Abstract
Expression of the Drosophila orphan nuclear receptor DHR78 is regulated by the steroid hormone ecdysone and is required for growth and viability during larval stages. In contrast to our understanding of its biological functions, however, relatively little is known about how DHR78 acts as a transcription factor. Here we show that DHR78 is an obligate partner for Moses (Middleman of seventy-eight signaling), a SAM (sterile α motif) domain-containing cofactor that requires DHR78 for its stability. Unlike other nuclear receptor cofactors, Moses has no obvious interaction domains and displays a unique binding specificity for DHR78. Moses acts as a corepressor, inhibiting DHR78 transcriptional activity independently of histone deacetylation. Consistent with their close association, DHR78 and Moses proteins are coexpressed during development and colocalize to specific genomic targets in chromatin. Moses mutants progress normally through early larval stages, like DHR78 mutants, but display an opposite overgrowth phenotype, with hypertrophy of adult tissues. Genetic interactions between DHR78 and moses result in a similar phenotype, suggesting that the relative dose of Moses and DHR78 regulates growth and prevents cancer. The tight functional association between DHR78 and Moses provides a new paradigm for understanding the molecular mechanisms by which cofactors modulate nuclear receptor signaling pathways.
Keywords: Nuclear receptors, cofactors, transcriptional regulation, hormone signaling, cancer
Nuclear receptors (NRs) play a critical role at the interface between chemical signaling and transcriptional control of growth, metabolism, and development in higher organisms. They are defined by a highly conserved DNA-binding domain (DBD) that consists of two zinc fingers and a less-conserved ligand-binding domain (LBD) that provides a ligand-dependent platform for regulatory interactions. Integral to LBD function is the C-terminal activation function-2 domain (AF-2), which is often required for direct interaction with protein cofactors that modulate NR activity (Rosenfeld et al. 2006). Although studies over the past decade have identified a number of ligands for NRs, many receptors remain orphans with no known ligand.
The fruit fly, Drosophila melanogaster, provides an ideal model system for dissecting the mechanisms of NR regulation in the context of a developing animal (King-Jones and Thummel 2005). The Drosophila genome encodes 18 canonical NRs that span all major receptor subfamilies, as defined by phylogenetic studies of multiple Metazoan species (Laudet 1997; Thornton 2003). Furthermore, in contrast to the complexity of vertebrate hormone signaling pathways, Drosophila has only one known physiologically active steroid hormone, 20-hydroxyecdysone (referred to here as ecdysone), which directs the major developmental transitions in the life cycle, including molting and puparium formation (Riddiford 1993; Thummel 1996). Ecdysone initiates transcriptional hierarchies through binding and activating an NR heterodimer comprised of the Ecdysone Receptor (EcR) and its RXR partner, Ultraspiracle (USP) (for review, see Riddiford et al. 2000). Although 16 of the 18 fly receptors remain orphans (Koelle et al. 1991; Reinking et al. 2005), approximately half of the Drosophila NRs are transcriptionally regulated by a high-titer pulse of ecdysone at the end of the third instar (L3), the final larval stage (Sullivan and Thummel 2003). This ecdysone pulse triggers puparium formation and the prepupal stage of development, terminating the juvenile growth phase and initiating maturation during metamorphosis.
Here, we characterize one of these ecdysone-regulated orphan NRs, DHR78 (NR2D1), which has two vertebrate orthologs, TR2 (NR2C1) and TR4 (NR2C2). DHR78 protein binds to a subset of EcR/USP-binding sites in vitro, suggesting that it may inhibit ecdysone responses through binding site competition (Fisk and Thummel 1995). Cotransfection assays have supported this model by demonstrating that DHR78 can inhibit the ecdysone-dependent induction of a reporter gene (Zelhof et al. 1995). DHR78-null mutants display growth defects, dying as small L3 (Fisk and Thummel 1998). Interestingly, a similar defect is seen in TR4 mutant mice that display early lethality and significant growth retardation (Collins et al. 2004). The mechanisms that underlie the biological functions of DHR78 or its vertebrate orthologs, however, remain unknown.
Ectopic expression of DHR78 throughout development has no effects on the animal, suggesting that DHR78 is regulated post-transcriptionally (Fisk and Thummel 1998). The most parsimonious explanation for this level of control is a ligand or a specific cofactor that regulates DHR78 activity. Comprehensive screens of natural and synthetic lipophilic compounds and hormones using a GAL4-DHR78 LBD protein in transient transfection assays revealed no candidate ligands (Baker 2002). In contrast, a specific DHR78-interacting protein was identified by a yeast two-hybrid screen and designated Moses, for Middleman of seventy-eight signaling.
In this study we report our initial characterization of Moses. Biochemical evidence demonstrates that Moses exhibits remarkable binding specificity for DHR78. This is confirmed in vivo, where we observe precise colocalization of the two proteins. Biochemical and genetic data indicate that Moses acts as a corepressor for DHR78 and that DHR78 is both necessary and sufficient for Moses protein stability. In addition, the activity of a GAL4-DHR78 fusion protein is sensitive to the genetic dose of moses, implying that levels of Moses protein regulate DHR78 function. In support of this model, lethal hypomorphic alleles of moses exhibit a cancerous phenotype that can be recapitulated by a genetic interaction between moses and DHR78. Taken together, our data indicate that Moses is a dedicated corepressor for DHR78 that regulates both receptor activity and stability in a dose-dependent manner, maintaining appropriate growth during development.
Results
Identification of a specific DHR78-interacting protein
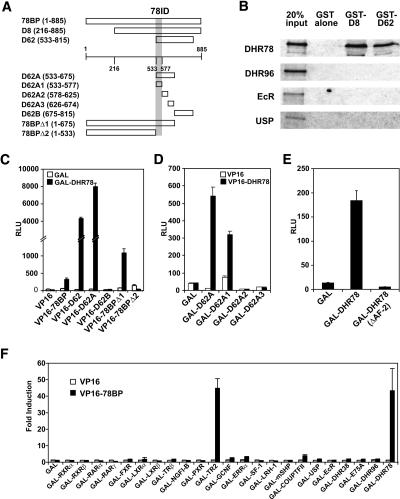
To identify interacting proteins that may regulate DHR78 activity, a mid-L3 cDNA library was subjected to a yeast two-hybrid screen using the DHR78 LBD as bait. Two independent clones were identified, designated D8 and D62, which encode overlapping regions of the same novel protein (Fig. 1A). As expected, both D8 and D62 proteins bind to DHR78 in vitro when assayed by GST pulldown (Fig. 1B). No interaction, however, was observed with three other Drosophila NRs, DHR96, EcR, and USP, suggesting that this interaction is specific to DHR78 (Fig. 1B).
Figure 1.
Identification of a specific DHR78-binding protein. (A) Schematic representation of yeast two-hybrid clones (D8 and D62), full-length version of D8 (78BP), D62A (amino acids 533–675), D62A1 (533–577), D62A2 (578–625), D62A3 (626–674), D62B (675–815), and two C-terminal truncations of 78BP, 78BPΔ1 (1–675) and 78BPΔ2 (1–533), with numbers representing amino acid positions. The DHR78 interaction domain (78ID) is pictured as a gray bar. (B) GST pulldowns show that GST-D8 and GST-D62, but not GST alone, bind to [35S]Met-labeled DHR78 and show no interaction with DHR96, EcR, or USP. (C–F) Mammalian two-hybrid interaction assays in HEK293 cells. (C) GAL or GAL-DHR78 were tested for their ability to interact with a VP16 control, VP16–78BP, VP16–D62, and VP16–D62 constructs as depicted. (D) VP16–DHR78 interaction with GAL-D62A1 but not GAL-D62A2 or GAL-D62A3 defines the 78ID between amino acids 533 and 577. (E) The C-terminally truncated GAL-DHR78ΔAF2 fails to associate with VP16–78BP. (F) A survey of mammalian and Drosophila GAL-LBD fusions shows that VP16–78BP interacts significantly with only DHR78 and TR2.
Using RT–PCR, we isolated 1.2 kb of 5′ sequences upstream of the D8 coding region. Adding these sequences to D8 resulted in a single long ORF encoding an 885-amino-acid protein, designated the 78-binding protein (78BP) (Fig. 1A; Supplementary Fig. 1). To delimit the region of interaction between 78BP and DHR78, a GAL-DHR78 LBD fusion protein was tested for its ability to interact with varying segments of 78BP fused to the VP16 transcriptional activation domain using a mammalian two-hybrid assay. Consistent with our original findings, the DHR78 LBD associated with 78BP and D62 (Fig. 1C). An interaction was also observed between the DHR78 LBD and 78BPΔ1 (missing the C-terminal 210 amino acids), but not with 78BPΔ2 (lacking the C-terminal 353 amino acids) or D62B (encompassing amino acids 675–815). This narrowed the DHR78-interacting domain to a region between 533 and 675 amino acids, consistent with the ability of D62A to bind to the DHR78 LBD (Fig. 1C). These results were confirmed and refined by switching the two components in the mammalian two-hybrid assay using VP16–DHR78 and different segments of 78BP fused to GAL. This study mapped the interaction domain to a region between amino acids 533 and 577, as demonstrated by the ability of D62A1, but not D62A2 or D62A3, to bind to the DHR78 LBD (Fig. 1D). Conversely, deletion of the C-terminal 13 amino acids of DHR78, which contains the canonical AF-2 domain, eliminates DHR78 LBD binding to 78BP, defining this as a conventional AF-2-dependent cofactor interaction (Fig. 1E). Finally, to identify other NRs that could interact with 78BP, we surveyed a panel of GAL-fused LBDs derived from 17 vertebrate and six Drosophila NRs using a mammalian two-hybrid assay. Surprisingly, we only observed a significant association of 78BP with DHR78 and its human ortholog, TR2, suggesting that this novel cofactor is specific to the DHR78 subclass of NRs (Fig. 1F).
The DHR78-binding protein acts as a corepressor
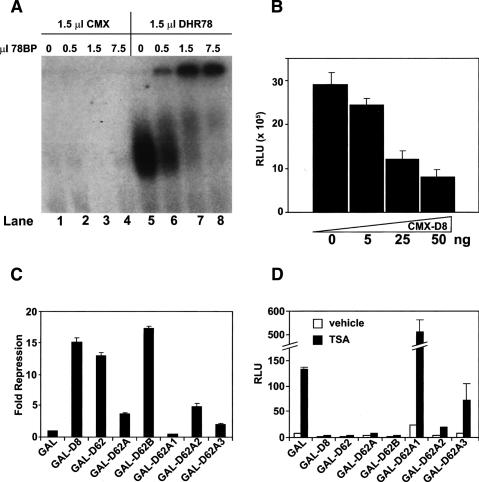
A DNA mobility-shift assay was performed to determine whether 78BP can interact with the DNA-bound form of DHR78. As expected, 78BP does not bind to a CRBPII element that contains two canonical DR1 NR-binding sites (Fig. 2A, lanes 1–4; Mangelsdorf et al. 1991). DHR78, however, can bind to this element (Fig. 2A, lane 5), as reported previously (Zelhof et al. 1995). In addition, 78BP can efficiently bind to the DHR78–CRBPII complex, significantly reducing the electrophoretic mobility of the bound form of DHR78 (Fig. 2A, lanes 6–8). To test whether this interaction affects the transcriptional activity of DHR78, increasing amounts of a CMX-D8 expression construct were cotransfected into HEK293 cells along with a VP16–DHR78 expression construct and a tk-CRBPII-luc reporter gene (Fig. 2B). The D8 protein effectively repressed this activated form of DHR78, indicating that 78BP can act as a corepressor. Mapping studies revealed that the repressive function of 78BP lies within the D62 C-terminal region (Figs. 1A, 2C). The majority of this function maps between amino acids 675 and 815 (D62B), with a second, less-effective repression domain mapping between amino acids 578 and 625 (D62A2) (Fig. 2C). Interestingly, this repressive function of 78BP is not blocked by the addition of trichostatin A (TSA), an inhibitor of type I and type II histone deacetylases (Fig. 2D). This result indicates that 78BP functions differently from conventional NR corepressors such as NCoR and SMRT, and appears to act independently of these deacetylase activities.
Figure 2.
The DHR78-binding protein acts as a corepressor. (A, lanes 1–4) Electrophoretic mobility shift assay showing that in the absence of DHR78, increasing amounts of 78BP fail to associate with a 32P-labeled CRPBII element. In contrast, DHR78 alone binds to the CRBPII element (lane 5), and this complex does not migrate into the gel upon the addition of increasing amounts of 78BP (lanes 6–8). Microliters of in vitro translated protein are shown. (B) VP16–DHR78 activates a tk-CRBPII-luc reporter gene and this response is repressed upon adding increasing amounts of a D8 expression plasmid (CMX-D8). (C) Cotransfection of HEK293 cells showing that GAL-fused constructs of 78BP mediate transcriptional repression through at least two separate domains, D62B and D62A2. (D) GAL-fused constructs of 78BP mediate transcriptional repression both in the absence (open bars) and presence (black bars) of 1 μM TSA in HEK293 cells.
The DHR78-binding protein corresponds to Moses, a SAM (sterile α motif) domain protein
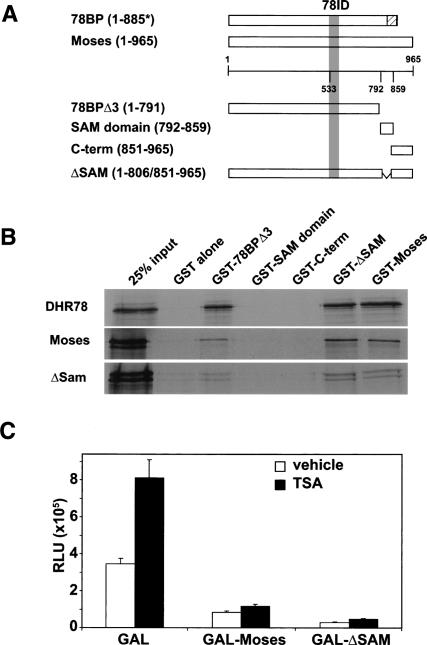
Consistent with a mechanism of action that is distinct from that of other NR corepressors such as NCoR and SMRT, 78BP does not contain any of the conserved protein domains that define these cofactors (Privalsky 2004; Rosenfeld et al. 2006). Rather, it has only a single recognizable feature, a C-terminal SAM domain, although this domain is truncated at the C terminus of 78BP. Comparison of the 78BP sequence with the Drosophila genomic databases revealed that the corresponding annotated genome sequence and EST clones contain an additional 37 base pairs two-thirds of the way through sequences that encode a full-length SAM domain (Grumbling and Strelets 2006). RT–PCR using RNA isolated from several Drosophila strains, including the stock from which the yeast two-hybrid library was derived, failed to detect the sequence change present in the 78BP cDNA. Thus, the full-length DHR78-interacting protein is 965 amino acids in length, with a canonical C-terminal 68-amino-acid SAM domain (Fig. 3A; Supplementary Fig. 1; Qiao and Bowie 2005). We have designated this protein as Moses. The Moses SAM domain is one of six encoded by the Drosophila genome that are designated as SPM domains, based on sequence identity (Peterson et al. 2004). Interestingly, the other characterized Drosophila proteins in this subfamily act as transcriptional repressors. As expected, Moses retains the ability to efficiently bind DHR78, as demonstrated by a GST-pulldown assay (Fig. 3B). Moses also acts as a repressor that is resistant to TSA (Fig. 3C), consistent with the presence of both repression domains, located between amino acids 675 and 815 and 578 and 625 (Fig. 2C; Supplementary Fig. 1). In addition, Moses can bind to itself, although this binding occurs independently of its SAM domain (Fig. 3B). The SAM domain also does not contribute to the repressive function of Moses (Fig. 3C).
Figure 3.
Moses binds to itself and DHR78, and acts as a transcriptional repressor. (A) Schematic representation of full-length Moses (amino acids 1–965), C-terminally truncated Moses (78BPΔ3, amino acids 1–791), the Moses SAM domain (amino acids 792–859), an N-terminal Moses deletion (C-term, amino acids 851–965), and the SAM domain deletion (ΔSAM, deletion of amino acids 806–851). The hatched box on 78BP (amino acids 835–885) represents where its sequence differs from that of Moses. The DHR78 interaction domain (78ID) is pictured as a gray bar. (B) GST pulldowns showing that GST-78BPΔ3, GST-ΔSAM, and GST-Moses, but not GST, GST-SAM, or GST-C-term, bind to [35S]Met-labeled DHR78, Moses, and ΔSAM. (C) GAL-Moses and GAL-ΔSAM inhibit basal transcription over GAL alone, in the absence (open bars) or presence (black bars) of 1 μM TSA in HEK293 cells.
Moses and DHR78 are coexpressed during development
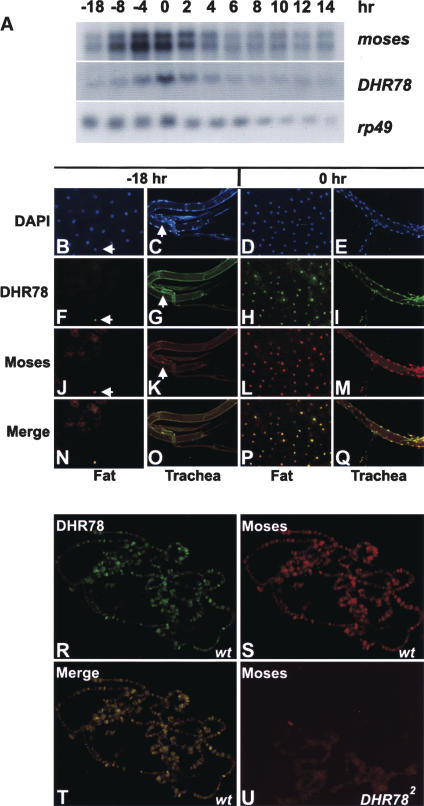
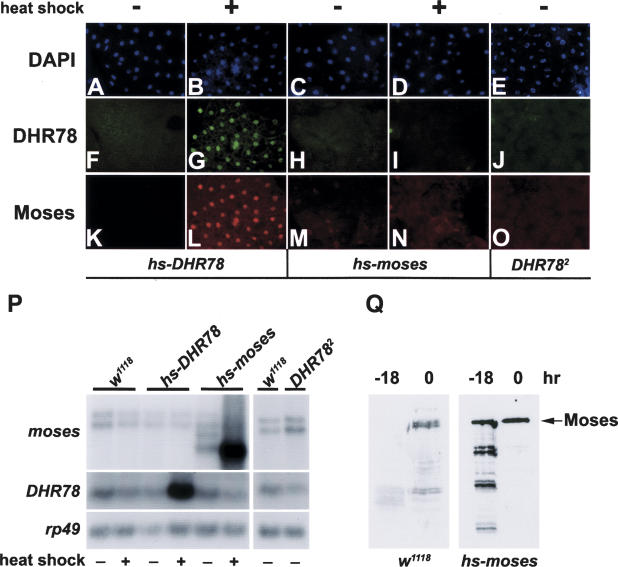
The identification of Moses as a specific interacting factor for DHR78 raises the possibility that these proteins might be coexpressed in vivo. As a first step toward addressing this issue, Northern blot hybridization was used to determine the temporal patterns of DHR78 and moses transcription from the mid-L3, when DHR78 mutants arrest development, through prepupal stages (Fig. 4A). Both moses and DHR78 transcripts are detectable throughout this time course, with peaks of expression centered around puparium formation (0 h in Fig. 4). The overall fold change in DHR78 transcript levels, however, is higher than that of moses during this time course. Similar results were seen when tissues dissected from mid-L3 (−18 h) or newly formed prepupae (0 h) were stained with affinity-purified polyclonal antibodies specific for Moses or DHR78 (Fig. 4B–Q). The spatial expression pattern of Moses follows that of DHR78 described earlier (Fisk and Thummel 1998). Curiously, however, rather than seeing widespread low levels of Moses and DHR78 protein in mid-L3, relatively high levels of protein accumulation are detected in only a few nuclei in each tissue (Fig. 4F,G,J,K, arrows). Similarly, although DHR78 and Moses are more widespread in tissues from newly formed prepupae, their distribution and apparent abundance continue to track precisely with one another (Fig. 4H,I,L,M,P,Q). This colocalization was also seen at specific DHR78-binding sites in the giant larval salivary gland polytene chromosomes (Fig. 4R–T). Importantly, the binding of Moses to chromatin is absolutely dependent on DHR78, in that this binding is not seen in a DHR78 mutant animal (Fig. 4U), consistent with our in vitro mobility shift assays (Fig. 2A).
Figure 4.
Spatial and temporal expression profiles of Moses and DHR78. (A) Northern blot analysis to detect moses and DHR78 transcripts of w1118 animals staged as mid-L3 (−18 and −8 h), late L3 (−4 h), newly formed prepupae (0 h), prepupae staged at 2-h intervals (2, 4, 6, 8, and 10 h), and pupae (12 and 14 h). Hours are relative to puparium formation. Hybridization to detect rp49 mRNA was included as a control for loading and transfer. (B–Q) Fat body and trachea from either −18-h mid-L3 or 0-h prepupae were stained with DAPI (B–E), DHR78 antibodies (F–I), or Moses antibodies (J–M). (N–Q) A merge of the antibody patterns. White arrows mark nuclei that contain DHR78 and Moses protein. (R–T) Antibody stains of giant salivary gland polytene chromosomes from late L3 (−4 h) w1118 animals (wt) to detect DHR78 (R), Moses (S), and the merge of the two (T) bound to chromatin. (U) Antibody stains of Moses in hs-DHR78/hs-DHR78; DHR782/DHR782 mutants that have been rescued to late L3 (−4 h) by transient expression of a DHR78 transgene during embryogenesis for 30 min at 37°C and maintained at 18°C through development, showing loss of chromatin localization.
DHR78 protein stabilizes Moses
The precise colocalization of Moses and DHR78 raised the possibility that Moses protein stability might depend upon its association with DHR78. Consistent with this hypothesis, ectopic overexpression of DHR78 in mid-L3 fat body (Fig. 5G), where endogenous protein levels of both DHR78 and Moses are normally low (Fig. 4F,J), results in high levels of Moses protein (Fig. 5L). Conversely, removal of DHR78 from the fat body of newly formed prepupae (Fig. 5J), where DHR78 and Moses are both relatively abundant (Fig. 4H,L), results in a loss of Moses protein (Fig. 5O). Remarkably, even high levels of heat-induced Moses expression in mid-L3 fat bodies (hs-moses) results in no significant Moses protein accumulation or effects on DHR78 protein levels (Fig. 5I,N). In all cases, only minor changes were seen in levels of endogenous moses or DHR78 mRNA under these different experimental conditions (Fig. 5P). Thus, the stability of Moses protein appears to be absolutely dependent upon its DHR78 partner. This conclusion is supported by Western blot analyses of protein extracts from wild-type and hs-moses animals. In control w1118 animals, full-length Moses protein is only detectable when DHR78 levels are high (0 h), with degraded fragments visible when DHR78 levels are low (−18 h) (Fig. 5Q). This effect is pronounced in heat-treated hs-moses animals, where apparent degradation products of Moses are only seen at −18 h (Fig. 5Q). Taken together, these data indicate that DHR78 is an obligate partner for Moses.
Figure 5.
Moses protein is stabilized by DHR78. Fat bodies stained with DAPI (A–E), or antibodies directed against DHR78 (F–J) or Moses (K–O), from either mid-L3 (A–D,F–I,K–N) or 0-h prepupae (E,J,O). Either hs-DHR78 transformants, hs-moses transformants, or DHR782 mutants were examined, as indicated. The DHR782 mutants were rescued to later stages as described in the legend for Figure 4U. Animals were heat treated (+) or not (−) for 1 h at 37°C and then allowed to develop for 3 h prior to collection. (P) Northern blot analysis of w1118, hs-DHR78, hs-moses animals, and DHR782 mutants, staged and treated as described above, showing that ectopic expression of DHR78 and moses, and the DHR782 point mutation, do not significantly affect the levels of endogenous moses and DHR78 mRNA. Hybridization to detect rp49 mRNA was included as a control. (Q) Western blot analysis of protein extracts from heat-treated w1118 and the hs-moses transformant at the indicated times relative to pupariation, using antibodies against Moses, showing that Moses is unstable at a time when DHR78 protein is low (−18 h), but not when DHR78 is high (0 h). The relative level of protein assayed is 10-fold higher for w1118 lanes (equivalent to 0.5 animal per lane) than hs-moses lanes (equivalent to 0.05 animal per lane).
moses mutants display massive overgrowth
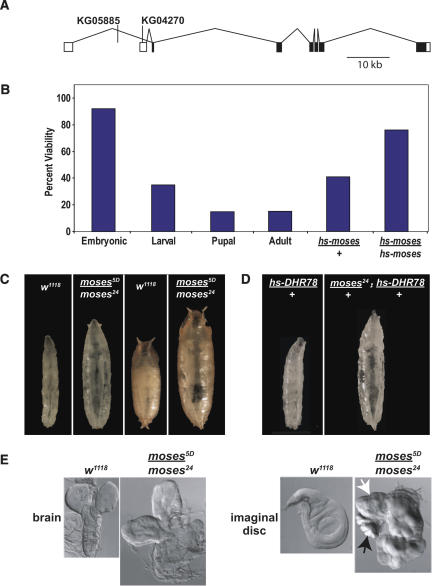
If Moses activity is linked to that of DHR78, then we would expect to see functional interactions between the genes that encode these two proteins. As a first step toward testing this possibility, we used imprecise excision of two viable P-element insertions located in the 5′ untranslated region (UTR) of moses, KG05885, and KG04270, to create moses mutant alleles (Fig. 6A). Two mutations were selected for further characterization: moses24 derived from KG05885 and moses5D derived from KG04270. These mutations fail to complement each other in trans as well as a deficiency for the region, Df(2L)J39. Antibody stains reveal that Moses protein levels are significantly reduced, but not absent, in moses5D and moses24 mutants (data not shown). In addition, PCR and DNA sequencing reveal that the lesions associated with these mutations are located in the introns, leaving the coding region intact in both mutants. The moses5D and moses24 mutations thus appear to represent hypomorphic alleles of moses. Trans-heterozygous moses5D/moses24 mutants die as late L3 or early prepupae, with 15% of the animals escaping to adulthood with no apparent defects (Fig. 6B). The number of adult survivors is significantly increased by introducing one or two copies of a heat-inducible wild-type moses transgene (hs-moses) into the mutant background (Fig. 6B), demonstrating that the mutant alleles specifically disrupt moses function. In addition, the dose-dependent rescue of moses lethality suggests that the amount of moses gene product may be important for its activity. Overexpression of Moses, either with or without the SAM domain, has no effect on development, similar to ectopic DHR78 overexpression (Fisk and Thummel 1998). Also like DHR78, moses mutants progress normally through L1 and L2. In contrast to the growth defects seen in DHR78 mutant larvae, however, moses mutants display an opposite overgrowth phenotype (Fig. 6C). These animals are characterized by marked delays of up to 10–14 d in pupariation, along with severe hemolymph-induced bloating, fat clearing, and hyperplasic overgrowth of the imaginal discs and brain (Fig. 6C,E). Similar overgrowth phenotypes are seen in trans-heterozygous moses5D/moses24 mutants or in animals carrying either moses5D or moses24 in combination with Df(2L)J39.
Figure 6.
Mutations in moses are lethal and lead to uncontrolled growth. (A) Schematic representation of the moses locus on chromosome 2 shown from 5′ (left) to 3′ (right), with UTRs (white boxes) and protein-coding regions (black boxes) as well as the sites of two P-element insertions used for imprecise excisions. (B) Graph showing the viability of staged moses5D/moses24 animals relative to internal control heterozygotes from a total of 800 embryos that were followed through development. The lethality of moses5D/moses24 mutants can be rescued to adulthood with increasing levels of moses (hs-moses/+ and hs-moses/hs-moses, n = 400 for each). (C) The phenotypes of w1118 control or moses5D/moses24 mutants are shown as late L3 (left two panels) or early pupae (right two panels). moses mutants pictured are 7–10 d older than the controls in order to show the overgrowth phenotype. All moses mutants that arrest as late L3 or prepupae display overgrowth and disc hyperplasia. (D) Heat-treated moses24/+;hs-DHR78/+ L3 (right panel), but not hs-DHR78/+ control L3 (left panel), display an overgrowth phenotype indistinguishable from moses5D/moses24 mutants. (E) Late L3 brains and wing imaginal discs from control w1118 or moses5D/moses24 mutants. Note the fused haltere (black arrow) and leg (white arrow) imaginal discs in mutants.
Moses levels regulate DHR78 activity in vivo
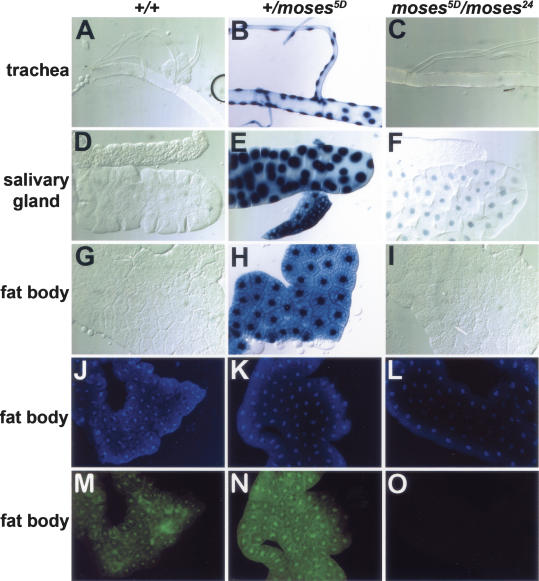
We used the GAL4-LBD activation system to determine whether Moses is acting as a corepressor for DHR78 in vivo. Transgenic lines were constructed that carry a heat-inducible gene encoding the yeast GAL4 DBD fused to the DHR78 LBD (hsp70-GAL4-DHR78), in combination with a GAL4-dependent UAS-nlacZ reporter gene that directs the synthesis of nuclear-localized β-galactosidase. This system has been shown to accurately reflect the spatial patterns of NR LBD activation at different stages of development (Kozlova and Thummel 2002; Palanker et al. 2006). Consistent with Moses acting as a transcriptional corepressor for DHR78, no β-galactosidase activity was detected in wild-type L3 (Fig. 7A,D,G). In contrast, reducing the levels of Moses by introducing a single mutant copy of the gene resulted in strong GAL4-DHR78 activation (Fig. 7B,E,H). Surprisingly, however, further reducing Moses levels by using the transheterozygous moses mutant resulted in a loss of β-galactosidase activity (Fig. 7C,F,I). This loss of activity can be explained by an absence of the GAL4-DHR78 fusion protein in moses5D/moses24 mutants (Fig. 7O), whereas high levels of heat-induced protein were readily detectable in both wild-type and moses5D/+ animals (Fig. 7M,N). Similarly, heat-induced GAL4-DHR78 protein can only be detected in tissues of wild-type animals where Moses is expressed (data not shown). Thus, the genetic dose of moses is critical for both GAL4-DHR78 transcriptional activity and protein stability.
Figure 7.
Moses levels affect GAL4-DHR78 activity and stability. Trachea (A–C), salivary glands (D–F), and fat bodies (G–I) from hs-GAL4-DHR78/UAS-nlacZ late L3 that are either wild type for moses (+/+), or with one (+/moses5D) or two (moses5D/moses24) mutant copies of moses. Animals were heat treated at 37°C for 30 min to induce GAL4-DHR78 expression prior to staging and collection. Nuclear β-galactosidase was detected by X-gal staining. Fat bodies from animals described above were stained with DAPI (J–L) or anti-GAL4 antibodies to detect the GAL4-DHR78 fusion protein (M–O). Similar results to those shown for +/moses5D were seen in +/moses24 animals (data not shown).
Functional interactions between DHR78 and Moses
The genetic rescue experiments (Fig. 6B) and GAL4-LBD activation studies (Fig. 7) each indicate that moses acts in a dose-dependent manner. If this is true, and if moses acts exclusively through DHR78, then we should be able to observe functional interactions by altering the relative genetic dose of DHR78 and moses. To test this, we overexpressed DHR78 in moses24/+ animals. Control animals that carry one mutant copy of moses (moses24/+), or heat-treated hs-DHR78/+ transformants, display no developmental defects, surviving normally (Fig. 6D; Fisk and Thummel 1998). In contrast, ∼3% of heat-treated moses24/+; hs-DHR78/+ animals (n = 400) display overgrowth phenotypes that are indistinguishable from those of moses mutants (Fig. 6D). This genetic interaction, combined with the DHR78 and moses mutant phenotypes, suggests that DHR78 and Moses normally act together to regulate growth and suppress cancer.
Discussion
This study describes the isolation and initial characterization of a novel cofactor, Moses, which appears to be dedicated to the DHR78 subclass of NRs. We show that DHR78 is an obligate partner for Moses and that Moses acts as a corepressor in a dose-dependent manner. Below, we discuss the unique features of Moses that set it apart from other NR cofactors and present a model for understanding how this factor acts together with DHR78 to regulate growth during development.
Moses is a novel SAM domain-containing NR corepressor
Corepressors are well known to play a central role in NR regulation. The best characterized of these are SMRT and NCoR, which share significant blocks of sequence similarity and act through a wide range of NRs (for reviews, see Privalsky 2004; Rosenfeld et al. 2006). These corepressors function as platforms that recruit higher-order protein complexes, including histone deacetylases that establish and maintain a repressed chromatin state. A Drosophila homolog of SMRT has been identified, SMRTER, which shares limited sequence identity with its vertebrate counterpart, including a SANT domain, as well as some functional characteristics (Tsai et al. 1999). A novel corepressor has also been identified in Drosophila, designated Alien, although, like NCoR and SMRT, it interacts with multiple mammalian NRs and exerts its repressive effect through histone deacetylation (Dressel et al. 1999).
Moses distinguishes itself from these more conventional corepressors in several ways. First, Moses displays an unusual degree of binding specificity, showing selective interactions with DHR78 and its mammalian ortholog TR2, suggesting that its function is restricted to this small subfamily of NRs (Fig. 1F). The observation that Moses protein stability is absolutely dependent on DHR78 in newly formed prepupae, when at least half of the other Drosophila NRs are expressed (Sullivan and Thummel 2003), supports the proposal that this binding specificity also occurs in vivo (Fig. 5). Second, Moses exerts its repressive effect in the presence of TSA, suggesting that it does not act through type I or type II histone deacetylases like other known corepressors (Figs. 2D, 3C). Third, Moses protein stability is absolutely dependent on DHR78, suggesting that the receptor is its obligate partner and that all Moses functions proceed through DHR78. This can be seen in the precise colocalization of DHR78 and Moses protein in vivo as well as at the level of specific binding sites in the genome, where Moses binding overlaps that of DHR78 and is dependent on its NR partner (Figs. 4, 5). To our knowledge, no other cofactor has been shown to have such a tight functional association with a NR. Fourth, Moses regulates DHR78 activity in a novel dose-dependent manner. This suggests that Moses acts as a repressor by recruiting itself to the complex, and that increasing amounts of Moses exert more repressive function, as discussed in more detail below. Finally, Moses does not contain any conserved motifs with SMRT or NCoR. Rather, Moses is the first SAM domain-containing protein that has been shown to act as an NR cofactor. Like other SAM domain-containing proteins, such as Polyhomeotic (PH) and TEL, Moses acts as a repressor (for review, see Qiao and Bowie 2005). It does so, however, independently of its SAM domain, indicating that it does not repress transcription through SAM-mediated homopolymerization, as has been proposed for PH and TEL (Kim et al. 2001, 2002). In addition, the sequences between amino acids 533 and 577 that are sufficient and necessary for Moses binding to DHR78 do not contain a canonical LXXLL motif or its known variants that normally provide this function (Privalsky 2004; Rosenfeld et al. 2006). Taken together, our studies of Moses provide a new paradigm for NR–corepressor interactions, defining SAM domain proteins as a new member of this regulatory class and showing that dose-dependent NR–cofactor interactions can modulate receptor activity.
A model for the regulatory interactions between DHR78 and Moses
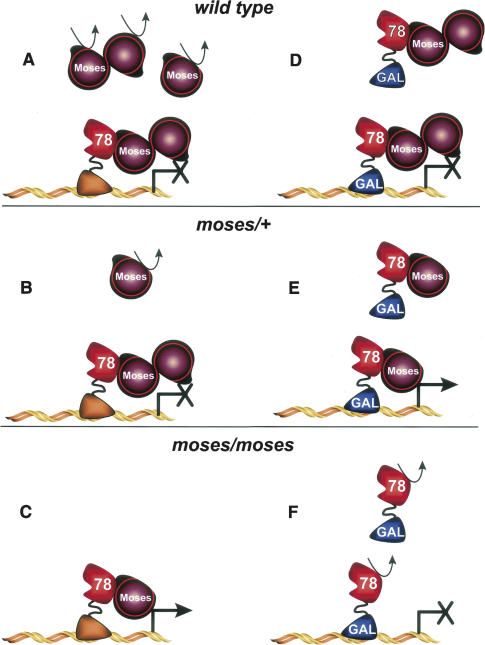
The precise colocalization of DHR78 and Moses indicates that DHR78 function cannot be understood except in the context of its corepressor, Moses. Although the mechanisms by which Moses modulates DHR78 activity remain unclear, many of our observations can be explained by a model for DHR78–Moses functional associations (Fig. 8). We propose that in wild-type animals, Moses protein that is bound to DHR78 forms a stable complex, with multiple copies of Moses repressing DHR78 transcriptional activity through an as-yet-unknown mechanism (Fig. 8A). Normal steady-state levels of Moses result from the combination of its stable association with DHR78 and the rapid degradation of newly translated unbound Moses protein. Lowering the dose of Moses by introducing a single mutant allele (moses/+) provides sufficient Moses protein to maintain DHR78-mediated repression, although the overall steady-state level of Moses protein is reduced (Fig. 8B). Upon combining two hypomorphic moses mutant alleles, however, Moses protein falls below a threshold level that is required to maintain DHR78 repression. As a result, DHR78-regulated genes are ectopically activated, resulting in overgrowth and cancer (Fig. 8C).
Figure 8.
A model for functional interactions between DHR78 and Moses. Either endogenous DHR78 (left half) or heat-induced GAL4-DHR78 fusion protein (right half) are depicted, along with different doses of Moses protein. Wild-type (A,D), moses/+ (B,E), or moses/moses mutant (C,F) genetic backgrounds are shown. The curved arrow represents the degradation of unstable protein. See text for a detailed discussion of the model.
This model can also be used to explain our results with the GAL4-LBD activation study. We propose that heat-induced GAL4-DHR78 protein can only associate with free Moses protein, stabilizing that protein and forming a repressive complex on the UAS-lacZ promoter (Fig. 8D). The Moses protein that is bound to endogenous DHR78 remains in that stable complex, unavailable for association with GAL4-DHR78. In the presence of a single moses mutant allele, although enough free Moses protein is available to stabilize the heat-induced GAL4-DHR78 protein, it is no longer sufficient to act as a corepressor, leading to high levels of GAL4-LBD activity (Fig. 8E). Finally, in moses hypomorphic mutants, Moses protein is primarily in a stable association with endogenous DHR78, leaving no free protein to stabilize the heat-induced GAL4-DHR78 fusion, which is rapidly turned over (Fig. 8F).
A prediction of this model is that DHR78 and Moses form an obligate complex, with one factor dependent upon the other for their stable accumulation. Although this has been demonstrated for Moses binding to DHR78 (Fig. 5), only the GAL4-DHR78 fusion protein has been shown to be dependent on Moses for its stable accumulation (Fig. 7). Attempts to examine DHR78 protein levels in moses5D/moses24 mutants revealed uniformly reduced levels of DHR78 protein, correlating with the lower level of Moses that is present in these mutants (data not shown). This is consistent with our model, in that sufficient Moses may be present to maintain DHR78 stability, although this complex is abnormally activated, directing overgrowth (Fig. 8C). One future direction for our research is to isolate a moses-null mutant, with the prediction that DHR78 protein should be absent in this genetic background. Intensive efforts to recover such mutants by P-element imprecise excision were unsuccessful, indicating that other methods, such as gene targeting, will be required to achieve this goal.
Finally, our studies leave us with the question of how the DHR78–Moses complex normally controls target gene transcription during development. When and where is the repression of DHR78 by Moses overcome, and what facilitates this regulation? One clue arises from our GAL4-DHR78 activation study, in which the activity we observe is spatially restricted to those tissues where DHR78 and Moses are normally expressed (data not shown). This suggests that another factor is required to activate the DHR78–Moses complex, possibly a specific coactivator. Further molecular screens with DHR78 and DHR78–Moses may allow us to identify this putative additional component of the DHR78 transcriptional complex.
Our work also raises the possibility that a SAM domain protein may control TR2 and TR4 activity in vertebrates. Relatively little is known about the regulatory properties of these two orphan NRs, although TR2 appears to function as a repressor (Chinpaisal et al. 1998; Franco et al. 2001). Specific phosphorylation can increase the stability of TR2 and lead to recruitment of the P/CAF coactivator in cultured cells (Khan et al. 2005, 2006), while the phosphorylation state of the TR4 AF-1 domain can regulate its transcriptional activity (Huq et al. 2006). A vertebrate corepressor for TR4 has been identified, designated TIP27, characterized by two zinc-finger motifs (Nakajima et al. 2004). Its mechanism of action and in vivo functions, however, remain undefined. Scanning of the human protein sequence databases with Moses reveals several proteins with significant sequence identity that is restricted to the SAM domain. Although it would be interesting to test whether these SAM domain proteins could act as TR2 or TR4 cofactors, the restricted sequence identity suggests that this activity will not be evident through cross-species sequence comparisons, as was shown for the Drosophila SMRTER corepressor (Tsai et al. 1999). Rather, discovery of the functional mammalian counterpart for Moses is likely to require direct molecular screens for TR2/TR4 cofactor binding and function.
DHR78 and Moses act together to regulate growth and prevent cancer
Studies in Drosophila have led to key insights regarding fundamental biological mechanisms that control cancer, such as the discovery and characterization of the Notch and Wnt signaling pathways (for recent reviews, see Brumby and Richardson 2005; Vidal and Cagan 2006). No ties, however, have been made between cancer in Drosophila and steroid hormone signaling or NR function—central players in several forms of human cancer, including breast cancer (for reviews, see Koehler et al. 2005; Lonard and O’Malley 2006). This study describes the first evidence of such a link, providing a basis for using Drosophila to further our understanding of this critical human disease. While DHR78-null mutants progress normally through early development, they show marked defects in growth at the end of the juvenile larval growth phase and fail to initiate maturation during metamorphosis. Remarkably, TR4 mutant mice display a similar phenotype. These mutants are indistinguishable from their wild-type siblings at the time of birth, but fail to grow at a normal rate during juvenile stages (Collins et al. 2004). The cause of this growth defect is unknown, although preliminary studies indicate that it is not due to feeding behavior or defective growth hormone secretion (Collins et al. 2004). Moses mutants also progress normally through early stages and stall in their development at the end of larval development. Interestingly, however, their terminal phenotype is opposite that of DHR78 mutants, displaying overgrowth, with tumors forming in the brains and imaginal discs. These hypertrophic imaginal tissues gradually fuse with one another, forming sheets of cells that are indistinguishable from those seen in Drosophila tumor suppressor mutants (Gateff 1978; Gateff et al. 1993). These opposing phenotypes fit with a role for Moses in repressing DHR78 activity and predict that DHR78 is normally required to promote growth, with Moses acting to restrict that function. This conclusion is consistent with the low frequency of overgrowth observed in heat-treated moses24/+; hs-DHR78/+ animals (Fig. 6D), where overexpressed DHR78 may escape the dose-dependent repression by Moses in a few animals, promoting unrestricted growth.
Both TR2 and TR4 are expressed in breast-cancer cell lines and have been proposed to contribute to this form of cancer. In MCF-7 cells, TR2 or TR4 can suppress estrogen-receptor (ER) signaling as well as inhibit estrogen-induced cell proliferation, suggesting possible cross-talk with ER in breast cancer (Hu et al. 2002; Shyr et al. 2002). Similar to the reported ability of DHR78 to compete for binding to sites normally recognized by the EcR/USP ecdysteroid receptor (Zelhof et al. 1995), TR2 and TR4 inhibit ER DNA binding, although this interaction is achieved by heterodimerization rather than binding site competition (Hu et al. 2002). Direct roles for TR2 and TR4 in cancer, however, remain to be defined. The observation that moses mutants, and genetic interactions between DHR78 and moses, lead to cancer provides an invertebrate genetic model system to pursue these studies.
One candidate for mediating the effects of DHR78/Moses on growth is the Moses SAM domain. This domain is not required for Moses homophilic interactions or repressive functions, yet SAM domains have been well defined as critical sites for interactions with other SAM domain proteins (Qiao and Bowie 2005). A possible candidate for interacting with Moses to regulate growth is the protein encoded by lethal(3)malignant brain tumor (l(3)mbt) (Gateff et al. 1993). Mutants in l(3)mbt display phenotypes that are essentially identical to those seen in hypomorphic moses mutants, including late L3/early prepupal lethality, increased size, and brain and imaginal disc neoplasms (Gateff et al. 1993). An attractive model is that the DHR78/Moses complex regulates growth through its association with L(3)MBT, mediated by SAM–SAM heterophilic interactions. Experiments are in progress to test this possibility.
The identification of growth defects as a result of DHR78 and moses mutations provides an ideal opportunity to exploit Drosophila genetics for defining the roles of NRs and their cofactors in cancer. We anticipate that further characterization of the functional interactions between DHR78 and Moses will provide a mechanistic framework for understanding how the DHR78/TR2/TR4 class of NRs function in normal development and disease, as well as a foundation for understanding how NR–cofactor interactions can regulate growth.
Materials and methods
Plasmids
Yeast two-hybrid libraries were screened with DHR78 (amino acids 118–599) using pVJL11 as a vector (Jullien-Flores et al. 1995). CMX-GAL (Umesono and Evans 1989) and CMX-VP16 (Forman et al. 1995) were used for cotransfections. CMX-GAL-DHR78, CMX-GAL-DHR96, CMX-GAL-EcR, and CMX-GAL-USP are described (Baker et al. 2000). CMX-GAL-DHR78ΔAF2 was constructed by deleting coding sequences for the C-terminal 13 amino acids of DHR78 (587–599) from CMX-GAL-DHR78 by PCR. CMX-GAL-Moses and CMX-GAL-ΔSAM encode for amino acids 1–965 and 1–806/851–965, respectively, of Moses. CMX-GAL-D8, CMX-GAL-D62, CMX-GAL-D62A, CMX-GAL-D62A1, CMX-GAL-D62A2, CMX-GAL-D62A3, CMX-GAL-D62B, and CMX-D8 encode for amino acids 216–885, 533–815, 533–577, 578–625, 626–674, 675–815, and 216–885, respectively. GAL-RXRα, GAL-RXRβ, GAL-RARα, GAL-RARγ, GAL-FXR, GAL-LXRα, GAL-TRβ, and GAL-LRH-1 are described (Giguere et al. 1987; Ishikawa et al. 1990; Mangelsdorf et al. 1990, 1992; Forman et al. 1995; Janowski et al. 1996; Makishima et al. 1999; Lu et al. 2000). GAL-LXRβ, GAL-NGFI-B, GAL-PXR, GAL-TR2, GAL-GCNF, GAL-ERRα, GAL-SF-1, GAL-mSHP, and GAL-COUPTFII were constructed and encode for amino acids 155–461, 333–598, 108–434, 179–483, 122–476, 145–423, 79–461, 1–260, and 145–414, respectively. CMX-VP16–78BP, CMX-VP16–DHR78, CMX-VP16–D62, CMX-VP16–D62A, CMX-VP16–D62B, CMX-VP16–78BPΔ1, and CMX-VP16–78BPΔ2 encode for amino acids 1–885, 1–599, 533–815, 533–675, 675–815, 1–675, and 1–532 of 78BP, respectively. For GST pulldowns, cDNAs corresponding to 78BP, Moses, ΔSAM, DHR78, DHR96, EcR, and USP (amino acids 1–885, 1–965, 1–806/851–965, 1–599, 1–723, 1–849, 1–508, respectively) were each cloned into pBS(SK−) (Stratagene). For protein expression, D8, D62, 78BPΔ3, SAM domain, C-term, ΔSAM, and Moses (amino acids 216–885 and 533–815 of 78BP, respectively, and amino acids 1–791, 792–859, 851–965, 1–806/851–965, and 1–965 of Moses, respectively) were cloned into pGEX-4T-1 or pGEX-5X-1 (Amersham Biosciences).
Yeast two-hybrid screen
A male Drosophila cDNA library, constructed from wandering L3, kindly provided by M. Kuroda (Harvard Medical School, Boston, MA), was screened for interactions using the bait plasmid pVJL11-DHR78 LBD essentially as described (Hama et al. 1999). The library uses the λACT phage in conjunction with the BNN 132 Escherichia coli host bacteria strain (Durfee et al. 1993). Isolated clones in pACT were PCR-amplified and sequenced for identification.
Rapid amplification of cDNA ends (5′ RACE) and RT–PCR
5′ RACE was used to extend the 5′ sequences of the D8 cDNA. RNA isolated from SL2 cells (TriPure, Roche) was used as a template along with an oligonucleotide derived from the 5′ 100 base pairs (bp) of the D8 clone and the poly(dA) primer provided in the 5′ RACE kit (Roche). For cloning the 3′ end of moses, RNA was isolated from 0-h w1118 prepupae (TriPure, Roche) and used as a template for an RT–PCR reaction (Takara) with the following primers: (5′–3′) GGTTAACTCCTGGTCTGTG GACG and GGACAGGATCGCTGCTAACTAGC.
Protein purification, GST pulldowns, and electrophoretic mobility shift assay (EMSA)
pGEX vectors were transformed into BL21-DE3-p bacteria (Invitrogen) and grown in LB-Amp to an OD600 of 0.6. Cultures were induced with 0.5 mM IPTG (Sigma-Aldrich) for 35–75 min. Cultures were pelleted and washed in PBS, resuspended in ice-cold PBS (5 mL/g bacteria) with 0.1% Tween 20 (Mallinckrodt Baker) containing protease inhibitor cocktail tablets (Roche), 100 μg/mL lysozyme (Sigma-Aldrich), 50 U/mL DNase I (Amersham), and 1 mM dithiothreitol (Sigma-Aldrich). Cells were lysed by sonication and the lysate was cleared by centrifugation at 43,000g for 30 min. Protein was bound and washed by the batch method to Glutathione Sepharose 4B according to the manufacturer’s protocol (Amersham). GST pulldowns were performed essentially as described (Lu et al. 2000) with 2 μg of purified GST or GST-fused protein. EMSA were performed essentially as described (Willy et al. 1995).
Cell culture and cotransfection assays
HEK293 cells were maintained and transfected as described (Lu et al. 2000). TSA was solubilized in ethanol, diluted 1:1000 in medium, and added to cells 8 h after transfection. Cells were harvested 24–26 h after transfection and assayed for luciferase and β-galactosidase activities. For experiments in Figures 1 and 2, assays were done as described (Baker et al. 2003). For data shown in Figure 3C, 75 μL per well (of 100 μL total lysate) was read for luciferase activity using the Bright-Glo Luciferase Assay System (Promega). This was followed by transferring 40 μL per well to a new 96-well plate, adding 125 μL per well ONPG buffer (Baker et al. 2003), and incubating at 37°C for color development. Light units and OD420 were read on a POLARstar Optima (BMG Labtech). All data represent the mean ± SD of at least three independent assays.
Lethal phase analysis and staging of animals
For lethal phase analysis, animals were maintained on agar plates supplemented with yeast paste and assayed every 24 h for developmental progression. Transheterozygous moses mutants were identified by their lack of GFP expression associated with the balancer chromosome. Third instar larvae were staged by the addition of 0.5% bromophenol blue to the food as previously described, with wandering blue gut mid-L3 referred to as −18 h relative to pupariation and clear gut late L3 as −4 h (Andres and Thummel 1994). Prepupae and pupae were staged in hours after puparium formation.
Northern blot hybridization
Total RNA was isolated using TriPure Isolation Reagent (Roche) from staged animals, fractionated by formaldehyde gel electrophoresis, transferred to nylon membranes, and probed with radioactively labeled probes (Beckstead et al. 2005). rp49 was used as a loading and transfer control. Two probes (A-B and C-D) were used simultaneously to detect moses transcripts. The following primers were used to generate these probes: for A-B, (5′–3′) CGATCGCTGCTGCTCAGCGGATATG and CCATC GGTGCATCCTTATGATTGTC; and for C-D, GCACTGCT GACACTGGCGCGTG and CGAGTAGATCAGAGCGGGT GTTAC. The probe for DHR78 was as described (Fisk and Thummel 1998).
Western blot and antibody staining and GAL4 activation assay
Larval tissues were isolated by dissection and fixed in 4% formaldehyde/1× PBS on ice for 20 min. Salivary-gland polytene chromosomes were fixed and squashed according to a standard protocol (Andrew and Scott 1994). DAPI (4′,6-Diamidino-2-phenyindole, dilactate) (Sigma-Aldrich) was added to Vectashield (Vector Laboratories) at 0.2 mg/mL final concentration. For Westerns, larvae were homogenized in PBS and boiled for 5 min in SDS loading buffer. Samples were subjected to SDS-PAGE and transferred to a nitrocellulose membrane for analysis. Antibody binding was detected by SuperSignal reagent according to the supplied protocol (Pierce Biotechnology). For GAL4 activation assays, fixed tissue was stained in 0.2% X-GAL (Roche) for 15 min at 37°C. To generate the Moses antibody, a DNA fragment corresponding to amino acids 553–813 was cloned into the pMAL-c2X expression vector (New England BioLabs). This region encompasses both the DHR78 interaction domain and the repressive function of Moses. The purified fusion protein was injected into guinea pigs (Rockland Immunochemicals). Antiserum was affinity-purified as described (Carroll and Laughon 1987), using a Moses protein containing amino acids 216–815. Affinity-purified anti-Moses antibodies were used at 1:1000 dilution for Western blot studies and at 1:50 for immunofluorescence. Other antibodies used were as follows: rabbit anti-DHR78 (Fisk and Thummel 1998), Cy3-conjugated donkey anti-guinea pig IgG, Cy2-conjugated anti-rabbit IgG (Jackson ImmunoResearch), and rabbit anti-GAL4 DBD IgG (Santa Cruz Biotechnology). Animals were imaged on a Leica MZ12.5 dissecting microscope, equipped with a CoolSNAP-Pro cf COLOR camera (Media Cybernetic). Tissues were imaged on an Zeiss Axioskop 2 Plus microscope using the same camera.
Drosophila stocks and mutants
Drosophila stocks were obtained from the following sources: w1118, P[w+mCEcol/lacZScer/UAS.T:SV40/nls2] = UAS-nlacZ (Bloomington Stock Center); P[SUPor-P]KG04270, P[SUPor-P]KG05885 (Bellen et al. 2004); Gal4-DHR78 (Palanker et al. 2006); P[hsDHR78-9] = hs-DHR78, DHR782 (Fisk and Thummel 1998); and y w cin/FM6/y+ Y (source of RNA for yeast two-hybrid library, DiBenedetto et al. 1987). A full-length moses cDNA cloned into pCaSpeR-hs (Thummel and Pirrotta 1992) was used to establish a transformant line carrying the P element on the third chromosome (designated hs-moses). moses24 and moses5D mutants were maintained over the balancer chromosome CyO, P[w+mC = GAL4-Kr.C]DC4, P[w+mC = UAS-GFP.S65T]DC8 (Casso et al. 1999).
Acknowledgments
We thank Mitzi Kuroda for supplying the yeast two-hybrid library and the parental fly stock from which it was made; Ann Uberecken, Heather Lawrence, Yu Shi, and Brett Welch for excellent technical assistance; Michael Kay for supplying his tissue culture facility and luminometer; and Makoto Makishima for helpful discussions and guidance. K.D.B. was supported by an NIH NRSA Postdoctoral Fellowship and R.B.B. was supported by an NIH Multidisciplinary Cancer Research Training Program (T32 CA0093247). This research was supported by the Robert A. Welch Foundation (#I-1275), Howard Hughes Medical Institute, and the NIH.
Footnotes
Supplemental material is available at http://www.genesdeve.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1519007
References
- Andres A.J., Thummel C.S., Thummel C.S. Methods for quantitative analysis of transcription in larvae and prepupae. In: Goldstein L.S.B., Fyrberg E.A., Fyrberg E.A., editors. Drosophila melanogaster: Practical uses in cell and molecular biology. Academic Press; New York: 1994. pp. 565–573. [DOI] [PubMed] [Google Scholar]
- Andrew D.J., Scott M.P., Scott M.P. Immunological method for mapping protein distributions on polytene chromosomes. In: Goldstein L.S.B., Fyrberg E.A., Fyrberg E.A., editors. Drosophila melanogaster: Practical uses in cell and molecular biology. Academic Press; New York: 1994. pp. 353–370. [DOI] [PubMed] [Google Scholar]
- Baker K.D. “Characterization of a second receptor for ecdysteroids in Drosophila and the isolation of a novel orphan receptor-binding protein.” Ph.D. thesis, University of Texas Southwestern Medical Center, Dallas, TX. 2002. [Google Scholar]
- Baker K.D., Warren J.T., Thummel C.S., Gilbert L.I., Mangelsdorf D.J., Warren J.T., Thummel C.S., Gilbert L.I., Mangelsdorf D.J., Thummel C.S., Gilbert L.I., Mangelsdorf D.J., Gilbert L.I., Mangelsdorf D.J., Mangelsdorf D.J. Transcriptional activation of the Drosophila ecdysone receptor by insect and plant ecdysteroids. Insect. Biochem. Mol. Biol. 2000;30:1037–1043. doi: 10.1016/s0965-1748(00)00075-8. [DOI] [PubMed] [Google Scholar]
- Baker K.D., Shewchuk L.M., Kozlova T., Makishima M., Hassell A., Wisely B., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Shewchuk L.M., Kozlova T., Makishima M., Hassell A., Wisely B., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Kozlova T., Makishima M., Hassell A., Wisely B., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Makishima M., Hassell A., Wisely B., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Hassell A., Wisely B., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Wisely B., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Caravella J.A., Lambert M.H., Reinking J.L., Krause H., Lambert M.H., Reinking J.L., Krause H., Reinking J.L., Krause H., Krause H., et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/s0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- Beckstead R.B., Lam G., Thummel C.S., Lam G., Thummel C.S., Thummel C.S. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Hiesinger P.R., Schulze K.L., Rubin G.M., Schulze K.L., Rubin G.M., Rubin G.M., et al. The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby A.M., Richardson H.E., Richardson H.E. Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- Carroll S.B., Laughon A., Laughon A. Production and purification of polyclonal antibodies to the foreign segment of β-galactosidase fusion proteins. In: Glover D.M., editor. DNA cloning. IRL Press; Oxford, UK: 1987. pp. 89–111. [Google Scholar]
- Casso D., Ramirez-Weber F.A., Kornberg T.B., Ramirez-Weber F.A., Kornberg T.B., Kornberg T.B. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 1999;88:229–232. doi: 10.1016/s0925-4773(99)00174-4. [DOI] [PubMed] [Google Scholar]
- Chinpaisal C., Lee C.H., Wei L.N., Lee C.H., Wei L.N., Wei L.N. Mechanisms of the mouse orphan nuclear receptor TR2-11-mediated gene suppression. J. Biol. Chem. 1998;273:18077–18085. doi: 10.1074/jbc.273.29.18077. [DOI] [PubMed] [Google Scholar]
- Collins L.L., Lee Y.F., Heinlein C.A., Liu N.C., Chen Y.T., Shyr C.R., Meshul C.K., Uno H., Platt K.A., Chang C., Lee Y.F., Heinlein C.A., Liu N.C., Chen Y.T., Shyr C.R., Meshul C.K., Uno H., Platt K.A., Chang C., Heinlein C.A., Liu N.C., Chen Y.T., Shyr C.R., Meshul C.K., Uno H., Platt K.A., Chang C., Liu N.C., Chen Y.T., Shyr C.R., Meshul C.K., Uno H., Platt K.A., Chang C., Chen Y.T., Shyr C.R., Meshul C.K., Uno H., Platt K.A., Chang C., Shyr C.R., Meshul C.K., Uno H., Platt K.A., Chang C., Meshul C.K., Uno H., Platt K.A., Chang C., Uno H., Platt K.A., Chang C., Platt K.A., Chang C., Chang C. Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc. Natl. Acad. Sci. 2004;101:15058–15063. doi: 10.1073/pnas.0405700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBenedetto A.J., Lakich D.M., Kruger W.D., Belote J.M., Baker B.S., Wolfner M.F., Lakich D.M., Kruger W.D., Belote J.M., Baker B.S., Wolfner M.F., Kruger W.D., Belote J.M., Baker B.S., Wolfner M.F., Belote J.M., Baker B.S., Wolfner M.F., Baker B.S., Wolfner M.F., Wolfner M.F. Sequences expressed sex-specifically in Drosophila melanogaster adults. Dev. Biol. 1987;119:242–251. doi: 10.1016/0012-1606(87)90225-9. [DOI] [PubMed] [Google Scholar]
- Dressel U., Thormeyer D., Altincicek B., Paululat A., Eggert M., Schneider S., Tenbaum S.P., Renkawitz R., Baniahmad A., Thormeyer D., Altincicek B., Paululat A., Eggert M., Schneider S., Tenbaum S.P., Renkawitz R., Baniahmad A., Altincicek B., Paululat A., Eggert M., Schneider S., Tenbaum S.P., Renkawitz R., Baniahmad A., Paululat A., Eggert M., Schneider S., Tenbaum S.P., Renkawitz R., Baniahmad A., Eggert M., Schneider S., Tenbaum S.P., Renkawitz R., Baniahmad A., Schneider S., Tenbaum S.P., Renkawitz R., Baniahmad A., Tenbaum S.P., Renkawitz R., Baniahmad A., Renkawitz R., Baniahmad A., Baniahmad A. Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol. Cell. Biol. 1999;19:3383–3394. doi: 10.1128/mcb.19.5.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J., Becherer K., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J., Chen P.L., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J., Yeh S.H., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J., Yang Y., Kilburn A.E., Lee W.H., Elledge S.J., Kilburn A.E., Lee W.H., Elledge S.J., Lee W.H., Elledge S.J., Elledge S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Fisk G.J., Thummel C.S., Thummel C.S. Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc. Natl. Acad. Sci. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk G.J., Thummel C.S., Thummel C.S. The DHR78 nuclear receptor is required for ecdysteroid signaling during the onset of Drosophila metamorphosis. Cell. 1998;93:543–555. doi: 10.1016/s0092-8674(00)81184-8. [DOI] [PubMed] [Google Scholar]
- Forman B.M., Umesono K., Chen J., Evans R.M., Umesono K., Chen J., Evans R.M., Chen J., Evans R.M., Evans R.M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- Franco P.J., Farooqui M., Seto E., Wei L.N., Farooqui M., Seto E., Wei L.N., Seto E., Wei L.N., Wei L.N. The orphan nuclear receptor TR2 interacts directly with both class I and class II histone deacetylases. Mol. Endocrinol. 2001;15:1318–1328. doi: 10.1210/mend.15.8.0682. [DOI] [PubMed] [Google Scholar]
- Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- Gateff E., Loffler T., Wismar J., Loffler T., Wismar J., Wismar J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: Developmental studies and molecular localization of the gene. Mech. Dev. 1993;41:15–31. doi: 10.1016/0925-4773(93)90052-y. [DOI] [PubMed] [Google Scholar]
- Giguere V., Ong E.S., Segui P., Evans R.M., Ong E.S., Segui P., Evans R.M., Segui P., Evans R.M., Evans R.M. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- Grumbling G., Strelets V., Strelets V. FlyBase: Anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Tall G.G., Horazdovsky B.F., Tall G.G., Horazdovsky B.F., Horazdovsky B.F. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 1999;274:15284–15291. doi: 10.1074/jbc.274.21.15284. [DOI] [PubMed] [Google Scholar]
- Hu Y.C., Shyr C.R., Che W., Mu X.M., Kim E., Chang C., Shyr C.R., Che W., Mu X.M., Kim E., Chang C., Che W., Mu X.M., Kim E., Chang C., Mu X.M., Kim E., Chang C., Kim E., Chang C., Chang C. Suppression of estrogen receptor-mediated transcription and cell growth by interaction with TR2 orphan receptor. J. Biol. Chem. 2002;277:33571–33579. doi: 10.1074/jbc.M203531200. [DOI] [PubMed] [Google Scholar]
- Huq M.M., Gupta P., Tsai N.P., Wei L.N., Gupta P., Tsai N.P., Wei L.N., Tsai N.P., Wei L.N., Wei L.N. Modulation of testicular receptor 4 (TR4) activity by MAP-kinase mediated phosphorylation. Mol. Cell. Proteomics. 2006;5:2072–2082. doi: 10.1074/mcp.M600180-MCP200. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Umesono K., Mangelsdorf D.J., Aburatani H., Stanger B.Z., Shibasaki Y., Imawari M., Evans R.M., Takaku F., Umesono K., Mangelsdorf D.J., Aburatani H., Stanger B.Z., Shibasaki Y., Imawari M., Evans R.M., Takaku F., Mangelsdorf D.J., Aburatani H., Stanger B.Z., Shibasaki Y., Imawari M., Evans R.M., Takaku F., Aburatani H., Stanger B.Z., Shibasaki Y., Imawari M., Evans R.M., Takaku F., Stanger B.Z., Shibasaki Y., Imawari M., Evans R.M., Takaku F., Shibasaki Y., Imawari M., Evans R.M., Takaku F., Imawari M., Evans R.M., Takaku F., Evans R.M., Takaku F., Takaku F. A functional retinoic acid receptor encoded by the gene on human chromosome 12. Mol. Endocrinol. 1990;4:837–844. doi: 10.1210/mend-4-6-837. [DOI] [PubMed] [Google Scholar]
- Janowski B.A., Willy P.J., Devi T.R., Falck J.R., Mangelsdorf D.J., Willy P.J., Devi T.R., Falck J.R., Mangelsdorf D.J., Devi T.R., Falck J.R., Mangelsdorf D.J., Falck J.R., Mangelsdorf D.J., Mangelsdorf D.J. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Jullien-Flores V., Dorseuil O., Romero F., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J.H., Dorseuil O., Romero F., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J.H., Romero F., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J.H., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J.H., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J.H., Berger R., Tavitian A., Gacon G., Camonis J.H., Tavitian A., Gacon G., Camonis J.H., Gacon G., Camonis J.H., Camonis J.H. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- Khan S.A., Park S.W., Huq M., Wei L.N., Park S.W., Huq M., Wei L.N., Huq M., Wei L.N., Wei L.N. Protein kinase C-mediated phosphorylation of orphan nuclear receptor TR2: Effects on receptor stability and activity. Proteomics. 2005;5:3885–3894. doi: 10.1002/pmic.200402062. [DOI] [PubMed] [Google Scholar]
- Khan S.A., Park S.W., Huq M.D., Wei L.N., Park S.W., Huq M.D., Wei L.N., Huq M.D., Wei L.N., Wei L.N. Ligand-independent orphan receptor TR2 activation by phosphorylation at the DNA-binding domain. Proteomics. 2006;6:123–130. doi: 10.1002/pmic.200500068. [DOI] [PubMed] [Google Scholar]
- Kim C.A., Phillips M.L., Kim W., Gingery M., Tran H.H., Robinson M.A., Faham S., Bowie J.U., Phillips M.L., Kim W., Gingery M., Tran H.H., Robinson M.A., Faham S., Bowie J.U., Kim W., Gingery M., Tran H.H., Robinson M.A., Faham S., Bowie J.U., Gingery M., Tran H.H., Robinson M.A., Faham S., Bowie J.U., Tran H.H., Robinson M.A., Faham S., Bowie J.U., Robinson M.A., Faham S., Bowie J.U., Faham S., Bowie J.U., Bowie J.U. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.A., Gingery M., Pilpa R.M., Bowie J.U., Gingery M., Pilpa R.M., Bowie J.U., Pilpa R.M., Bowie J.U., Bowie J.U. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 2002;9:453–457. doi: 10.1038/nsb802. [DOI] [PubMed] [Google Scholar]
- King-Jones K., Thummel C.S., Thummel C.S. Nuclear receptors—A perspective from Drosophila. Nat. Rev. Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Koehler K.F., Helguero L.A., Haldosen L.A., Warner M., Gustafsson J.A., Helguero L.A., Haldosen L.A., Warner M., Gustafsson J.A., Haldosen L.A., Warner M., Gustafsson J.A., Warner M., Gustafsson J.A., Gustafsson J.A. Reflections on the discovery and significance of estrogen receptor-β. Endocr. Rev. 2005;26:465–478. doi: 10.1210/er.2004-0027. [DOI] [PubMed] [Google Scholar]
- Koelle M.R., Talbot W.S., Segraves W.A., Bender M.T., Cherbas P., Hogness D.S., Talbot W.S., Segraves W.A., Bender M.T., Cherbas P., Hogness D.S., Segraves W.A., Bender M.T., Cherbas P., Hogness D.S., Bender M.T., Cherbas P., Hogness D.S., Cherbas P., Hogness D.S., Hogness D.S. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- Kozlova T., Thummel C.S., Thummel C.S. Spatial patterns of ecdysteroid receptor activation during the onset of Drosophila metamorphosis. Development. 2002;129:1739–1750. doi: 10.1242/dev.129.7.1739. [DOI] [PubMed] [Google Scholar]
- Laudet V. Evolution of the nuclear receptor superfamily: Early diversification from an ancestral orphan receptor. J. Mol. Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- Lonard D.M., O’Malley B.W., O’Malley B.W. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Lu T.T., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., Mangelsdorf D.J., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., Mangelsdorf D.J., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., Mangelsdorf D.J., Schoonjans K., Kerr T.A., Auwerx J., Mangelsdorf D.J., Kerr T.A., Auwerx J., Mangelsdorf D.J., Auwerx J., Mangelsdorf D.J., Mangelsdorf D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Makishima M., Okamoto A.Y., Repa J.J., Tu H., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Okamoto A.Y., Repa J.J., Tu H., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Repa J.J., Tu H., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Tu H., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B., Lustig K.D., Mangelsdorf D.J., Shan B., Mangelsdorf D.J., Shan B., Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Ong E.S., Dyck J.A., Evans R.M., Ong E.S., Dyck J.A., Evans R.M., Dyck J.A., Evans R.M., Evans R.M. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Umesono K., Kliewer S.A., Borgmeyer U., Ong E.S., Evans R.M., Umesono K., Kliewer S.A., Borgmeyer U., Ong E.S., Evans R.M., Kliewer S.A., Borgmeyer U., Ong E.S., Evans R.M., Borgmeyer U., Ong E.S., Evans R.M., Ong E.S., Evans R.M., Evans R.M. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell. 1991;66:555–561. doi: 10.1016/0092-8674(81)90018-0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Borgmeyer U., Heyman R.A., Zhou J.Y., Ong E.S., Oro A.E., Kakizuka A., Evans R.M., Borgmeyer U., Heyman R.A., Zhou J.Y., Ong E.S., Oro A.E., Kakizuka A., Evans R.M., Heyman R.A., Zhou J.Y., Ong E.S., Oro A.E., Kakizuka A., Evans R.M., Zhou J.Y., Ong E.S., Oro A.E., Kakizuka A., Evans R.M., Ong E.S., Oro A.E., Kakizuka A., Evans R.M., Oro A.E., Kakizuka A., Evans R.M., Kakizuka A., Evans R.M., Evans R.M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes & Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Fujino S., Nakanishi G., Kim Y.S., Jetten A.M., Fujino S., Nakanishi G., Kim Y.S., Jetten A.M., Nakanishi G., Kim Y.S., Jetten A.M., Kim Y.S., Jetten A.M., Jetten A.M. TIP27: A novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acids Res. 2004;32:4194–4204. doi: 10.1093/nar/gkh741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L., Necakov A.S., Sampson H.M., Ni R., Hu C., Thummel C.S., Krause H.M., Necakov A.S., Sampson H.M., Ni R., Hu C., Thummel C.S., Krause H.M., Sampson H.M., Ni R., Hu C., Thummel C.S., Krause H.M., Ni R., Hu C., Thummel C.S., Krause H.M., Hu C., Thummel C.S., Krause H.M., Thummel C.S., Krause H.M., Krause H.M. Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development. 2006;133:3549–3562. doi: 10.1242/dev.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A.J., Mallin D.R., Francis N.J., Ketel C.S., Stamm J., Voeller R.K., Kingston R.E., Simon J.A., Mallin D.R., Francis N.J., Ketel C.S., Stamm J., Voeller R.K., Kingston R.E., Simon J.A., Francis N.J., Ketel C.S., Stamm J., Voeller R.K., Kingston R.E., Simon J.A., Ketel C.S., Stamm J., Voeller R.K., Kingston R.E., Simon J.A., Stamm J., Voeller R.K., Kingston R.E., Simon J.A., Voeller R.K., Kingston R.E., Simon J.A., Kingston R.E., Simon J.A., Simon J.A. Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics. 2004;167:1225–1239. doi: 10.1534/genetics.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalsky M.L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Qiao F., Bowie J.U., Bowie J.U. The many faces of SAM. Sci. STKE. 2005;2005 doi: 10.1126/stke.2862005re7. re7. [DOI] [PubMed] [Google Scholar]
- Reinking J., Lam M.M., Pardee K., Sampson H.M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Lam M.M., Pardee K., Sampson H.M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Pardee K., Sampson H.M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Sampson H.M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Yang P., Williams S., White W., Lajoie G., Edwards A., Williams S., White W., Lajoie G., Edwards A., White W., Lajoie G., Edwards A., Lajoie G., Edwards A., Edwards A., et al. The Drosophila nuclear receptor E75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Riddiford L.M. Hormones and Drosophila development. In: Bate M., Martinez-Arias A., Martinez-Arias A., editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 899–939. [Google Scholar]
- Riddiford L.M., Cherbas P., Truman J.W., Cherbas P., Truman J.W., Truman J.W. Ecdysone receptors and their biological actions. Vitam. Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M.G., Lunyak V.V., Glass C.K., Lunyak V.V., Glass C.K., Glass C.K. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Shyr C.R., Hu Y.C., Kim E., Chang C., Hu Y.C., Kim E., Chang C., Kim E., Chang C., Chang C. Modulation of estrogen receptor-mediated transactivation by orphan receptor TR4 in MCF-7 cells. J. Biol. Chem. 2002;277:14622–14628. doi: 10.1074/jbc.M110051200. [DOI] [PubMed] [Google Scholar]
- Sullivan A.A., Thummel C.S., Thummel C.S. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol. Endocrinol. 2003;17:2125–2137. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- Thornton J.W. Nonmammalian nuclear receptors: Evolution and endocrine disruption. Pure Appl. Chem. 2003;75:1827–1839. [Google Scholar]
- Thummel C.S. Files on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- Thummel C.S., Pirrotta V., Pirrotta V. New pCasper P-element vectors. Drosoph. Inf. Serv. 1992;71:150. [Google Scholar]
- Tsai C.C., Kao H.Y., Yao T.P., McKeown M., Evans R.M., Kao H.Y., Yao T.P., McKeown M., Evans R.M., Yao T.P., McKeown M., Evans R.M., McKeown M., Evans R.M., Evans R.M. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Umesono K., Evans R.M., Evans R.M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Vidal M., Cagan R.L., Cagan R.L. Drosophila models for cancer research. Curr. Opin. Genet. Dev. 2006;16:10–16. doi: 10.1016/j.gde.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Willy P.J., Umesono K., Ong E.S., Evans R.M., Heyman R.A., Mangelsdorf D.J., Umesono K., Ong E.S., Evans R.M., Heyman R.A., Mangelsdorf D.J., Ong E.S., Evans R.M., Heyman R.A., Mangelsdorf D.J., Evans R.M., Heyman R.A., Mangelsdorf D.J., Heyman R.A., Mangelsdorf D.J., Mangelsdorf D.J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes & Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Zelhof A.C., Yao T.P., Evans R.M., McKeown M., Yao T.P., Evans R.M., McKeown M., Evans R.M., McKeown M., McKeown M. Identification and characterization of a Drosophila nuclear receptor with the ability to inhibit the ecdysone response. Proc. Natl. Acad. Sci. 1995;92:10477–10481. doi: 10.1073/pnas.92.23.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]