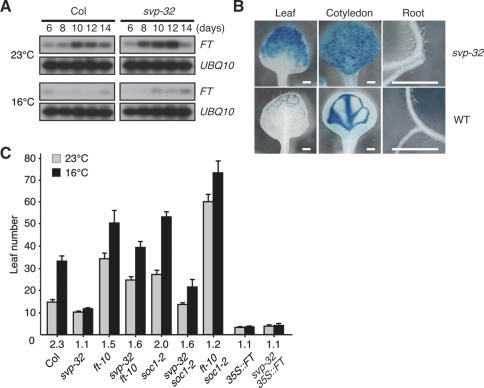
The conclusion that SVP functions as a floral repressor (Hartmann et al. 2000) raises an important question: On which flowering time gene does SVP exert its negative effects in the transduction of ambient temperature signaling? An analysis of the expression levels of known flowering time genes in the svp mutants revealed that the expression levels of FT (Kardailsky et al. 1999; Kobayashi et al. 1999), a floral integrator, were substantially elevated in the svp-32 mutants at both 23°C and 16°C (Fig. 3A). A similar up-regulation of FT in the svp-32 mutants was observed at a series of defined growth stages (Supplementary Fig. 5; Boyes et al. 2001). These observations indicated that the thermosensory signaling pathway functions, at least in part, via FT (Blázquez et al. 2003; Halliday et al. 2003). A reporter assay, carried out to confirm the negative regulation of FT expression effected by SVP, revealed profound ectopic pFT∷GUS expression in both the leaves and vascular root tissues of the svp-32 mutants (Fig. 3B). This suggests that SVP is required for the stable repression of FT in the ground tissues of the leaves of wild-type plants. Considering that FT is the major output of CO (Schmid et al. 2003; Wigge et al. 2005; Yoo et al. 2005) and that FT mRNA is an important component of the long-distance signaling mechanism that triggers flowering (Huang et al. 2005), the early flowering phenotypes observed in the svp-32 mutants can be explained as follows: The absence of SVP activity induces the accumulation of FT mRNA in the leaf transportable to the shoot apex, thereby triggering floral development. Consistent with a role of FT downstream from SVP, the loss of FT function partially suppressed the early flowering of the svp-32 mutants, the constitutive expression of FT masked the phenotype in the svp-32 mutants (Fig. 3C), and FT expression was significantly reduced in 35S∷SVP plants (Supplementary Fig. 2). Importantly, svp-32 ft-10 double mutants showed a weak temperature response, as did ft-10 mutants (flowering time ratio = 1.6 vs. 1.5, respectively), although svp-32 single mutants showed temperature insensitivity. Similar phenotypic masking by temperature-sensitive mutants has been observed in fca flc and fve flc mutants (Blázquez et al. 2003; Balasubramanian et al. 2006). One possible scenario explaining why svp-32 ft-10 mutants were more responsive than svp-32 single mutants is that svp-32 mutants display a temperature-insensitive phenotype as the result of increased FT activity, the floral-promoting effects of which are more profound at 16°C. When FT function is absent in the double mutants, the floral-promoting effect by FT at 16°C is not present and, therefore, temperature sensitivity may be restored to a level similar to that found in ft-10 single mutants. The observation that svp-32 ft-10 mutants were—albeit weakly—temperature sensitive indicates that the ambient temperature signaling mechanism of SVP requires FT and an additional downstream target(s). One possible target candidate is SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), since the soc1-2 mutation additively reduced the temperature sensitivity of ft-10 mutants (flowering time ratio of ft-10 soc1-2 double mutants = 1.2) (Fig. 3C), although soc1-2 single mutants responded to temperature changes. Consistent with this redundant role of FT and SOC1, neither the ft-10 nor soc1-2 single mutations completely suppressed the early flowering phenotypes of the svp-32 plants (Fig. 3C). Rather, the early flowering of the svp-32 mutants is masked, in large part, by the ft soc1 double mutation (T. Mizoguchi, pers. comm.). Nevertheless, we cannot exclude the possibility that TWIN SISTER OF FT (TSF) (Yamaguchi et al. 2005) and FD (Abe et al. 2005; Wigge et al. 2005) may be also the target of SVP.
Figure 3.
Role of SVP as an FT repressor. (A) Time-course expression of FT in wild-type (Col) and svp-32 plants at 23°C and 16°C. FT expression level was monitored in 6-, 8-, 10-, 12-, and 14-d-old seedlings. (B) pFT∷GUS expression patterns in 10-d-old seedlings of wild-type (Col) and svp-32 plants at 23°C. Bars, 500 μm. (C) Flowering time of svp-32 ft-10, svp-32 35S∷FT, and ft-10 soc1-2 double mutants at 23°C and 16°C. The numbers listed below denote the flowering time ratios (16°C/23°C).
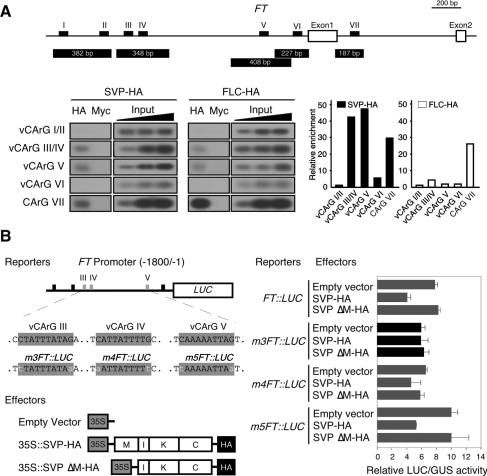
SVP is a member of the MADS-box proteins, which function as transcriptional regulators via their DNA-binding motifs (Riechmann and Meyerowitz 1997). As such, it appears likely that the negative regulation of FT expression by the SVP protein can be achieved via direct binding to the FT sequence. This hypothesis was bolstered by the findings that the 1.8-kb promoter region of FT harbors six variants of CArG motifs (vCArG) (Fig. 4A; Tang and Perry 2003), the consensus binding sequences of the MADS-box proteins, and that the first intron of FT harbors a CArG motif to which FLC proteins directly bind (Helliwell et al. 2006; Searle et al. 2006). Chromatin immunoprecipitation (ChIP) assays using Arabidopsis protoplasts were carried out to evaluate this hypothesis. Using chromatin immunoprecipitated with HA antibodies, we detected amplified products from fragments harboring vCArG III/IV, vCArG V, and CArG VII (Fig. 4A), indicating that SVP and FLC proteins bind to these motifs in vivo. The vCArG III/IV and vCArG V motifs were more efficiently precipitated by SVP-HA. The CArG VII motif, which is present in the first intron of FT, was strongly enriched by FLC-HA proteins, which is consistent with previous findings (Helliwell et al. 2006; Searle et al. 2006). This motif was also precipitated by SVP-HA proteins, but SVP’s binding affinity appeared to be weaker than that of FLC. It therefore appears likely that SVP preferentially binds to the vCArG motifs of the FT promoter and that FLC preferentially binds to the CArG VII of the first intron of FT. As the vCArG III/IV and V motifs were observed to bind efficiently, we verified the direct binding of the SVP proteins to these motifs in vivo by conducting a transient expression assay in protoplasts transfected with SVP-HA proteins and FT-promoter-driven luciferase (LUC) reporters (Fig. 4B). An abundance of SVP protein (35S∷SVP-HA) effected a reduction of FT∷LUC activity. This reduction disappeared when SVP protein was used without its MADS domain (35S∷SVP ΔM-HA), thereby indicating that the reduction in luciferase activity was induced by the binding of SVP to the FT promoter via the MADS domain. A subsequent assay aimed at assessing the ability of SVP-HA to repress the activity of an FT promoter harboring a mutation in the vCArG motif revealed that SVP-HA failed to reduce the expression of FT∷LUC harboring mutations in vCArG III (m3FT∷LUC). This result suggests that vCArG III is required for the SVP-mediated negative regulation of FT expression. Coupled with the mapping of the SVP-binding site in the FT promoter, our findings support our hypothesis that SVP binds directly to CArG motifs, thereby regulating FT expression to modulate flowering time in response to ambient temperature changes.
Figure 4.
Binding of SVP protein to the vCArG III in the FT promoter. (A) A ChIP assay using protoplasts transfected with SVP-HA and FLC-HA constructs. The location of six vCArG motifs (vCArG I to vCArG VI) identified in a 1.8-kb FT promoter and the different fragments analyzed by PCR are represented. A CArG motif to which FLC is known to bind within the first intron of FT (Helliwell et al. 2006; Searle et al. 2006) is designated as CArG VII. A fourfold dilution series of the input DNA was used as a semiquantitative standard. Relative enrichment indicates the amplified signal value normalized against that of input DNA. The value of enrichment in vCArG I/II was set to 1 for SVP-HA and FLC-HA. Similar results were obtained from five independent experiments. (Input) Total input chromatin DNA; (HA) DNA selected using HA antibodies; (Myc) DNA selected using Myc antibodies. (B) The effects of SVP-HA protein on the FT promoter activities. A schematic representation of the reporters and the effectors used in this assay is shown. vCArG motifs are shaded in gray and mutations introduced in vCArG motifs are indicated in lowercase. m3FT∷LUC indicates the FT∷LUC construct harboring a mutated vCArG III. Luciferase activities were normalized by GUS activities. This experiment was repeated five times, with similar results.
In conclusion, based on the results reported here, ambient temperature signaling in Arabidopsis is mediated by SVP, which functions within the thermosensory pathway, but only partially within the FT pathway (Blázquez et al. 2003; Wigge et al. 2005). SVP represses FT expression via direct binding to the vCArG III motif in the FT promoter. The SVP-mediated control of FT gene expression (Fig. 4) may represent a mechanism used by the plant to adjust the timing of flower development under fluctuating temperature conditions, although we cannot dismiss the possibility that altered interactions between MADS-box proteins, including SVP, FLC, and FLM (de Folter et al. 2005; Balasubramanian et al. 2006; Helliwell et al. 2006; Searle et al. 2006), effect the adjustment of flowering times at different ambient temperatures. We propose that the genetic evidence reported here is a valuable supplement to current knowledge on the manner in which plants integrate environmental signals to modulate development.
Materials and methods
Plant materials, growth conditions, and measurement of flowering time
All mutations used in this study were in the Columbia (Col) background, unless otherwise noted. svp-31 (SALK_026551) and svp-32 (SALK_ 072930), both T-DNA insertion lines of SVP, were obtained from the Arabidopsis Biological Resource Center (ABRC) (Alonso et al. 2003). To confirm the T-DNA insertion sites of these alleles, we sequenced the PCR products amplified using left border primers and gene-specific primers. SVP overexpressor plants obtained from H. Sommer (Max Planck Institut, Köln, Germany) have been described previously (Masiero et al. 2004). The mutant lines used in this study are described in Supplementary Table S1. The plants were grown in soil or MS medium at 23°C or 16°C under long-day (LD) conditions (16 h light/8 h dark) with light provided at an intensity of 120 μmol m−2 sec−1. The homozygosity of the double mutants was verified via PCR genotyping. The details of the genotyping procedures are available on request. The flowering times of the plants are expressed as the total number of primary leaves of at least 12 plants.
Expression analysis
Expression levels of the flowering time genes were determined via semiquantitative reverse transcriptase-meditated PCR or real-time PCR. Total RNA was extracted using Trizol reagent (Invitrogen), and 1 μg of total RNA was used to synthesize the complementary DNA. The primer sequences and amplification conditions are available on request. The real-time PCR analysis was performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems), and expression levels were normalized against that of tubulin. For the histochemical GUS analysis, we generated a SVP∷GUS translational fusion construct. The 4.9-kb SVP genomic region was amplified using JH2929 (5′-GTGGTCGACACTTT TTATTTTACTCTGG-3′) and JH2985 (5′-GGATCCGCACCACCATA CGGTAAGCTGC-3′), and then fused with the GUS reporter gene. FT∷GUS plants (Takada and Goto 2003) were obtained from K. Goto (Research Institute for Biological Sciences, Okayama, Japan). SVP cDNA-GFP chimeric constructs were used as a reporter to examine the localization pattern of SVP. To generate the 35S promoter-driven SVP cDNA-GFP construct, the GFP sequence was in-frame-fused to the C-terminal region of a 35S∷SVP chimeric plasmid. A particle bombardment system (PDS-1000/He; Bio-Rad) was utilized for the delivery of DNA-coated tungsten particles into onion epidermal cells. After 24 h of incubation at 23°C or 16°C, the subcellular localization pattern was observed under a fluorescence microscope (Carl Zeiss).
ChIP assay
ChIP assays were conducted as described (Tang and Perry 2003) with minor modifications. The Arabidopsis protoplasts were transfected with either SVP cDNA fused to HA tags or FLC cDNA fused to HA tags and then incubated for 24 h at room temperature. The expressions of the SVP-HA and FLC-HA proteins were determined by protein blots using extracts from the protoplasts. After formaldehyde fixation, the chromatin of the protoplasts was isolated and sheared via sonication. Mouse anti-HA antibodies (Santa Cruz Biotechnology) or anti-Myc 9B11 antibodies (Cell Signaling Technology) were used to immunoprecipitate the genomic fragments. Five sequence fragments spanning six vCArG motifs within the promoter and a CArG motif in the first intron of FT were amplified from the immunoprecipitated genomic DNA. PCR products were visualized after 35 cycles using DNA purified from chromatin immunoprecipitated with antibodies against HA or Myc. Nonselected input DNA and Myc antibody-selected DNA were used as PCR templates for the positive and negative controls, respectively. Quantitation of the enrichment of CArG motifs by the SVP-HA and FLC-HA proteins were performed on PhosphorImager plates (Fujifilm BAS 2500; Fuji). The primers used for the ChIP assays are described in Supplementary Table S2. Detailed descriptions of the protocols of these experiments are available on request.
Luciferase reporter assay
To generate the FT∷LUC construct, we amplified 1.8 kb of the FT promoter fragment using JH3096 (5′-TGAACACTAACATGATTGAATGA CA-3′) and JH2865 (5′-GATCTTGAACAAACAGGTGGT-3′) and fused this to luciferase. The luciferase reporter constructs harboring the mutated vCArG motifs within the FT promoter were used as reporters to examine the effects of the vCArG motifs on the specific binding of SVP to the FT promoter. Site-directed mutagenesis was utilized to generate the FT∷LUC constructs harboring the mutated vCArG motifs, using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene), in accordance with the manufacturer’s instructions. The primers used in this mutagenesis protocol are shown in Supplementary Table S3. Mutations introduced into the vCArG motifs in these constructs were verified via sequencing. SVP—with or without its MADS domain (35S∷SVP-HA and 35S∷SVP ΔM-HA, respectively)—was used as an effector. A reporter and an effector were cotransfected into the protoplasts. The 35S∷GUS construct was used as an internal control. Luciferase activities were normalized by GUS activities. Detailed descriptions of the protocols are available upon request.
Acknowledgments
We thank J. Alonso, R. Amasino, S. Michaels, T. Araki, C. Dean, J. Ecker, M. Koornneef, H. Lee, I. Lee, J.M. Martinez-Zapater, K.H. Paek, C.M. Park, P. Huijser, S.K. Yoo, S.Y. Yoo, D. Weigel, the Arabidopsis stock center, the Pioneer Hi-Bred International Inc., and the Plant Bioscience Limited for material, and G. Choi for the measurement of hypocotyl lengths. Financial support was provided by the Crop Functional Genomics Center of the 21C Frontier Program (J.S.L.), and by BioGreen 21 Program and the Plant Signaling Network Research Center of Korea University (J.H.A.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1518407
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T., Ichinoki H., Notaguchi M., Goto K., Araki T., Notaguchi M., Goto K., Araki T., Goto K., Araki T., Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Zimmerman J., Barajas P., Cheuk R., Barajas P., Cheuk R., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Atkin O.K., Tjoelker M.G., Tjoelker M.G. Thermal acclimation and the dynamic response of plant respiration to temperature. Trends Plant Sci. 2003;8:343–351. doi: 10.1016/S1360-1385(03)00136-5. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D., Sureshkumar S., Lempe J., Weigel D., Lempe J., Weigel D., Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Ahn J.H., Weigel D., Ahn J.H., Weigel D., Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Gorlach J., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Gorlach J., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Gorlach J., McCaskill A.J., Hoffman N.E., Davis K.R., Gorlach J., Hoffman N.E., Davis K.R., Gorlach J., Davis K.R., Gorlach J., Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland G., Prat Monguio S., Prat Monguio S. Cell signalling and gene regulation signalling mechanisms in plants: Examples from the present and the future. Curr. Opin. Plant Biol. 2005;8:457–461. doi: 10.1016/j.pbi.2005.07.016. [DOI] [PubMed] [Google Scholar]
- de Folter S., Immink R.G., Kieffer M., Parenicova L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Immink R.G., Kieffer M., Parenicova L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Kieffer M., Parenicova L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Parenicova L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Busscher M., Kooiker M., Colombo L., Kater M.M., Kooiker M., Colombo L., Kater M.M., Colombo L., Kater M.M., Kater M.M., et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter A.H., Fitter R.S., Fitter R.S. Rapid changes in flowering time in British plants. Science. 2002;296:1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- Gendall A.R., Levy Y.Y., Wilson A., Dean C., Levy Y.Y., Wilson A., Dean C., Wilson A., Dean C., Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Guy C.L. Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1990;41:187–223. [Google Scholar]
- Halliday K.J., Salter M.G., Thingnaes E., Whitelam G.C., Salter M.G., Thingnaes E., Whitelam G.C., Thingnaes E., Whitelam G.C., Whitelam G.C. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 2003;33:875–885. doi: 10.1046/j.1365-313x.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- Hartmann U., Hohmann S., Nettesheim K., Wisman E., Saedler H., Huijser P., Hohmann S., Nettesheim K., Wisman E., Saedler H., Huijser P., Nettesheim K., Wisman E., Saedler H., Huijser P., Wisman E., Saedler H., Huijser P., Saedler H., Huijser P., Huijser P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S., Wood C.C., Robertson M., James Peacock W., Dennis E.S., Robertson M., James Peacock W., Dennis E.S., James Peacock W., Dennis E.S., Dennis E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- Huang T., Bohlenius H., Eriksson S., Parcy F., Nilsson O., Bohlenius H., Eriksson S., Parcy F., Nilsson O., Eriksson S., Parcy F., Nilsson O., Parcy F., Nilsson O., Nilsson O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science. 2005;309:1694–1696. doi: 10.1126/science.1117768. [DOI] [PubMed] [Google Scholar]
- Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P., Renzoni A., Chiaruttini C., Springer M., Cossart P., Chiaruttini C., Springer M., Cossart P., Springer M., Cossart P., Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D., Nguyen J.T., Chory J., Harrison M.J., Weigel D., Chory J., Harrison M.J., Weigel D., Harrison M.J., Weigel D., Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T., Kaya H., Goto K., Iwabuchi M., Araki T., Goto K., Iwabuchi M., Araki T., Iwabuchi M., Araki T., Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Levy Y.Y., Mesnage S., Mylne J.S., Gendall A.R., Dean C., Mesnage S., Mylne J.S., Gendall A.R., Dean C., Mylne J.S., Gendall A.R., Dean C., Gendall A.R., Dean C., Dean C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science. 2002;297:243–246. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- Masiero S., Li M.A., Will I., Hartmann U., Saedler H., Huijser P., Schwarz-Sommer Z., Sommer H., Li M.A., Will I., Hartmann U., Saedler H., Huijser P., Schwarz-Sommer Z., Sommer H., Will I., Hartmann U., Saedler H., Huijser P., Schwarz-Sommer Z., Sommer H., Hartmann U., Saedler H., Huijser P., Schwarz-Sommer Z., Sommer H., Saedler H., Huijser P., Schwarz-Sommer Z., Sommer H., Huijser P., Schwarz-Sommer Z., Sommer H., Schwarz-Sommer Z., Sommer H., Sommer H. INCOMPOSITA: A MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development. 2004;131:5981–5990. doi: 10.1242/dev.01517. [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M., Amasino R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M., Meyerowitz E.M. MADS domain proteins in plant development. Biol. Chem. 1997;378:1079–1101. [PubMed] [Google Scholar]
- Samach A., Wigge P.A., Wigge P.A. Ambient temperature perception in plants. Curr. Opin. Plant Biol. 2005;8:483–486. doi: 10.1016/j.pbi.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G., Schwarz-Sommer Z., Yanofsky M.F., Coupland G., Yanofsky M.F., Coupland G., Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U., Demar M., Bressan R., Weigel D., Lohmann J.U., Bressan R., Weigel D., Lohmann J.U., Weigel D., Lohmann J.U., Lohmann J.U. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Krober S., Amasino R.A., Coupland G., He Y., Turck F., Vincent C., Fornara F., Krober S., Amasino R.A., Coupland G., Turck F., Vincent C., Fornara F., Krober S., Amasino R.A., Coupland G., Vincent C., Fornara F., Krober S., Amasino R.A., Coupland G., Fornara F., Krober S., Amasino R.A., Coupland G., Krober S., Amasino R.A., Coupland G., Amasino R.A., Coupland G., Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Sharma N., Deswal R., Sharma N., Deswal R., Deswal R. The molecular biology of the low-temperature response in plants. Bioessays. 2005;27:1048–1059. doi: 10.1002/bies.20307. [DOI] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S., Edwards J.A., Peacock W.J., Dennis E.S., Peacock W.J., Dennis E.S., Dennis E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R.M., Amasino R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Sung S., Amasino R.M., Amasino R.M. Remembering winter: Toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 2005;56:491–508. doi: 10.1146/annurev.arplant.56.032604.144307. [DOI] [PubMed] [Google Scholar]
- Takada S., Goto K., Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Perry S.E., Perry S.E. Binding site selection for the plant MADS domain protein AGL15: An in vitro and in vivo study. J. Biol. Chem. 2003;278:28154–28159. doi: 10.1074/jbc.M212976200. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D., Busch W., Schmid M., Lohmann J.U., Weigel D., Schmid M., Lohmann J.U., Weigel D., Lohmann J.U., Weigel D., Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T., Kobayashi Y., Goto K., Abe M., Araki T., Goto K., Abe M., Araki T., Abe M., Araki T., Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H., Yoo S.Y., Lee J.S., Ahn J.H., Lee J.S., Ahn J.H., Ahn J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Xu Y., Tan E.L., Kumar P.P., Xu Y., Tan E.L., Kumar P.P., Tan E.L., Kumar P.P., Kumar P.P. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. 2002;99:16336–16341. doi: 10.1073/pnas.212624599. [DOI] [PMC free article] [PubMed] [Google Scholar]