Abstract
Fission yeast shugoshin Sgo1 is meiosis specific and cooperates with protein phosphatase 2A to protect centromeric cohesin at meiosis I. The other shugoshin-like protein Sgo2, which requires the heterochromatin protein Swi6/HP1 for full viability, plays a crucial role for proper chromosome segregation at both mitosis and meiosis; however, the underlying mechanisms are totally elusive. We here demonstrate that, unlike Sgo1, Sgo2 is dispensable for centromeric protection of cohesin. Instead, Sgo2 interacts with Bir1/Survivin and promotes Aurora kinase complex localization to the pericentromeric region, to correct erroneous attachment of kinetochores and thereby enable tension-generating attachment. Forced localization of Bir1 to centromeres partly restored the defects of sgo2Δ. This newly identified interaction of shugoshin with Survivin is conserved between mitosis and meiosis and presumably across eukaryotes. We propose that ensuring bipolar attachment of kinetochores is the primary role of shugoshin and the role of cohesion protection might have codeveloped to facilitate this process.
Keywords: Shugoshin, spindle checkpoint, chromosome segregation, Aurora
For proper partition of chromosomes to daughter cells, sister chromatids must be held together before mitotic chromosome segregation. Sister chromatid cohesion is established during DNA replication depending on the cohesin complex, comprising two SMC (structural maintenance of chromosome) family proteins, a kleisin subunit Scc1/Rad21 and an accessory subunit Scc3. Cohesion is maintained until metaphase when sister kinetochores attach to microtubules emanating from the opposite spindle poles (bipolar attachment). At the onset of anaphase, the anaphase-promoting complex (APC)-dependent degradation of securin Pds1/Cut2 allows the activation of a specific endopeptidase, separase Esp1/Cut1, which in turn cleaves Scc1/Rad21. Thereby, the cohesin complex is dissociated to release sister-chromatid cohesion, resulting in the separation of sister chromatids (Uhlmann 2003; Hirano 2005; Nasmyth and Haering 2005). In animal mitotic cells, however, most cohesin dissociates from chromosome arms during prophase (called “prophase pathway”), depending on the phosphorylation of cohesin by mitotic kinases such as Polo-like kinase (Losada et al. 2002; Hauf et al. 2005). However, centromeric cohesin is retained until metaphase by the function of shugoshin (Sgo/MEI-S332) (Lee et al. 2005; Watanabe 2005). This shugoshin-dependent centromeric protection is essential for mitotic chromosome segregation in mammalian cells but not in fly or yeast. In most organisms, shugoshin is required for meiotic chromosome segregation, where stepwise dissociation of cohesin is more essential (Miyazaki and Orr-Weaver 1994; Watanabe 2005). Recent studies indicated that, as a part of the protection machinery, shugoshin recruits protein phosphatase 2A to centromeres to prevent the phosphorylation of cohesin and thereby its dissociation (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006). Although shugoshin was well characterized as a protector of centromeric cohesin (Kitajima et al. 2004; Marston et al. 2004; Rabitsch et al. 2004), a study in budding yeast suggested that shugoshin was also involved in activating the spindle checkpoint that senses the lack of tension, but not of attachment, on mitotic chromosomes (Indjeian et al. 2005). However, the underlying mechanism of this potential link between shugoshin and the checkpoint remains elusive.
For proper chromosome segregation in mitosis, the sister kinetochores must attach spindle microtubules emanating from opposite spindle poles, which generates tension across centromeres. When sister kinetochores are misattached to microtubules during mitosis, Aurora kinase plays an essential role at centromeres in destabilizing erroneous attachment and thereby promoting bipolar attachment (Tanaka 2002; Hauf and Watanabe 2004; Pinsky and Biggins 2005; Cimini et al. 2006). A recent study suggests that the Aurora kinase complex itself acts as a sensor of tension (Sandall et al. 2006). The spindle checkpoint, another regulatory mechanism to achieve chromosome alignment, senses unattached kinetochores or a lack of tension and inhibits the premature entry to anaphase. Aurora is again required for activating the checkpoint in the absence of tension, at least in budding yeast and animal cells. One plausible model is that the destabilization of the tensionless kinetochore microtubule attachment executed by Aurora, results in the generation of unattached kinetochores, which consequently activates the spindle checkpoint (Pinsky et al. 2006).
Fission yeast possess two shugoshin-like proteins, Sgo1 and Sgo2, both of which localize at the pericentromeric heterochromatin region, the site enriched with cohesin and crucial for centromeric cohesion. Sgo1 is meiosis I-specific and plays a crucial role in protecting centromeric cohesin during meiosis I, whereas Sgo2 is ubiquitously expressed and required for proper chromosome segregation in both mitosis and meiosis. Unlike Sgo1, Sgo2 is dispensable for protecting cohesin at meiosis I but instead required for proper disjunction of homologous chromosomes (homologs) as well as monopolar attachment of unified sister kinetochores. Sgo2 also plays an important role in mitotic chromosome segregation (Kitajima et al. 2004; Rabitsch et al. 2004). Aurora B kinase forms a complex with INCENP and Survivin (and further with Borealin/Dasra in metazoans) and changes its localization from chromosomes or centromeres to the spindle midzone in mitosis (Morishita et al. 2001; Petersen and Hagan 2003; Vagnarelli and Earnshaw 2004). We now found that Sgo2 is required for the localization of the Aurora kinase complex at centromeres, in a redundant capacity with heterochromatin protein Swi6/HP1. Forced localization of Bir1/Survivin to centromeres in sgo2Δ swi6Δ cells restored growth. Sgo2 closely associates with Bir1, a component essential for the centromeric localization of the Aurora kinase complex. The overall phenotypes of sgo2Δ, not only during mitosis but also during meiosis, are explainable by the reduced activity of Aurora. Our results imply that, in addition to the protective function, shugoshin has a crucial role in promoting centromeric localization of the Aurora kinase complex and thereby enabling tension-generating attachment of kinetochores.
Results
Shugoshin has no role in protecting centromeric cohesion in fission yeast mitosis
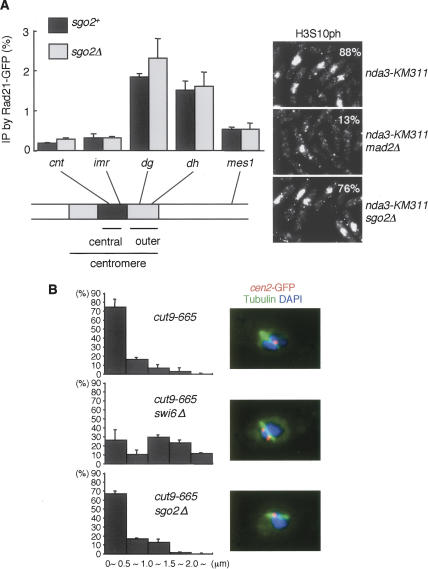
Fission yeast shugoshin Sgo1, which is expressed only in meiosis, plays an essential role in protecting meiotic cohesin containing Rec8 at centromeres, whereas ubiquitously expressed Sgo2 is dispensable for this protection (Kitajima et al. 2004; Rabitsch et al. 2004). Since Sgo2 is important for mitotic chromosome segregation as well, we explored whether Sgo2 is required for preserving mitotic cohesin containing Rad21 or centromeric cohesion in mitosis as is observed in animal SGO-defective cells. To synchronize the fission yeast cell cycle at prometaphase, we abolished the spindle formation by a cold-sensitive mutation of β-tubulin, nda3-KM311. Chromatin immunoprecipitation (ChIP) of the cohesin subunit Rad21 indicated that cohesins are enriched at the pericentromeric region in control (sgo2+) prometaphase cells (Fig. 1A; Tomonaga et al. 2000; Watanabe et al. 2001). Notably, sgo2Δ cells, which also arrested at prometaphase by the nda3-KM311 mutation, exhibited a virtually identical pattern and amount of cohesin localization around the centromere (Fig. 1A).
Figure 1.
Sgo2 does not protect centromeric cohesion in fission yeast mitosis. (A) nda3-KM311 rad21+-GFP cells carrying sgo2Δ or not were arrested at prometaphase and fixed for ChIP analysis. Rad21-GFP levels throughout the indicated chromosome sites were measured by ChIP analysis with anti-GFP. The quality of the prometaphase arrest was determined by staining for histone H3-Ser-10 phosphorylation (H3S10ph). Note that sgo2Δ cells as well as sgo2+ cells largely arrested at prometaphase by nda3-KM311 inactivation, whereas checkpoint-defective mad2Δ cells did not (also see Supplementary Fig. 1). Error bars represent the SD of duplicate PCRs. (B) The indicated strains cultured at 25°C were shifted to 36°C for 4 h (metaphase arrest), fixed, and stained for tubulin and cen2-GFP. The distance between cen2-GFP dots was measured. Error bars represent the SD of duplicate samples (each n > 50).
To explore centromeric cohesion directly, we arrested the cell cycle at metaphase by inactivating APC with the cut9-665 mutation and measured the distance of GFP-marked centromere (cen2-GFP) dots along the mitotic spindle (Fig. 1B), expecting that the pulling force of the spindle would challenge the centromeric cohesion, thereby exposing potential cohesion defects. We indeed detected an increased distance of cen2-GFP in swi6Δ cells, in which pericentromeric heterochromatin is poorly formed and thereby cohesin is not enriched at the pericentromeric region (Bernard et al. 2001; Nonaka et al. 2002). In contrast, sgo2Δ cells exhibited no increase in the separation of cen2-GFP dots (Fig. 1B). Taken together, these results indicate that Sgo2 plays little or no role in protecting cohesin. Since Sgo2 is the sole shugoshin expressed during mitosis, these results mean that, differently from animal cells, the role of shugoshin during mitosis is not the protection of centromeric cohesin in fission yeast.
Sgo2 is required for preventing cosegregation of sister chromatids and maintaining the spindle checkpoint in the absence of tension
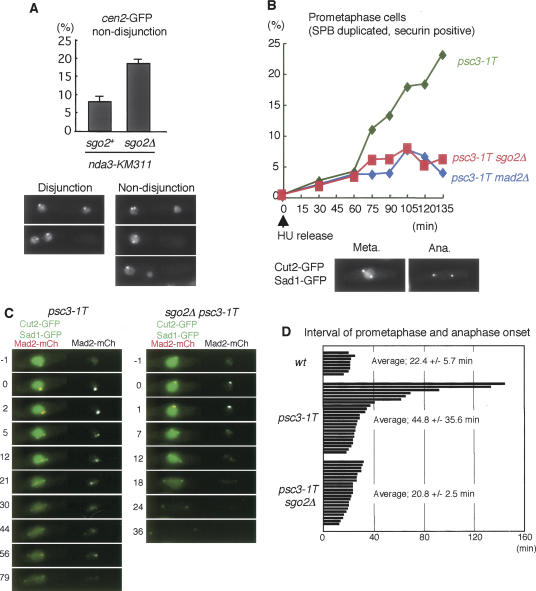
Since Sgo2 localizes at centromeres and sgo2Δ cells show hypersensitivity to the microtubule-destabilizing drug thiabendazole (TBZ) (Kitajima et al. 2004), we thought that sgo2Δ cells might have a defect in kinetochore function or the spindle checkpoint. In a normal cell cycle of fission yeast, kinetochores are clustered near the spindle pole body (SPB) and rapidly captured by microtubules emanating from the duplicated SPBs at prometaphase. When spindle microtubules are depolymerized by nda3-KM311 and reformed by shift to permissive temperature, the attachment of kinetochores to microtubules is more error-prone than under physiological conditions (Trautmann et al. 2004). We examined whether Sgo2 is important to correct erroneous attachment in this situation. We arrested the cells by nda3-KM311 at prometaphase and released them to observe cen2-GFP in anaphase cells. In contrast to sgo2+ cells, sgo2Δ cells showed increased cosegregation of sister chromatids (Fig. 2A). This indicates that Sgo2 plays an important role in establishing bipolar attachment of kinetochores possibly through promoting the correction of erroneous attachment.
Figure 2.
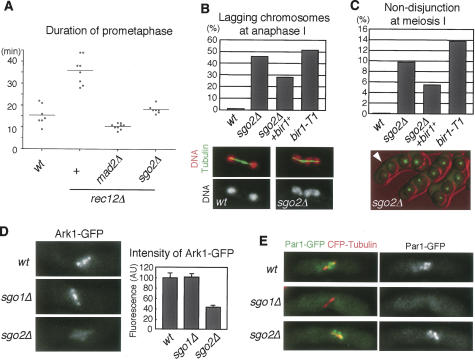
Sgo2 is required for preventing cosegregation of sister chromatids and activating the spindle checkpoint in the absence of tension. (A) Quantification of cen2-GFP nondisjunction in sgo2+ nda3-KM311 and sgo2Δ nda3-KM311 cells after block at 20°C and release to 36°C. Error bars represent the SD of duplicate samples (each n > 100). Note that some cells segregate the DNA within one cell compartment because of the displacement of the nucleus relative to the septum. (B) The indicated strains were arrested at G1/S phase by adding HU at 25°C and released to 36°C. Prometaphase (SPB duplicated and securin/Cut2-positive) cells were counted at each time point (n > 150). Examples of Cut2-GFP and Sad1-GFP images at metaphase and anaphase are shown. Note that Cut2-GFP, but not Sad1-GFP at SPBs, disappears from the anaphase cell. (C) Live imaging of the indicated strains expressing Mad2-mCherry, Cut2-GFP, and Sad1-GFP. The numbers indicate elapsed time (in minutes). (D) Prometaphase length of single cells was measured for each strain. One bar corresponds to a single cell.
We next examined whether Sgo2 also plays a role in the spindle checkpoint. sgo2Δ cells could arrest at prometaphase by abolishing spindle formation (Fig. 1A; Supplementary Fig. 1), implying that Sgo2 is dispensable for activating the spindle checkpoint in the absence of attachment. We asked whether Sgo2 is required for the checkpoint sensing the absence of tension. To generate a tension-less situation in mitosis, we inactivated sister-chromatid cohesion by using a temperature-sensitive mutation of a cohesin subunit, psc3-1T (Biggins and Murray 2001; Nonaka et al. 2002). Cells were arrested at G1/S phase by adding Hydroxyurea (HU) at a permissive temperature and then released to a restrictive temperature. At the following metaphase, the psc3-1T cells showed a premature separation of sister centromeres (data not shown) and a delay, as cells exhibiting duplicated SPBs and securin signals accumulated (Fig. 2B). Remarkably, this delay was abolished by the deletion of sgo2+ as well as the deletion of the checkpoint component mad2+. We further performed live image analysis of single cells. The duration of prometaphase was determined by measuring the time from the separation of SPBs to the disappearance of securin Cut2-GFP. We found that this time is much extended in 40% of psc3-1T cells, and this mitotic delay is abolished by deletion of sgo2+ (Fig. 2C,D). Moreover, we often detected Mad2 signals at kinetochores during the extended prometaphase of psc3-1T cells (Fig. 2C), suggesting that Mad2 localized at kinetochores of tension- or attachment-less chromosomes. Remarkably, all sgo2Δ psc3-1T cells generated initial signals of Mad2 as in sgo2+ psc3-1T cells (Fig. 2C,D). We further confirmed that the localization of Mad2 or Bub1 in cells arrested at prometaphase by nda3-KM311 was intact in sgo2Δ cells (data not shown). These results suggest that Sgo2 is dispensable for the initial activation of the checkpoint, which occurs at early prometaphase presumably prior to attachment, but instead is required for maintaining the checkpoint in the absence of tension.
Sgo2 facilitates Aurora kinase complex localization to centromeres
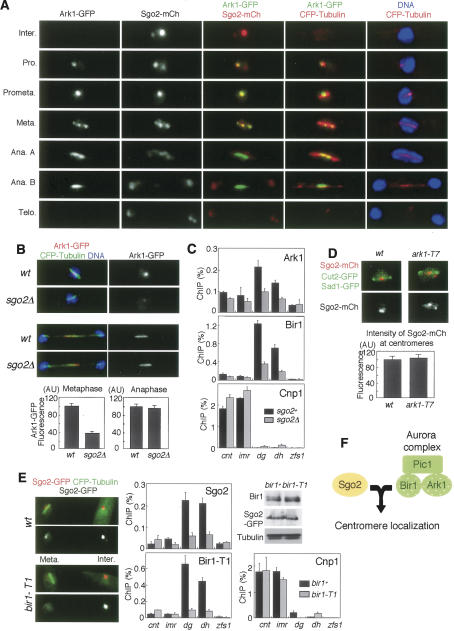
It is proposed that Aurora B kinase plays a crucial role in correcting erroneous microtubule-kinetochore attachment and activating the spindle checkpoint when tension is not properly generated at centromeres (Tanaka et al. 2002; Hauf et al. 2003; Pinsky and Biggins 2005). We demonstrate that the Aurora functions of establishing bipolar attachment and activating the spindle checkpoint are well conserved in fission yeast, and are similar to the Sgo2 functions (Supplementary Fig. 2). Therefore, we explored the possibility that Sgo2 is functionally related to Aurora in vivo. At first, we examined the localization of Sgo2 and Ark1 in combination with tubulin throughout the cell cycle (Fig. 3A). In interphase, Sgo2 is mostly localized as one to three dots within the nucleus, which all colocalize with a part of the telomere clusters but not with centromeres (Supplementary Fig. 3A). In early prometaphase, Sgo2 forms a dot near the SPB, and the signals sometimes split along the spindle at metaphase. These mitotic signals are closely associated with the centromere protein Cnp3 (Supplementary Fig. 3B). Thus, Sgo2 dramatically changes its cellular localization from telomere-associated regions to centromeres at entry to M phase. Remarkably, these Sgo2 dots at prometaphase and metaphase almost completely colocalized with the signals of Ark1 (Fig. 3A; Petersen et al. 2001). By ChIP, Aurora kinase complex, like Sgo2, localizes at the pericentromeric regions at prometaphase (Fig. 3C,E; Morishita et al. 2001). At the onset of anaphase when Ark1 relocates to the spindle midzone, Sgo2 mostly disperses from centromeres within the nucleus (Fig. 3A, anaphase A). A very small amount of Sgo2 colocalizes with Ark1 at the spindle midzone (Fig. 3A, anaphase B). These results suggest a close interaction of Sgo2 with the Aurora kinase complex especially at centromeres throughout prophase until metaphase.
Figure 3.
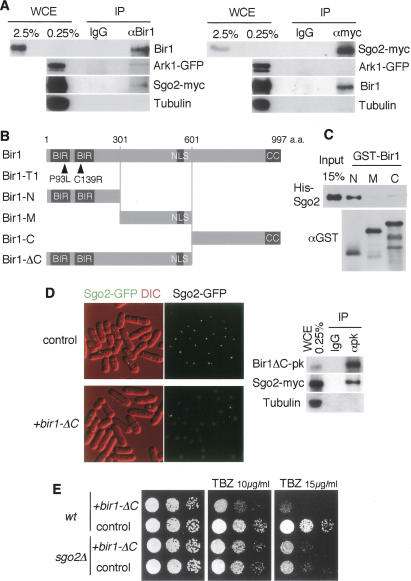
Sgo2 facilitates Aurora kinase complex localization to centromeres. (A) sgo2+-mCherry ark1+-GFP cells expressing CFP-atb2+ (α2-tubulin) were fixed and stained with DAPI (4,6-diamidino-2-phenylindole). Representative stainings of cells at interphase, prophase, prometaphase, metaphase, anaphase A, anaphase B, and telophase are shown. (B) Ark1-GFP was detected at metaphase and anaphase in wild-type and sgo2Δ cells expressing CFP-atb2+. DNA was stained with Hoechst 33342. Signal intensity of Ark1-GFP was measured. Error bars represent the SEM (n > 12). (C) ChIP analysis of Ark1-GFP, Bir1, and Cnp1 (CENP-A protein as a control) in sgo2Δ cells. Wild-type or sgo2Δ cells carrying the nda3-KM311 mutation were arrested at prometaphase by incubating for 10 h at 20°C and then analyzed by ChIP with anti-GFP, anti-Bir1, and anti-Cnp1 antibodies. Note that the localization of Bir1 and Ark1 at pericentromeric regions was largely reduced in sgo2Δ cells. Error bars represent the SD (n = 2). (D) Wild-type and ark1-T7 cells were arrested by HU for 3 h at 25°C and 1 h at 36°C, then released for 1 h at 36°C and detected for Sgo2-mCherry, Cut2-GFP (spindle and nuclear signals), and Sad1-GFP (SPB signals). DNA was stained by DAPI. The signal intensity of Sgo2-mCherry was measured. Error bars represent the SEM (n = 15). (E) Sgo2-GFP was observed in wild-type and bir1-T1 cells cultured at a permissive temperature, 25°C. Note that metaphase Sgo2-GFP signals are specifically reduced in bir1-T1 cells. A ChIP assay of Sgo2-GFP, Bir1, and Cnp1 was performed using bir1+ or bir1-T1 cells arrested at prometaphase at a permissive temperature (20°C). Error bars represent the SD (n > 2). The expression of Bir1 or Bir1-T1 and Sgo2-GFP in this condition was examined by Western blotting using anti-Bir1 and anti-GFP antibodies. (F) A schematic model illustrating how Sgo2 and Bir1 mutually require each other for their centromeric localization, on which centromeric localization of the Aurora kinase complex depends.
We next examined whether sgo2Δ influences the centromeric localization of the Aurora kinase complex. Remarkably, the localization of Ark1 was significantly reduced at centromeres but not at the spindle midzone in sgo2Δ cells (Fig. 3B). By ChIP assay, the enrichment of Ark1 and Bir1 at pericentromeric regions was largely reduced by sgo2Δ to 23% and 21% of the wild-type level, respectively (Fig. 3C; these reduction ratios were calculated as described in Materials and Methods). Conversely, we examined the localization of Sgo2 in cells carrying mutations in either ark1+, pic1+ (INCENP), or bir1+. At least at a permissive temperature (25°C), four mutants, ark1-T7, ark1-T8, bir1(cut17)-275, pic1-T269, preserved intact Sgo2 localization (Supplementary Fig. 4; data not shown); however, bir1-T1 largely lost centromeric Sgo2 signals at metaphase, but signals were comparable to bir1+ cells in anaphase and interphase (Fig. 3E; Supplementary Fig. 4). A ChIP assay confirmed the reduction of centromeric localization of Sgo2 as well as Bir1-T1 protein itself, albeit Bir1-T1 protein was expressed at normal levels (Fig. 3E). This is remarkable because bir1-T1 cells exhibit normal and better growth than bir1-275 cells at the permissive temperature (see Fig. 4B; data not shown). These results suggest a specific impairment of Bir1-T1 protein in localizing Sgo2 at centromeres. In contrast, Ark1 may be dispensable for Sgo2 localization, since either ark1-T7 or ark1-T8 cells showed little defect in Sgo2 localization even at a restrictive temperature (Fig. 3D; Supplementary Fig. 4A). Together with other results (Supplementary Fig. 4D; Morishita et al. 2001; Leverson et al. 2002; Vagnarelli and Earnshaw 2004), these results suggest a model that Sgo2 and Bir1 mutually require each other for their centromeric localization and Ark1 is loaded depending on Bir1 and Pic1 (Fig. 3F).
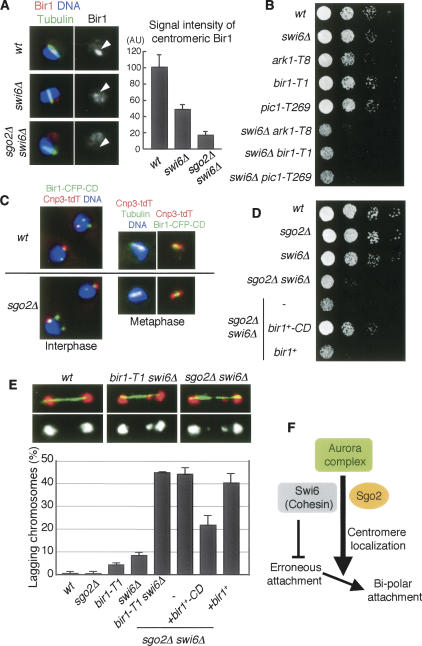
Figure 4.
Forced localization of Bir1 to pericentromeric regions restores sgo2Δ defects. (A) Cells of the indicated strains were stained for Bir1, tubulin, and DNA. Representative images of the indicated cells with short spindles (metaphase) are shown. The signal intensity of Bir1 at metaphase was measured. The error bars represent the SEM (n > 13). (B) Serial dilutions of the indicated strains were spotted onto a YEA plate and incubated at 25°C. (C) Cnp3-tdTomato and Bir1-CFP-CD were observed in wild-type and sgo2Δ cells. (D) Serial dilution of the indicated strains were spotted onto a YEA plate and incubated at 30°C. (E) The indicated strains were cultured at 25°C, fixed, and stained for tubulin and DNA. Examples are shown on top; (red) DNA, (green) tubulin. Frequencies of lagging chromosomes in anaphase cells (n > 100) were examined. The error bars represent the SD of two or three different experiments. (F) A schematic model illustrating how Sgo2 and Swi6 promote bipolar kinetochore–microtubule attachment. A centromeric cohesion defect caused by swi6Δ or cohesin mutation but not by sgo2Δ would provoke erroneous kinetochore–microtubule attachment, which can be restored depending on centromeric Aurora. Since Sgo2 promotes the Aurora localization at centromeres in redundant capacity with Swi6, sgo2Δ swi6Δ double but not either single mutant accumulates erroneous attachment, which is seen as lagging chromosomes.
Since cohesin mutations reportedly impair the centromeric localization of the Aurora kinase complex in fission yeast and animal cells (Morishita et al. 2001; Sonoda et al. 2001) and centromeric cohesion is intact in sgo2Δ cells (Fig. 1), the residual localization of the Aurora kinase complex in sgo2Δ cells could depend on this cohesion pathway. Supporting this scenario, we found that in sgo2Δ psc3-1T double mutants, the centromeric localization of Bir1 was abolished even more than in either sgo2Δ or psc3-1T single mutant at the restrictive temperature (Supplementary Fig. 5A,B).
Forced localization of Bir1 to pericentromeric regions restores colethality of sgo2Δ with swi6Δ
The fission yeast heterochromatin protein Swi6/HP1 plays an important role in strengthening centromeric cohesion by recruiting cohesin at centromeres (Bernard et al. 2001; Nonaka et al. 2002). Accordingly, we found that Bir1 (and Ark1) localization at centromeres is reduced in swi6Δ cells (Fig. 4A; Supplementary Fig. 5C), suggesting that Swi6 also contributes to the full localization of Aurora at centromeres, at least partly through the cohesin pathway. We previously reported that the deletion of sgo2+ in swi6Δ cells causes a marked growth defect (Kitajima et al. 2004; see also Fig. 4D), and this is also the case if mutations in the Aurora kinase complex are combined with swi6Δ (Fig. 4B), in either case conspicuously provoking lagging chromosomes at anaphase (Fig. 4E). Consistent with these observations, centromeric Bir1 localization was even more abolished in sgo2Δ swi6Δ cells than in swi6Δ cells (Fig. 4A). If Sgo2 was solely important for centromeric Bir1 recruitment, the lethality of sgo2Δ swi6Δ cells should be restored by ectopically localizing Bir1 complex at centromeres. To examine this prediction, we expressed in sgo2Δ swi6Δ cells Bir1 fused with CFP and the chromo domain (CD), which binds to Lys-9-methylated histone H3 largely locating at pericentromeric heterochromatic regions (Kitajima et al. 2006). The engineered protein Bir1-CD, indeed, localized at centromeric regions even without the aide of Sgo2 (Fig. 4C). Remarkably, the expression of Bir1-CD, but neither Bir1 nor CD alone, restored growth of sgo2Δ swi6Δ cells and partly suppressed the occurrence of lagging chromosome (Fig. 4E; Supplementary Fig. 6A). The TBZ sensitivity of sgo2Δ cells was partly suppressed by the Bir1-CD expression as well (Supplementary Fig. 6B). Taken all together, these results strengthen the conclusion that Sgo2 has an important role, in redundant capacity with Swi6, to localize Bir1 at centromeres (Fig. 4F).
Meiotic phenotype of sgo2Δ can be explained by a reduction of Aurora localization
Given that Sgo2 is required for activating the spindle checkpoint in the absence of tension in mitosis, we examined whether meiosis I also requires Sgo2 for activating the checkpoint when homologous chromosomes are not connected by linkages called chiasmata. Cells depleted of rec12+ (a homologous gene to budding yeast SPO11, which mediates the initiation of recombination reaction) indeed activated the checkpoint and delayed APC-dependent securin degradation (Fig. 5A). This delay was suppressed by deleting the checkpoint gene mad2+, verifying the above assumption. Remarkably, sgo2Δ also suppressed the checkpoint activation of rec12Δ cells. In addition to this checkpoint defect at meiosis, sgo2Δ cells show several chromosome segregation defects during anaphase I, including lagging chromosomes and nondisjunction of homologs, whereas meiosis II is rather normal (Fig. 5B,C; Kitajima et al. 2004; Rabitsch et al. 2004). We wondered whether these meiotic phenotypes of sgo2Δ might also be caused by reduced localization of Aurora at centromeres like in mitosis. Supporting this assumption, lagging chromosomes and nondisjunction of homologs at meiosis I were elevated in bir1-T1 cells at a semipermissive temperature (Fig. 5B,C). Thus, the meiotic defects of sgo2Δ cells are, at least partly, reproduced in the Aurora-deficient meiosis.
Figure 5.
Sgo2 is required for kinetochore localization of Ark1 in meiosis. (A) Time from SPB separation to securin (Cut2) degradation in meiosis I was measured in the indicated strains at 30°C using live cell microscopy. Each dot represents the measurement from one cell; the bars indicate the average time. (B) The frequency of lagging chromosomes at anaphase I was examined in the indicated strains by inducing meiosis at 30°C and staining tubulin and DNA (n > 100). Examples of tubulin and DNA staining are shown at the bottom; lagging chromosomes are observed in the sgo2Δ cell. (C) Both homologs marked with cen2-GFP in the indicated strains were monitored for segregation during meiosis at 30°C (n > 200). Examples of sgo2Δ cells are shown at the bottom; the absence of chromatids marked by cen2-GFP in one side of the zygote indicates that nondisjunction occurred at meiosis I (arrowhead). (D) Quantification of the signal intensity of Ark1-GFP in wild-type, sgo1Δ, and sgo2Δ cells. Cells were arrested at metaphase I by repressing APC activation (slp1+ and cut23+ expression) and observed microscopically. The intensity of Ark1-GFP at metaphase I was measured. Error bars represent the SEM (n = 30). Representative images of each strain are shown. (E) Fluorescence of Par1-GFP was examined in wild-type, sgo1Δ, and sgo2Δ cells arrested at metaphase I. The spindle was visualized by expressing CFP-atb2+. Par1-GFP was detected as dots in most (>95%) metaphase wild-type or sgo2Δ cells but never in sgo1Δ cells.
To further explore the above possibility, we examined the localization of the Aurora kinase complex during meiosis. As expected, centromeric localization of Bir1 was reduced in sgo2Δ cells during meiosis, although its localization at the spindle midzone was intact (Supplementary Fig. 7A). Accordingly, Ark1 localization at centromeres at meiosis I was also reduced in sgo2Δ cells, but this reduction was not observed in cells depleted of Sgo1, the other shugoshin required for protection of centromeric cohesin at this stage (Fig. 5D). In striking contrast, the localization of Par1/PP2A-B56, a factor cooperating with Sgo1 to protect cohesin (Kitajima et al. 2006; Riedel et al. 2006), was abolished in sgo1Δ cells but not in sgo2Δ cells, further stressing the functional divergence of Sgo1 and Sgo2 at meiosis I (Fig. 5E). Moreover, Sgo2 partly requires Bir1 for its centromeric localization in meiosis, whereas Sgo1 has little requirement for it (Supplementary Fig. 7B). These analyses underscore the specific interaction of Sgo2 with the Aurora kinase complex for their localization not only in mitosis but also in meiosis. Supporting this conclusion, we further found that overexpression of Bir1 alleviated meiotic defects of sgo2Δ cells (Fig. 5B,C).
Sgo2 associates with Bir1
To examine the interaction of Sgo2 and Bir1 in vivo, we performed an immunoprecipitation assay. Since Sgo2 and the Aurora kinase complex colocalize only at M phase, we synchronized cells at prometaphase by the nda3-KM311 mutation and prepared extracts. At first, we performed conventional immunoprecipitation but could not detect any interaction even among the components of the Aurora kinase complex (data not shown), suggesting that the Aurora kinase complex is not stable in fission yeast extracts as repeatedly reported (Morishita et al. 2001; Huang et al. 2005). We then used a cross-linker to detect loosely associated proteins. As a result, Bir1 coprecipitated Ark1. Remarkably, Sgo2 was also coprecipitated with Bir1 in the same experiment albeit with a low efficiency (Fig. 6A). By the reverse immunoprecipitation, Sgo2 coprecipitated Bir1 but not Ark1 (Fig. 6A), verifying the close interaction between Sgo2 and Bir1 in vivo.
Figure 6.
Sgo2 associates with Bir1. (A) Coimmunoprecipitation of Bir1 and Sgo2. Cells expressing Sgo2-myc and Ark1-GFP were arrested at prometaphase by incubating for 10 h at 20°C (by nda3-KM311) and cross-linked by 0.8% formaldehyde. Immunoprecipitates (IP) from cell extracts obtained by anti-Bir1 or anti-myc antibody as well as control IgG were analyzed by Western blot using anti-Bir1, anti-GFP, anti-myc, and anti-tubulin (control) antibodies. (WCE) Whole-cell extract used for immunoprecipitation. (B) Schematic drawing of the indicated mutant or truncated constructs of Bir1. (C) GST-Bir1-N, GST-Bir1-M, GST-Bir1-C, and His-Sgo2 recombinant proteins were expressed and purified from E. coli. His-Sgo2 was incubated with the indicated GST-tagged proteins bound to beads. Bead-bound fractions were separated by SDS-PAGE and analyzed by Western blotting using antibodies against His and GST. (D) sgo2+-GFP cells carrying a vector or plasmid expressing Bir1-ΔC were examined for the localization of Sgo2-GFP. Cell extracts prepared from sgo2+-myc cells expressing Bir1-ΔC tagged with pk were immunoprecipitated using control IgG and anti-pk antibody and detected by Western blot using anti-pk, anti-myc, and anti-tubulin (control) antibodies. (E) Wild-type or sgo2Δ cells expressing Bir1-ΔC or vector alone were spotted onto minimal medium plates containing 0, 10, or 15 μg/mL TBZ and incubated at 30°C.
To further examine the direct interaction between Sgo2 and Bir1, we performed in vitro binding assays using bacterially produced recombinant proteins. We found that Sgo2 specifically associates with the N terminus of Bir1, which contains two tandem copies of the conserved BIR domain (Fig. 6B,C). Further precise mapping of the Sgo2-binding domains indicated that Sgo2 binds to regions adjacent to the BIR domains but not within them (Supplementary Fig. 8). Interestingly, the bir1-T1 mutant, which exhibits a defect in centromeric localization of Sgo2 and Bir1 itself even at a permissive temperature (Fig. 3E), turned out to carry two point mutations within each of the tandem BIR domains (Fig. 6B), and we did not detect any significant difference in binding affinity of N-terminal peptides of wild-type and mutant Bir1 to Sgo2 (data not shown). In contrast, the bir1-275 mutant, which exhibits little defect in centromeric localization of Sgo2 (Supplementary Fig. 4B), carries a mutation near the C terminus (A990T) (Morishita et al. 2001). These results are consistent with a notion that the BIR domains and its adjacent Sgo2-binding regions cooperatively play an important role for the centromeric localization of Bir1 and Sgo2.
Bir1 contains a conserved coiled-coil region at the C terminus (Fig. 6B; Morishita et al. 2001; Huang et al. 2005). Consistent with the foregoing results, a truncated Bir1 (Bir1-ΔC) missing the C-terminal conserved region but retaining the N-terminal domains and the nuclear localization sequence (NLS) still preserved the ability to interact with Sgo2 in vivo (Fig. 6D). When Bir1-ΔC was overexpressed in addition to endogenous Bir1, we noticed that Sgo2 localization was abolished not only from centromeres at metaphase but also from noncentromeric sites at interphase (Fig. 6D). These results imply that, although Bir1 is normally required for centromeric localization of Sgo2 in M phase, Bir1-ΔC has the potential, if ectopically expressed, to dominantly influence Sgo2 localization. Interestingly, although Bir1-ΔC expression in wild-type cells markedly increased the sensitivity to TBZ, it was not additive with the sensitivity incurred by deletion of sgo2+ (Fig. 6E), indicating that Bir1-ΔC specifically impaired chromosome segregation through aberrant Sgo2 localization. In such cells with mislocalized Sgo2, centromeric localization of Ark1 was also impaired (data not shown). These results support the notion that Sgo2 and the N terminus of Bir1 interact in vivo and the intact complex of Sgo2 and Bir1 plays an essential role in loading the Aurora kinase complex to centromeres.
Human shugoshin hSgo1 interacts with Aurora kinase complex
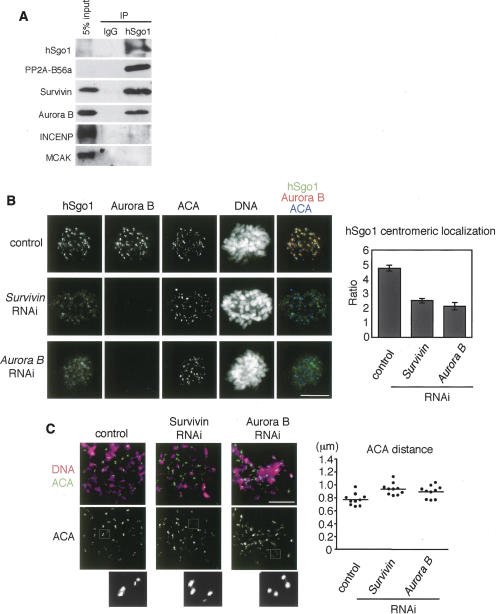
Since human shugoshin hSgo1 shows clear colocalization with the Aurora kinase complex at the inner centromeres in prometaphase and metaphase (Kitajima et al. 2005; McGuinness et al. 2005), we thought that the interaction between shugoshin and the Aurora kinase complex might be conserved in human cells. To test this possibility, we examined the physical association of hSgo1 with the Aurora kinase complex by immunoprecipitating hSgo1 from a solubilized chromatin fraction prepared from prometaphase-arrested cells and examined the precipitates for the existence of Aurora kinase complex components (Fig. 7A). In accordance with previous results (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006), PP2A-B56 subunit coprecipitated with hSgo1 most efficiently among the tested proteins. Importantly, Survivin and to a little less extent Aurora B were also coprecipitated with hSgo1, whereas INCENP and MCAK, another inner centromere-localizing protein, were hardly detected in the precipitates (Fig. 7A). These results suggest that hSgo1 associates with Aurora kinase proteins, in particular with Survivin and Aurora B. We did not detect such interaction between another human shugoshin hSgo2 and Aurora kinase complex proteins (data not shown).
Figure 7.
The human Aurora kinase complex functionally and physically interacts with hSgo1. (A) A chromatin extract of nocodazole-arrested mitotic HeLa cells was immunoprecipitated with anti-hSgo1 or control IgG. The extracts (input) and the immunoprecipitates (IP) were analyzed by Western blotting using antibodies against the indicated proteins. (B) HeLa cells were mock-transfected or transfected with siRNAs against Aurora B or Survivin, and stained with anti-hSgo1, anti-Aurora B, and ACA. DNA was counterstained with Hoechst 33342. Bar, 10 μm. Quantification of centromeric signals of hSgo1 is shown on the right. For each cell, the fluorescent intensities of hSgo1 at 10 centromeres were related to the intensities of the corresponding ACA signals. Error bars show the SEM (10 cells for each). (C) Mitotic cells after RNAi treatment were harvested by mitotic shake-off and then treated with nocodazole for 4 h. The prometaphase-arrested cells were spun on slides and stained with ACA and Hoechst 33342. Magnifications of representative ACA pairs are shown. Bar, 10 μm. The average distance between paired ACA signals is shown on the right. One dot represents the average distance of at least 20 pairs within a single cell. Ten cells were examined.
We next examined the interdependency of hSgo1 and Aurora kinase complex proteins for their localization. Remarkably, treatment with small interfering RNA (siRNA) of Survivin largely impaired the centromeric localization of hSgo1 but produced weak localization along the whole chromosome (Fig. 7B). Accordingly, centromeric cohesion was loosened in these cells since the sister kinetochore distance increased by ∼20% even in nocodazole-arrested cells (note that centromeres are not pulled outward in this condition). Aurora B RNA interference (RNAi) treatment caused identical defects (Fig. 7C). Conversely, the depletion of hSgo1 by RNAi did not cause obvious defect in the centromeric localization of Aurora B or Survivin (Supplementary Fig. 9). Thus, we conclude that the Aurora kinase complex plays an important role in localizing shugoshin to centromeres in human cells.
Discussion
Fission yeast Sgo1 was first identified as a protector of meiotic cohesin and shown to be conserved among eukaryotes, forming a protein family called shugoshin (Kitajima et al. 2004; Rabitsch et al. 2004). Studies in HeLa cells extended this concept to mitosis, where human shugoshin plays a crucial role in protecting centromeric cohesin (Salic et al. 2004; Kitajima et al. 2005; McGuinness et al. 2005). Recent studies revealed that these pathways share a common mechanism, in which shugoshin recruits PP2A to centromeres and antagonizes the phosphorylation of the cohesin complex, which facilitates cohesin dissociation (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006).
We describe here that fission yeast shugoshin Sgo2 is dispensable for protecting cohesin but instead required for promoting tension-generating bipolar attachment of kinetochores in mitosis and meiosis and inhibiting meiosis as long as tension-less attachments are present. The sole shugoshin in budding yeast, ScSgo1, which is required for protecting centromeric cohesin at meiosis, was identified in a screening for checkpoint components required for sensing the loss of tension in mitosis (Indjeian et al. 2005). Therefore, a role in the spindle checkpoint may be a conserved function of shugoshin. How does shugoshin function in the spindle checkpoint sensing the absence of tension? Since Xenopus Sgo1 was identified as a factor to bind to microtubules (Salic et al. 2004), one model proposed that shugoshin might bind at the connection between microtubule and kinetochore and thereby function as a mechanical sensor of tension at centromeres (Indjeian et al. 2005). We here reveal that shugoshin functions to localize the Aurora kinase complex at centromeres.
Recent studies in budding yeast and human cells suggest that Aurora plays a crucial role at centromeres in destabilizing tension-less kinetochore microtubule attachment, such as syntelic attachment (Tanaka et al. 2002; Hauf et al. 2003). The destabilization of attachment by Aurora would create unattached kinetochores, which simultaneously activates the spindle checkpoint (Pinsky et al. 2006). In Aurora-deficient cells, erroneous attachment is prematurely stabilized with the checkpoint being silenced, resulting in massive missegregation of chromosomes. Thus, it is proposed that Aurora is a crucial factor enabling tension-generating kinetochore attachment (Tanaka 2002; Pinsky and Biggins 2005). Studies of Aurora in fission yeast also support this view (S. Hauf and Y. Watanabe, unpubl.). Here, we demonstrate that fission yeast Sgo2 plays an important role in loading the Aurora kinase complex to centromeres based on the following evidence. The deletion of sgo2+ and partial loss of function of the Aurora kinase complex show very similar phenotypes in mitotic chromosome segregation and the spindle checkpoint sensing the loss of tension. Sgo2 and the Aurora kinase complex colocalize at centromeres during prometaphase and metaphase. Especially, Sgo2 and Bir1 directly interact in vitro and coprecipitate in extracts prepared in the presence of a weak cross-linker, indicating that they associate in vivo. Sgo2 and Bir1 mutually require each other to localize at centromeres; Ark1/Aurora loading to centromeres is dependent on this complex. The defects of sgo2Δ cells were partially suppressed by forcibly localizing Bir1 to centromeres, whereas Aurora defects were not suppressed by ectopically localizing Sgo2 to centromeres by the CD fusion (data not shown), implying that the Aurora kinase complex acts downstream from Sgo2. Finally, meiotic chromosome segregation defects in sgo2Δ cells were reproduced in bir1-T1 cells, and the sgo2Δ defects in meiosis were suppressed by overexpressing Bir1. Importantly, although any component of the Aurora kinase complex is essential for cell viability, sgo2Δ largely impairs the centromeric localization of Ark1 but preserves substantial viability. It is noteworthy that only ∼10% of the centromeric pool of the Aurora kinase complex is sufficient to sustain substantial viability in the bir1-T1 mutant (Fig. 3E). This explains the full viability of sgo2Δ cells, in which the Aurora kinase complex at centromeres reaches ∼20% of wild-type levels (Fig. 3C). Thus, the residual Aurora kinase complex at centromeres in sgo2Δ cells may support a minimum ability to correct the transient attachment errors in unperturbed mitosis, albeit it would be not enough to destabilize persistent tensionless attachment or to fully correct abnormal attachment caused by perturbed centromeres or spindles. Another explanation for the lethality of the Aurora kinase complex-deleted cells but not of sgo2Δ cells could be that the Aurora kinase complex has essential targets outside the centromere; for instance, in chromosome condensation (Morishita et al. 2001; Petersen and Hagan 2003), which is totally intact in sgo2Δ cells. Remarkably, sgo2Δ cells exhibit obvious defects in meiotic chromosome segregation even in unperturbed condition (Fig. 5B,C; Rabitsch et al. 2004), indicating that the Sgo2–Aurora pathway is more essential in meiosis.
Through the studies of Sgo2, we could unexpectedly find that heterochromatin defects and Aurora function are closely associated. Cells defective in pericentromeric heterochromatin like swi6Δ transiently produce lagging chromosomes or erroneous attachment, presumably because impaired centromere structures increase the chance of a single kinetochore being captured by microtubules emanating form both spindle poles (Fig. 4F; Pidoux et al. 2000). In unperturbed mitosis, however, these defects are mostly restored, since swi6Δ cells sustain substantial viability. Recent studies in mammalian cells as well as in fission yeast suggest that the function of Aurora is important for preventing erroneous attachment (Cimini et al. 2006; Knowlton et al. 2006; S. Hauf and Y. Watanabe, unpubl.). Consistently, we found that sgo2Δ as well as Aurora kinase complex mutations markedly enhance this swi6Δ defect while producing high incidence of lagging chromosomes and missegregation of chromosomes. These defects of sgo2Δ swi6Δ were substantially restored by forcibly localizing Bir1 to centromeres (Fig. 4). These results illuminate a crucial role of Sgo2 in correcting erroneous kinetochore attachment by loading the Aurora kinase complex to kinetochores.
We further demonstrate that human shugoshin hSgo1 associates with Survivin and Aurora and requires these components for its centromeric localization. Together with the recent finding in Drosophila that the Aurora kinase complex is required for centromeric localization of Sgo/Mei-S332 (Resnick et al. 2006), our studies suggest that the linkage between shugoshin and Aurora kinase complex is conserved among eukaryotes. Our studies in human cells present the strongest data to date indicating the existence of a complex including shugoshin and Survivin in vivo; hSgo1 could coprecipitate with Survivin better than Aurora in extracts prepared from chromatin fraction. This result fits with the immunoprecipitation using a cross-linker in fission yeast and with genetic results indicating that Sgo2 closely interacts with Bir1/Survivin for the centromeric localization. Although the linkage between shugoshin and the Aurora kinase complex is conserved across species, the precise manner of interaction has apparently diverged. The centromeric localization of Drosophila Mei-S332 reportedly requires phosphorylation by Aurora (Resnick et al. 2006); however, fission yeast Sgo2 does not require it, albeit Sgo2, like Mei-S332, is a good substrate of Ark1 in vitro (Supplementary Fig. 10). Whereas fission yeast shugoshin (Sgo2) is required for the localization and function of Aurora kinase complex at centromeres, Drosophila Mei-S332 as well as human Sgo1 is not required for the localization of the Aurora kinase complex (Resnick et al. 2006; Supplementary Fig. 9), albeit the centromeric function of the Aurora kinase complex might nevertheless be regulated by Mei-S332 (see below).
The sole shugoshin protein in budding yeast seems to play dual roles in protecting centromeric cohesin at meiosis I (but not at mitosis) as well as in establishing tension-generating attachment at mitosis (Katis et al. 2004; Kitajima et al. 2004; Marston et al. 2004; Indjeian et al. 2005). Drosophila SGO/MEI-S332 mutants show nondisjunction of homologs at meiosis I and a reduced ratio of meta/anaphase (but only slight or little defect in cohesion) in mitosis (Kerrebrock et al. 1992; LeBlanc et al. 1999). Therefore, we suggest that Mei-S332, the sole shugoshin of Drosophila, is also required for establishing tension-generating attachment, like fission yeast Sgo2. The localization of the Aurora kinase complex does not depend on Mei-S332; however, it is tenable that the activation of centromeric Aurora kinase complex may somewhat depend on Mei-S332 since they physically interact in vitro (Resnick et al. 2006). Similarly, fission yeast Sgo2 might play an additional role in activating centromeric Aurora rather than merely promoting its localization. Given that hSgo1 associates with Survivin (and Aurora) in HeLa cells (Fig. 7A), a similar functional link is conceivable also in human cells.
Studies in fission yeast enabled us to molecularly define two distinct shugoshin functions or pathways, which are carried out by two diverged shugoshins, Sgo1 and Sgo2; the former interacts with PP2A to protect cohesin, but the latter interacts with the Aurora kinase complex to facilitate centromeric Aurora function. We speculate that the ancestral shugoshin molecule played dual roles at kinetochores like in budding yeast or Drosophila; fission yeast shugoshin might have divided the labor to Sgo1 and Sgo2. Thus, our findings of a functional link between Sgo2 and the Aurora kinase complex open a new view that shugoshin in general may play a role in facilitating Aurora function at centromeres, thereby ensuring tension-generating kinetochore microtubule attachment. At the centromere, microtubule attachment is ensured by tension across centromeres, which is generated depending on the cohesion between sister chromatids. Therefore, cohesion and tension are two sides of a “coin” ensuring bipolar attachment of kinetochores. We suggest that the original role of shugoshin was to guarantee bipolar attachment rather than to protect cohesin, because fission yeast and presumably budding yeast, two primitive eukaryotes, exhibit only this role during mitosis. The protection role, once acquired, might facilitate the generation of tension by counteracting the spindle force, improving the fidelity of chromosome segregation. This function might have been modified to evolve meiosis, in which the requirement for centromeric protection is more essential and therefore has been preserved in all eukaryotes. Whatever the validity of this view, our finding of how Sgo2 acts will contribute to understand the fundamental regulation of eukaryotic chromosome segregation.
Materials and methods
Schizosaccharomyces pombe strain
Deletion and tagging of endogenous sgo2+, ark1+, bir1+, mad2+, and cnp3+ by GFP, 13 copies of Myc (abbreviated as myc), three copies of PK (pk), mCherry (mCh), and tdTomato (tdT) (Shaner et al. 2004) were performed using the PCR-based gene targeting method for S. pombe (Bahler et al. 1998). For generating temperature-sensitive mutants bir1-T1, ark1-T7, ark1-T8, and pic1-T269, we used a low-fidelity PCR method as previously reported (Nonaka et al. 2002). To express Bir1-CFP-CD, a sequence encoding CFP and two copies of the CD of Swi6 (amino acids 69–215) were fused to the C terminus of Bir1 and cloned under the promoter Padh81 (a weak version of the adh1+ promoter). The resulting plasmid was linearized and integrated at the lys1+ locus of chromosome I by using a hygr marker. To express Bir1, the bir1+ ORF was cloned under the promoter of Padh81 (for Fig. 4 experiments) or Padh41 (a mildly weak version of the adh1+ promoter, for Fig. 5 experiments), and the resultant plasmid was similarly integrated to the chromosome. To reduce APC activity during meiosis, we replaced the promoters of slp1+ and cut23+ (both are induced during meiosis and are required for APC activation) with that of rad21+, which is repressed during meiosis. All strains used are listed in Supplementary Table 1.
Culture
For prometaphase arrest, we used the nda3-KM311 mutation (Hiraoka et al. 1984) and cultured cells for 10 h at 20°C. For metaphase arrest, we used the cut9-665 mutation (Yamada et al. 1997) and cultured cells for 4 h at 36°C. For G1/S arrest, we used HU (Wako) at a final concentration of 12 mM, and cultured cells for 3 h at 25°C and for 1 h at 36°C (for inactivation of psc3-1T). General methods for induction of meiosis and monitoring chromosome segregation were as described previously (Kitajima et al. 2004).
Immunostaining
To stain phosphorylated S. pombe Histone-H3 (H3S10ph), we used anti-phospho-Histone H3 (Ser-10; 1:100; Upstate Biotechnology) and Cy3-tagged anti-rabbit antibody (1:2000; Chemicon). Tubulin was detected using the mouse anti-tubulin antibody TAT-1 (1:200; a gift from K. Gull) and Cy3-tagged anti-mouse antibody (1:2000; Chemicon). To detect GFP-tagged proteins, we used mouse anti-GFP antibody (1:100; Roche) and BODIPY FL-conjugated anti-mouse antibody (1:100; Molecular Probes). To detect Bir1, we used an anti-Bir1 polyclonal antibody (1:200) and BODIPY FL-conjugated anti-rabbit antibody (1:200; Molecular Probes).
Quantification of fluorescent signals
To quantify the centromeric fluorescent signals, in-focus images of Bir1, GFP-Bir1, Ark1-GFP, Sgo2-GFP, and Sgo2-mCh cells were taken with the use of MetaMorph imaging software (Universal Imaging). We measured the maximum intensity among the centromeric signals within the cells and subtracted the average of background intensity.
Quantification of centromeric enrichment and its reduction ratio of the Aurora kinase complex
After normalizing the immunoprecipitation (IP) ratio (percentage) between sgo2+ and sgo2Δ cells by using the control IP (percentage) of Cnp1, the IP ratio (percentage) at the pericentromeric (dg or dh) region was substracted by that at the arm (zfs1) regions in the Ark1 and Bir1 ChIP assays (Fig. 3D), giving the centromeric enrichment score. This score of sgo2Δ cells was divided by that of wild-type cells, giving the reduction ratio. These ratios at dg and dh were averaged, giving the “reduction ratio of centromeric enrichment” of the Aurora kinase complex in sgo2Δ cells.
Preparation of anti-Bir1 antibody
For the generation of polyclonal antibodies against Bir1, a cDNA fragment encoding for amino acids 1–300 of Bir1 was amplified and inserted in frame into plasmids pGEX4T-2 (Pharmacia Biotech) and pET-19b (Novagen) for the production of recombinant proteins GST-Bir1-N and His-Bir1-N, respectively. GST-Bir1-N was expressed in Escherichia coli strain BL21 and purified by Glutathione Sepharose (Amersham) according to the manufacturer’s instructions, followed by immunization of a rabbit. Antibodies were affinity-purified by His-Bir1-N conjugated to CNBr-activated Sepharose (Amersham).
ChIP assay
The procedure was carried out essentially as described previously (Yokobayashi et al. 2003). Anti-Bir1 polyclonal antibodies, anti-GFP polyclonal antibodies (Living Colors Full-length A.v. Polyclonal Antibody; BD Biosciences), and anti-Cnp1 polyclonal antibodies were used for immunoprecipitation. DNA prepared from whole-cell extracts or immunoprecipitated fractions was analyzed by quantitative PCR with the ABI PRISM7000 system (Applied Biosystems) using SYBR Premix Ex Taq (Perfect Real Time; Takara). The primers used for PCR were all described previously (Yokobayashi et al. 2003), except those for zfs1 (CCGGTTGAAAGGAATAGACT and TTTCTTGCCTGA GATACCGT). We included an untagged strain or control IgG immunoprecipitation in each experiment to account for nonspecific binding in ChIP fractions.
Coimmunoprecipitation from fission yeast extracts
Cultured cells were cross-linked by 0.8% formaldehyde. After washing with ChIP buffer 1 (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA at pH 7.5, 0.1% Triton X-100), cells were resuspended in buffer 1 containing 1 mM PMSF and complete protease inhibitors (Roche), 3 mM CaCl2, and 3 U/109 cells micrococcal nuclease (Sigma), and then lysed by a Multi-bead shocker (Yasui Kikai). Crude cell extracts were sonicated five times (15 sec each time), and the supernatant was collected after centrifugation. Complete digestion of DNA was confirmed by gel electrophoresis. Cell extracts were incubated with the indicated antibodies (rabbit IgG or anti-Bir1 for Bir1 IP, mouse IgG or anti-myc [9E10; Santa Cruz Biotechnology] for Sgo2 IP, and mouse IgG or anti-pk [MCA1360; Serotec] for Bir1-ΔC IP) for 1 h at 4°C. Protein A beads (Amersham) were added and incubation was continued for 2 h at 4°C. After washing with buffer 1 and buffer 1′ (50 mM HEPES-KOH at pH 7.5, 500 mM NaCl, 1 mM EDTA at pH 7.5, 0.1% Triton X-100), we analyzed the immunoprecipitates by SDS-PAGE and Western blotting with anti-Bir1 (1:10000), anti-GFP (1:800; Roche), anti-myc (1:1000; 9E10), anti-myc (1:1000; A14; Santa Cruz Biotechnology), anti-pk (1:1000; MCA1360), and TAT1 (1:5000) antibodies.
In vitro binding assay
Recombinant proteins GST-Bir1-N, GST-Bir1-M, GST-Bir1-C, and His-Sgo2 were expressed from E. coli. For each reaction, 0.1 μg of His-Sgo2 and 1 mg/mL BSA were added to HB buffer (25 mM MOPS at pH 7.2, 5 mM EGTA, 15 mM MgCl2, 0.2% NP-40, 10% glycerol, 500 mM KCl, 1 mM PMSF, Complete [Roche]) containing 1 μg of GST-tagged proteins bound to glutathione Sepharose beads. Samples were incubated for 40 min at 4°C, and the beads were washed three times with HB buffer. The bound fraction was separated by SDS-PAGE and analyzed by Western blotting using antibodies against His and GST.
RNAi and immunostaining of HeLa cells
Synthetic sense and antisense oligonucleotides for RNAi of hSgo1 (Kitajima et al. 2005), Survivin (Carvalho et al. 2003), and Aurora B (Andrews et al. 2004) were obtained from JbioS. siRNA transfection was performed as described (Kitajima et al. 2005). Immunofluorescent staining was performed as described (Liu et al. 2003) with anti-Sgo1 (1:1000) (Kitajima et al. 2005), anti-Survivin (1:1000; Novus Biologicals), anti-Aurora B (1:1000; BD Biosciences), and ACA (1:10000; a generous gift from Dr. Y. Takasaki). Secondary antibodies were Alexa Fluor 588 anti-rabbit antibodies (1:1000; Molecular Probes), Cy3-conjugated anti-mouse antibodies (1:1000; Chemicon), and Alexa Fluor 647 anti-human antibodies (1:1000; Molecular Probes). Images were captured by DeltaVision SoftWorx software (Applied Precision) and processed by deconvolution and z-stack projection. To measure the ACA distances, mitotic cells harvested by mitotic shake-off were treated with 330 nM nocodazole for 4 h, spun on glass slides by CytoSpin (Thermo Electron), and immunostained with ACA as described (Kitajima et al. 2006). The ACA distances were analyzed by SlideBook software (Intelligent Imaging Innovations).
Immunoprecipitation from human cell extracts
HeLa cells were harvested after treatment with 330 nM nocodazole for 12 h and lysed in extraction buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.2% Nonidet P-40, 10% glycerol, 1 mM NaF, 1 mM Na3VO4, 20 mM β-glycerophosphate, 10 mM β-mercaptoethanol, 0.2 mM PMSF, Complete Protease Inhibitor [Roche]) by two rounds of freezing and thawing. We collected the insoluble materials by centrifugation and solubilized chromatin proteins by treating with micrococcal nuclease (0.008 U/μL) in chromatin extraction buffer (10 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1.5 mM MgCl2, 0.25 M sucrose, 0.2 mM PMSF, Complete Protease Inhibitor) for 15 min at 30°C. The cleared chromatin extract was incubated with anti-hSgo1 or rabbit IgG coupled to protein A beads (Amersham) for 1 h at 4°C. After washing the beads with extraction buffer, we analyzed the immunoprecipitates by Western blotting with anti-hSgo1 (1:1000), anti-PP2A-B56α (1:1000; BD Biosciences), anti-Survivin (1:1000), anti-Aurora B (1:1000), anti-INCENP (1:1; a generous gift from Dr. T. Urano), and anti-MCAK (1:1000; a generous gift from Dr. P. Andrews).
Acknowledgments
We thank Masayuki Yamamoto, Mitsuhiro Yanagida, and the Yeast Genetic Resource Center (YGRC) for yeast strains; Takeshi Urano for INCENP antibodies; Paul D. Andrews for MCAK antibodies; Mohan Balasubramanian for the GFP-Bir1 strain; and Kayoko Tanaka and Roger Tsien for plasmids of mCherry and tdTomato. We also thank all members in our laboratory for their valuable support and particularly Yasuto Ando for help in measuring centromere distance in HeLa cells. S.H. acknowledges support by the Human Frontier Science Program (HFSP). This work was supported in part by the Toray Science Foundation, Uehara Memorial Foundation, and a Grant-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y.W.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1497307
References
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R., Duncan K., Wordeman L., Swedlow J.R., Wordeman L., Swedlow J.R., Swedlow J.R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Wu J., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R., Steever A.B., Wach A., Philippsen P., Pringle J.R., Wach A., Philippsen P., Pringle J.R., Philippsen P., Pringle J.R., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Genier S., Javerzat J.P., Allshire R.C., Javerzat J.P., Allshire R.C., Allshire R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A.W., Murray A.W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Carmena M., Sambade C., Earnshaw W.C., Wheatley S.P., Carmena M., Sambade C., Earnshaw W.C., Wheatley S.P., Sambade C., Earnshaw W.C., Wheatley S.P., Earnshaw W.C., Wheatley S.P., Wheatley S.P. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 2003;116:2987–2998. doi: 10.1242/jcs.00612. [DOI] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C.B., Salmon E.D., Wan X., Hirel C.B., Salmon E.D., Hirel C.B., Salmon E.D., Salmon E.D. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Hauf S., Watanabe Y., Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell. 2004;119:317–327. doi: 10.1016/j.cell.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M., Heckel A., van Meel J., Rieder C.L., Peters J.-M., van Meel J., Rieder C.L., Peters J.-M., Rieder C.L., Peters J.-M., Peters J.-M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Roitinger E., Koch B., Dittrich C.M., Mechtler K., Peters J.-M., Roitinger E., Koch B., Dittrich C.M., Mechtler K., Peters J.-M., Koch B., Dittrich C.M., Mechtler K., Peters J.-M., Dittrich C.M., Mechtler K., Peters J.-M., Mechtler K., Peters J.-M., Peters J.-M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes & Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Toda T., Yanagida M., Toda T., Yanagida M., Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Huang H.K., Bailis J.M., Leverson J.D., Gomez E.B., Forsburg S.L., Hunter T., Bailis J.M., Leverson J.D., Gomez E.B., Forsburg S.L., Hunter T., Leverson J.D., Gomez E.B., Forsburg S.L., Hunter T., Gomez E.B., Forsburg S.L., Hunter T., Forsburg S.L., Hunter T., Hunter T. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol. Cell. Biol. 2005;25:9000–9015. doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V.B., Stern B.M., Murray A.W., Stern B.M., Murray A.W., Murray A.W. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Katis V.L., Galova M., Rabitsch K.P., Gregan J., Nasmyth K., Galova M., Rabitsch K.P., Gregan J., Nasmyth K., Rabitsch K.P., Gregan J., Nasmyth K., Gregan J., Nasmyth K., Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W., Miyazaki W.Y., Birnby D., Orr-Weaver T.L., Miyazaki W.Y., Birnby D., Orr-Weaver T.L., Birnby D., Orr-Weaver T.L., Orr-Weaver T.L. The Drosophilamei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Kawashima S.A., Watanabe Y., Kawashima S.A., Watanabe Y., Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Hauf S., Ohsugi M., Yamamoto T., Watanabe Y., Hauf S., Ohsugi M., Yamamoto T., Watanabe Y., Ohsugi M., Yamamoto T., Watanabe Y., Yamamoto T., Watanabe Y., Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y., Iemura S., Natsume T., Kawashima S.A., Watanabe Y., Natsume T., Kawashima S.A., Watanabe Y., Kawashima S.A., Watanabe Y., Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Knowlton A.L., Lan W., Stukenberg P.T., Lan W., Stukenberg P.T., Stukenberg P.T. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- LeBlanc H.N., Tang T.T., Wu J.S., Orr-Weaver T.L., Tang T.T., Wu J.S., Orr-Weaver T.L., Wu J.S., Orr-Weaver T.L., Orr-Weaver T.L. The mitotic centromeric protein MEI-S332 and its role in sister-chromatid cohesion. Chromosoma. 1999;108:401–411. doi: 10.1007/s004120050392. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Hayashi-Hagihara A., Orr-Weaver T.L., Hayashi-Hagihara A., Orr-Weaver T.L., Orr-Weaver T.L. Roles and regulation of the Drosophila centromere cohesion protein MEI-S332 family. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:543–552. doi: 10.1098/rstb.2005.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson J.D., Huang H.K., Forsburg S.L., Hunter T., Huang H.K., Forsburg S.L., Hunter T., Forsburg S.L., Hunter T., Hunter T. The Schizosaccharomyces pombeAurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell. 2002;13:1132–1143. doi: 10.1091/mbc.01-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Hittle J.C., Jablonski S.A., Campbell M.S., Yoda K., Yen T.J., Hittle J.C., Jablonski S.A., Campbell M.S., Yoda K., Yen T.J., Jablonski S.A., Campbell M.S., Yoda K., Yen T.J., Campbell M.S., Yoda K., Yen T.J., Yoda K., Yen T.J., Yen T.J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003;5:341–345. doi: 10.1038/ncb953. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T., Hirano M., Hirano T., Hirano T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes & Dev. 2002;16:3004–3016. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A.L., Tham W.H., Shah H., Amon A., Tham W.H., Shah H., Amon A., Shah H., Amon A., Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- McGuinness B.E., Hirota T., Kudo N.R., Peters J.-M., Nasmyth K., Hirota T., Kudo N.R., Peters J.-M., Nasmyth K., Kudo N.R., Peters J.-M., Nasmyth K., Peters J.-M., Nasmyth K., Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki W.Y., Orr-Weaver T.L., Orr-Weaver T.L. Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet. 1994;28:167–168. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Morishita J., Matsusaka T., Goshima G., Nakamura T., Tatebe H., Yanagida M., Matsusaka T., Goshima G., Nakamura T., Tatebe H., Yanagida M., Goshima G., Nakamura T., Tatebe H., Yanagida M., Nakamura T., Tatebe H., Yanagida M., Tatebe H., Yanagida M., Yanagida M. Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells. 2001;6:743–763. doi: 10.1046/j.1365-2443.2001.00459.x. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y., Yamamoto M., Grewal S.I., Watanabe Y., Grewal S.I., Watanabe Y., Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Petersen J., Hagan I.M., Hagan I.M. S. pombe Aurora kinase/Survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 2003;13:590–597. doi: 10.1016/s0960-9822(03)00205-7. [DOI] [PubMed] [Google Scholar]
- Petersen J., Paris J., Willer M., Philippe M., Hagan I.M., Paris J., Willer M., Philippe M., Hagan I.M., Willer M., Philippe M., Hagan I.M., Philippe M., Hagan I.M., Hagan I.M. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 2001;114:4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L., Uzawa S., Perry P.E., Cande W.Z., Allshire R.C., Uzawa S., Perry P.E., Cande W.Z., Allshire R.C., Perry P.E., Cande W.Z., Allshire R.C., Cande W.Z., Allshire R.C., Allshire R.C. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 2000;113:4177–4191. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Biggins S., Biggins S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kung C., Shokat K.M., Biggins S., Kung C., Shokat K.M., Biggins S., Shokat K.M., Biggins S., Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Erratum. Nat. Cell Biol. Nat. Cell Biol. 2006;88:78–83. 100. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P., Gregan J., Schleiffer A., Javerzat J.P., Eisenhaber F., Nasmyth K., Gregan J., Schleiffer A., Javerzat J.P., Eisenhaber F., Nasmyth K., Schleiffer A., Javerzat J.P., Eisenhaber F., Nasmyth K., Javerzat J.P., Eisenhaber F., Nasmyth K., Eisenhaber F., Nasmyth K., Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Resnick T.D., Satinover D.L., MacIsaac F., Stukenberg P.T., Earnshaw W.C., Orr-Weaver T.L., Carmena M., Satinover D.L., MacIsaac F., Stukenberg P.T., Earnshaw W.C., Orr-Weaver T.L., Carmena M., MacIsaac F., Stukenberg P.T., Earnshaw W.C., Orr-Weaver T.L., Carmena M., Stukenberg P.T., Earnshaw W.C., Orr-Weaver T.L., Carmena M., Earnshaw W.C., Orr-Weaver T.L., Carmena M., Orr-Weaver T.L., Carmena M., Carmena M. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev. Cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C.G., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., Katou Y., Mori S., Itoh T., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., Mori S., Itoh T., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., Itoh T., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., Galova M., Petronczki M., Gregan J., Cetin B., Petronczki M., Gregan J., Cetin B., Gregan J., Cetin B., Cetin B., et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Salic A., Waters J.C., Mitchison T.J., Waters J.C., Mitchison T.J., Mitchison T.J. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I.X., Yates I.J.R., Oegema K., Hyman A., Desai A., Severin F., McLeod I.X., Yates I.J.R., Oegema K., Hyman A., Desai A., McLeod I.X., Yates I.J.R., Oegema K., Hyman A., Desai A., Yates I.J.R., Oegema K., Hyman A., Desai A., Oegema K., Hyman A., Desai A., Hyman A., Desai A., Desai A. A Bir1–Sli5 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y., Giepmans B.N., Palmer A.E., Tsien R.Y., Palmer A.E., Tsien R.Y., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Matsusaka T., Morrison C., Vagnarelli P., Hoshi O., Ushiki T., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Matsusaka T., Morrison C., Vagnarelli P., Hoshi O., Ushiki T., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Morrison C., Vagnarelli P., Hoshi O., Ushiki T., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Vagnarelli P., Hoshi O., Ushiki T., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Hoshi O., Ushiki T., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Ushiki T., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Nojima K., Fukagawa T., Waizenegger I.C., Peters J.M., Fukagawa T., Waizenegger I.C., Peters J.M., Waizenegger I.C., Peters J.M., Peters J.M., et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 2002;14:365–371. doi: 10.1016/s0955-0674(02)00328-9. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Galova M., Schiebel E., Stark M.J., Nasmyth K., Schiebel E., Stark M.J., Nasmyth K., Stark M.J., Nasmyth K., Nasmyth K. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N.A., Mumby M.C., Yu H., Shu H., Qi W., Mahmood N.A., Mumby M.C., Yu H., Qi W., Mahmood N.A., Mumby M.C., Yu H., Mahmood N.A., Mumby M.C., Yu H., Mumby M.C., Yu H., Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tomonaga T., Nagao K., Kawasaki Y., Furuya K., Murakami A., Morishita J., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Nagao K., Kawasaki Y., Furuya K., Murakami A., Morishita J., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Kawasaki Y., Furuya K., Murakami A., Morishita J., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Furuya K., Murakami A., Morishita J., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Murakami A., Morishita J., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Morishita J., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Yuasa T., Sutani T., Kearsey S.E., Uhlmann F., Sutani T., Kearsey S.E., Uhlmann F., Kearsey S.E., Uhlmann F., Uhlmann F., et al. Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes & Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Rajagopalan S., McCollum D., Rajagopalan S., McCollum D., McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell. 2004;7:755–762. doi: 10.1016/j.devcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. Chromosome cohesion and separation: From men and molecules. Curr. Biol. 2003;13:R104–R114. doi: 10.1016/s0960-9822(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P., Earnshaw W.C., Earnshaw W.C. Chromosomal passengers: The four-dimensional regulation of mitotic events. Chromosoma. 2004;113:211–222. doi: 10.1007/s00412-004-0307-3. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Shugoshin: Guardian spirit at the centromere. Curr. Opin. Cell Biol. 2005;17:590–595. doi: 10.1016/j.ceb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yokobayashi S., Yamamoto M., Nurse P., Yokobayashi S., Yamamoto M., Nurse P., Yamamoto M., Nurse P., Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Yamada H., Kumada K., Yanagida M., Kumada K., Yanagida M., Yanagida M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J. Cell Sci. 1997;110:1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S., Yamamoto M., Watanabe Y., Yamamoto M., Watanabe Y., Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]