Abstract
Aldosterone plays a major role in the regulation of salt balance and the pathophysiology of cardiovascular and renal diseases. Many aldosterone-regulated genes — including that encoding the epithelial Na+ channel (ENaC), a key arbiter of Na+ transport in the kidney and other epithelia — have been identified, but the mechanisms by which the hormone modifies chromatin structure and thus transcription remain unknown. We previously described the basal repression of ENaCα by a complex containing the histone H3 Lys79 methyltransferase disruptor of telomeric silencing alternative splice variant a (Dot1a) and the putative transcription factor ALL1-fused gene from chromosome 9 (Af9) as well as the release of this repression by aldosterone treatment. Here we provide evidence from renal collecting duct cells and serum- and glucocorticoid-induced kinase–1 (Sgk1) WT and knockout mice that Sgk1 phosphorylated Af9, thereby impairing the Dot1a-Af9 interaction and leading to targeted histone H3 Lys79 hypomethylation at the ENaCα promoter and derepression of ENaCα transcription. Thus, Af9 is a physiologic target of Sgk1, and Sgk1 negatively regulates the Dot1a-Af9 repressor complex that controls transcription of ENaCα and likely other aldosterone-induced genes.
Introduction
The renin-angiotensin-aldosterone system plays a major role in the control of blood pressure, extracellular fluid volume, and electrolyte balance, largely through the regulation of urinary Na+ excretion. The aldosterone-sensitive distal nephron (ASDN), composed of the late distal convoluted tubule, connecting tubule, and cortical and medullary collecting ducts, is the final arbiter of renal Na+ excretion. In the ASDN, transepithelial Na+ absorption occurs by apical Na+ entry via the epithelial Na+ channel (ENaC) and basolateral Na+ exit via the Na+,K+-ATPase. ENaC, composed of 3 subunits (α, β, and γ), constitutes the rate-limiting step in this process, and changes in its activity and/or plasma membrane abundance constitute key regulatory steps. Aldosterone increases transepithelial Na+ transport in the ASDN in large part through ENaCα induction in this region (1). Aldosterone increases ENaC function in 2 phases: an early phase involving upregulation of preexisting transport machinery and aldosterone-induced regulatory proteins, notably serum- and glucocorticoid-induced kinase–1 (Sgk1), which regulates the plasma membrane abundance of ENaC in part through phosphorylation of the ubiquitin ligase Nedd4-2 (2); and a delayed phase of aldosterone action involving de novo synthesis of ENaC, either from the liganded mineralocorticoid receptor directly binding hormone response elements in the ENaCα promoter to activate transcription (1) or through indirect mechanisms involving other proteins (3, 4). Sgk1 also participates in this late phase, presumably by phosphorylation of specific, as-yet undefined transcription factors, which then stimulate ENaCα transcription (5). Aldosterone administration or hyperaldosteronism induced by salt restriction increases ENaCα gene transcription in the ASDN without increasing β or γ subunit expression (3, 6) or altering ENaCα mRNA turnover (7).
Recently, in the mouse inner medullary collecting duct cell line mIMCD3, we identified a novel aldosterone signaling network governing ENaCα transcription that involves the putative transcription factor ALL1-fused gene from chromosome 9 (Af9) and the histone H3 Lys79 methyltransferase disruptor of telomeric silencing alternative splice variant a (Dot1a), which form a nuclear repressor complex that tonically associates with chromatin and hypermethylates histone H3 Lys79 associated with specific regions of the ENaCα promoter, thereby repressing ENaCα transcription under basal conditions (8). We also discovered that aldosterone inhibits Dot1a and Af9 expression, leading to hypomethylation of targeted histone H3 Lys79 and release of the basal transcriptional repression of ENaCα (8). This effect appears to be generalized to at least some other aldosterone-regulated genes, including those encoding connecting tissue growth factor, period homolog, and preproendothelin (8). A central question remaining in this model is how aldosterone signals to the Dot1a-Af9 complex. Given the multiple roles of Sgk1 in controlling ENaCα expression and action and our identification of a highly conserved consensus Sgk1 phosphorylation site (Ser435; Figure 1A) in the Af9 aa sequence, we tested the hypothesis that Sgk1-mediated Af9 phosphorylation regulates the Dot1a-Af9 complex (and therefore chromatin) and releases the repressive effects of the complex on ENaCα transcription. We found that aldosterone relieves the basal Dot1a-Af9 repression of ENaCa by promoting Sgk1 phosphorylation of Af9 and consequent disruption of the repression complex.
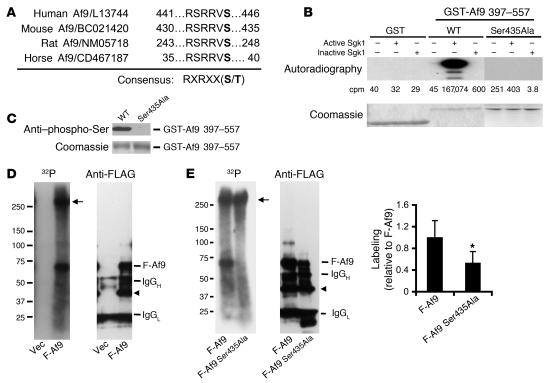
Figure 1. Af9 Ser435 is phosphorylated by Sgk1 in vitro and is a major phosphorylation site in mIMCD3 cells.
(A) Af9 possesses a highly conserved putative phosphorylation site, Ser435, for Sgk1. Shown are aa sequences surrounding Ser435 of Af9 (in bold) from different species. (B) Sgk1 phosphorylates Af9 in vitro. GST, WT GST-Af9 397–557, or GST-Af9 397–557 Ser435Ala purified from E. coli was phosphorylated in vitro in the presence of [γ-32P]ATP, separated by SDS-PAGE, stained (bottom), and autoradiographed (top). Mean 32P values (in cpm) of the corresponding bands are shown, with SE less than 25% in all cases (n = 3). (C) Af9 phosphorylation by Sgk1 requires Ser435. As in B, except for omission of [γ-32P]ATP (n = 3). (D) Af9 is phosphorylated in mIMCD3 cells. mIMCD3 cells transfected with pCMV500 vector (Vec) or pFLAG-Af9 (F-Af9) were labeled with [32P]orthophosphate and analyzed by IP-IB with the FLAG M2 antibody. A representative autoradiograph (left) and corresponding IB (right) are shown (n = 3). Arrow denotes unknown protein; arrowhead denotes possibly degraded Af9 fusions. (E) Af9 Ser435 is a major phosphorylation site in mIMCD3 cells. As in D, except that pFLAG-Af9 Ser435Ala construct (F-Af9 Ser435Ala) replaced pCMV500. A weak band migrating at the position of FLAG-Af9 Ser435Ala was visible after exposure of 12 more hours. Plot of the 32P signal normalized to the corresponding protein abundance of each Af9 fusion, with WT Af9 labeling assigned as 1 (n = 5). *P < 0.05.
Results
Af9 Ser435 is phosphorylated by Sgk1 in vitro and is a major phosphorylation site in mIMCD3 cells.
Because purification of the glutathione-S-transferase (GST) fusion protein containing full-length Af9 is impractical (8), GST-Af9 397–557 was used as substrate for in vitro phosphorylation to determine directly whether Sgk1 phosphorylates Af9. Neither inactive nor active Sgk1 phosphorylated GST alone, whereas active, but not inactive, Sgk1 heavily phosphorylated GST-Af9 397–557 (Figure 1B). As hypothesized, the Ser435Ala mutation abolished this phosphorylation (Figure 1, B and C). Af9 phosphorylation was also evident in anti-FLAG antibody IPs of mIMCD3 cells that had been transfected with pFLAG-Af9 and metabolically labeled with [32P]orthophosphate (Figure 1D). An intense, approximately 60-kDa band corresponding to FLAG-Af9 was observed in autoradioraphs from pFLAG-Af9–transfected cells but not controls (Figure 1D). A protein of greater than 250 kDa was also coimmunoprecipitated with FLAG-Af9 and strongly phosphorylated, but it was not recognized by the anti-FLAG antibody when subjected to IB (Figure 1D). Sequential reprobing of the same blot with antibodies against Dot1a or Sgk1 did not detect either band (data not shown); therefore, the identity and function of the larger protein in Af9 phosphorylation are unknown. Although FLAG-Af9 and FLAG-Af9 Ser435Ala were comparably immunoprecipitated, the mutation reduced overall Af9 phosphorylation to about 50% that of WT Af9 (Figure 1E).
Aldosterone coordinately induces Sgk1 expression and Af9 Ser435 phosphorylation in mIMCD3 cells.
Because aldosterone rapidly induces Sgk1 expression in multiple systems, including mIMCD3 cells (9, 10), we expected aldosterone to increase Af9 phosphorylation. Accordingly, we generated and characterized (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI29850DS1) an antibody specific for Af9 phosphorylated at Ser435 (referred to as anti–phospho-Af9) and performed IB analyses with whole-cell lysates of mIMCD3 cells treated with 1 μM aldosterone or vehicle for 1, 1.5, or 2 hours. Aldosterone induced Sgk1 immunoreactivity slightly at 1 hour and significantly at 1.5 and 2 hours compared with controls (Figure 2A). As expected, Af9 phosphorylation was significantly enhanced at 1 hour and peaked at 1.5 hours of aldosterone treatment (Figure 2A). No significant changes in β-actin expression were detected (Figure 2A). We previously reported that 1.5 hours of aldosterone treatment markedly reduces Af9 mRNA and protein levels in mIMCD3 cells (8). Unfortunately, because the anti-Af9 antibody (11) used in our earlier report was destroyed, we were unable to detect total Af9 protein levels in the current experiments. Nonetheless, these data are consistent with the hypothesis that Sgk1 contributes to aldosterone-stimulated Af9 phosphorylation in mIMCD3 cells.
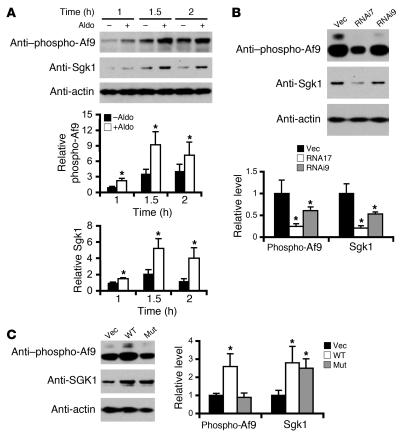
Figure 2. Af9 phosphorylation directly correlates with Sgk1 expression in mIMCD3 cells.
(A) Aldosterone coordinately induces Sgk1 expression and Af9 Ser435 phosphorylation. mIMCD3 cells were treated with ethanol or aldosterone (Aldo; 1 μM) for the indicated times and examined by IB with the indicated antibodies. Af9 phosphorylation was apparently affected by both ethanol and aldosterone. The Sgk1 or phosphorylated Af9 abundance (mean ± SE; n = 6) was normalized to the corresponding actin and expressed relative to vehicle-treated cells at the 1-hour time point (assigned as 1). *P < 0.05 versus corresponding –aldo. (B) RNAi knockdown of Sgk1 decreases Af9 phosphorylation. mIMCD3 cells stably transfected with pSilencer-4.1–CMV–Neo negative control vector (Vec), pSgk1-RNAi7 (RNAi7), or pSgk1-RNAi9 (RNAi9) were examined by IB with the indicated antibodies. The same set of stably transfected cells were repeatedly examined as in A. Data were analyzed essentially as in A (n = 4). (C) Overexpression of WT Sgk1, but not the kinase-dead mutant Sgk1, promotes Af9 phosphorylation. mIMCD3 cells were transiently transfected with pcDNA3.1 vector (Vec) or its derivatives encoding WT or kinase-dead mutant Sgk1 (Lys127Met; Mut) and analyzed by IB probed with the indicated antibodies. Data were analyzed similarly as in A (n = 4). *P < 0.05 versus vector control.
Af9 phosphorylation directly correlates with Sgk1 expression in mIMCD3 cells.
We sought additional evidence that Sgk1 phosphorylates Af9 by examining stable mIMCD3 cell lines carrying negative control vector or Sgk1-specific RNAi constructs pSgk1–RNA interference 7 (pSgk1-RNAi7) or pSgk1-RNAi9 for Sgk1 protein and Af9 phosphorylation levels. Sgk1 protein abundance and Af9 phosphorylation were coordinately decreased to 21% and 25%, respectively, in cells transfected with pSgk1-RNAi7 construct compared with vector control cells (Figure 2B). Similar, though less dramatic, results were obtained by transfection of pSgk1-RNAi9, with Sgk1 protein and Af9 phosphorylation levels reduced to approximately 50% and 40%, respectively. Levels of β-actin were invariant (Figure 2B), indicating equal sample loading and specificity of RNAi-mediated Sgk1 knockdown. In reciprocal experiments, transient overexpression in mIMCD3 cells of Sgk1, but not its kinase-dead mutant, increased Af9 phosphorylation by approximately 160% without affecting β-actin expression (Figure 2C).
Sgk1-mediated Af9 phosphorylation decreases the Dot1a-Af9 interaction in vitro and in mIMCD3 cells at the ENaCα promoter.
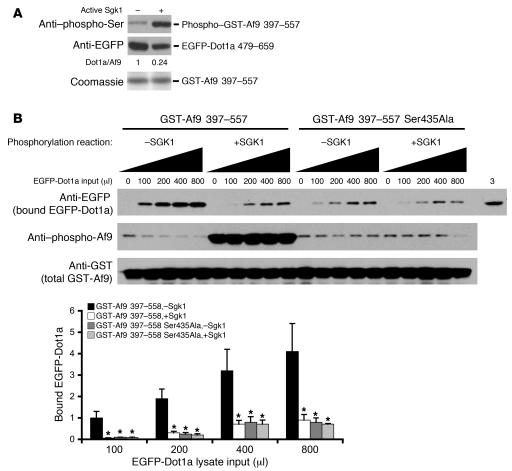
Because aa 397–557 in Af9 and aa 479–659 in Dot1a are the domains important for the Dot1a-Af9 interaction (8), and because Af9 Ser435 is located within the Dot1a-interacting domain, we hypothesized that Sgk1-mediated Af9 Ser435 phosphorylation regulates the Dot1a-Af9 interaction. Consistent with our hypothesis, Sgk1-phosphorylated GST-Af9 397–557 retained only 24% of the enhanced GFP–Dot1a 479–659 (EGFP-Dot1a 479–659) in lysates of transiently transfected mIMCD3 cells, compared with equal amounts of the nonphosphorylated GST-Af9 fusion (Figure 3A). Increasing the input amount of the EGFP-Dot1a–containing lysates, ranging 100–800 μl, generally resulted in more EGFP-Dot1a fusion retained by the fixed amount of WT or mutated GST-Af9 fusions, with or without phosphorylation by Sgk1 (Figure 3B). No effect was observed with the negative control, GST-Af9 397–557 Ser435Ala. Moreover, GST-Af9 397–557 exhibited approximately 4- to 20-fold higher affinity for EGFP-Dot1a, depending on the amount of added lysate, in the absence of Sgk1 than in the presence of Sgk1. Conversely, the ability to retain EGFP-Dot1a fusion and the sensitivity to Sgk1 were significantly reduced by the Ser435Ala mutation (Figure 3B). These results suggest that Sgk1-mediated Af9 phosphorylation decreases but does not fully abolish the Dot1a-Af9 interaction and that Ser435 plays an important role in binding to Dot1a. Phosphorylation of Ser435 by Sgk1 or Ser435Ala mutation presumably causes unfavorable conformational changes for Af9 interacting with Dot1a. Similar results have been reported for phosphorylation and Ala mutants of putative Ser phosphoacceptors in other protein-protein interactions (see Discussion).
Figure 3. Sgk1 impairs Af9 interaction with Dot1a in vitro.
(A) GST pulldown assay showing impaired ability of Sgk1-phosphorylated Af9 to retain the EGFP-Dot1a fusion. As in Figure 1C, GST-Af9 397–557 was phosphorylated by Sgk1 and incubated with mIMCD3 cell lysates containing EGFP-Dot1a 479–659. Bound proteins were examined by IB with the indicated antibodies (n = 3). (B) As in A, except for inclusion of GST-Af9 Ser435Ala as control, increasing input volumes (μl) of EGFP-Dot1a 479–659 lysate and the indicated antibodies. The same mIMCD3 cell lysates (3 μl) were loaded in the far right lane. The amount of bound EGFP-Dot1a 479–659 was normalized to that of total GST-Af9 fusions to calculate the relative binding efficiency and were plotted, with the efficiency of the reaction GST-Af9 397–557, –Sgk1, at 100 μl of input lysate assigned as 1 (n = 3). *P < 0.05 versus corresponding GST-Af9 397–557, –Sgk1.
In order to demonstrate further that Sgk1 regulates the Dot1a-Af9 interaction in mIMCD3 cells, we exploited our recent chromatin immunoprecipitation (ChIP) findings that endogenous Af9 or FLAG-Af9 associate with specific subregions R0–R3, but not subregion Ra (Figure 4A), of the ENaCα promoter (8). If Sgk1 phosphorylation of Af9 negatively regulates the association of Af9 with Dot1a, we hypothesized that Sgk1 would associate with the ENaCα promoter, decrease the association of the Dot1a-Af9 complex, and thus limit histone H3 Lys79 methylation at the ENaCα promoter. We performed ChIP experiments with anti-FLAG antibody on mIMCD3 cells transiently transfected with FLAG-tagged Sgk1 or the kinase-dead mutant Sgk1 in order to determine whether either is associated with the ENaCα promoter. Significant PCR amplicons at R0–R3, but not at Ra, were generated from immunoprecipitated chromatin derived from the cells expressing WT or mutant FLAG-Sgk1 fusions (Figure 4B). Vector-transfected cells generated only background signals. These data indicate that Sgk1, like Af9, specifically associates with the R0–R3, but not the Ra, subregions of the ENaCα promoter in mIMCD3 cells and that the mutation abolishing the kinase activity of Sgk1 does not significantly affect its association with the ENaCα promoter (see Discussion).
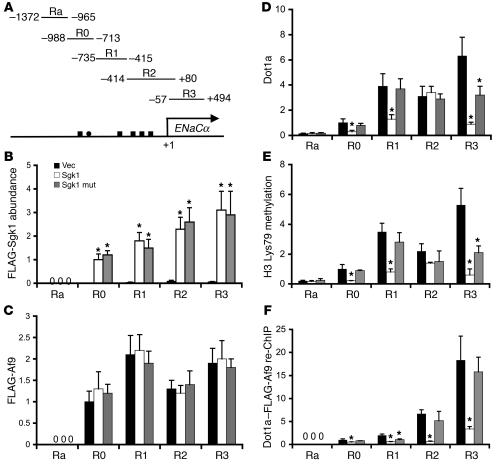
Figure 4. Sgk1 associates with and downregulates Dot1a-Af9 interaction at the ENaCα promoter.
(A) Diagram of the ENaCα promoter (8). (B) ChIP assay showing association of WT and kinase-dead mutant Sgk1 with the R0–R3, but not Ra, subregions of the ENaCα promoter. Chromatin was immunoprecipitated by anti-FLAG antibody from mIMCD3 cells transfected with pCMV500, WT pFLAG-Sgk1, or mutant pFLAG-Sgk1 Lys127Met, followed by quantitative real-time PCR with primers amplifying the Ra and R0–R3 subregions of ENaCα promoter as shown in A (n = 3). FLAG-Sgk1 abundance in R0 from the FLAG-Sgk1–transfected cells was assigned as 1. (C–F) ChIP and sequential ChIP assays demonstrating that overexpression of Sgk1 differentially affected the abundance of FLAG-Af9 (C), Dot1a (D), histone H3 Lys79 methylation (E), and FLAG-Af9 interaction with Dot1a (F) at the ENaCα promoter. mIMCD3 cells were cotransfected with pFLAG-Af9 and pcDNA3.1 or its derivatives expressing Sgk1 or its kinase-dead mutant, followed by ChIP with the indicated antibodies. For sequential ChIP (re-ChIP), chromatin was sequentially immunoprecipitated with anti-FLAG and anti-Dot1a antibodies. Relative ChIP or sequential ChIP efficiency was defined as the amount of (re)immunoprecipitated material compared with that of the initial input sample and was assigned as 1 in R0 from vector-transfected cells (n = 3–6). No Ra expression was detected in B, C, or F. *P < 0.05 versus vector control.
Because Af9 associates with the ENaCα promoter at the specific subregions (8) and Dot1a displays apparently non–sequence-specific DNA binding activity (8), Dot1a and Af9 might exist alone or as a Dot1a-Af9 complex at the ENaCα promoter. Sgk1 might regulate Dot1a and Af9 abundance and thus the level of histone H3 Lys79 methylation at the ENaCα promoter by regulating their DNA-binding activities, the Dot1a-Af9 interaction, or both. To evaluate these possibilities, we performed ChIP and sequential ChIP assays with mIMCD3 cells cotransfected with pFLAG-Af9 along with an empty vector or a construct expressing the untagged WT or kinase-dead Sgk1. FLAG-Af9 was used because the anti–phospho-Af9 antibody preferentially, if not exclusively, recognizes phosphorylated Af9 (Supplemental Figure 1A), and phosphorylation of Af9 impairs the Dot1a-Af9 interaction in vitro (Figure 3). Therefore, sequential ChIP data generated with that antibody would be difficult to interpret. As shown in Figure 4C and consistent with our previous observations (8), the association of FLAG-Af9 was only detected in the R0–R3, but not the Ra, subregions of the ENaCα promoter. Overexpression of WT or mutant Sgk1 did not significantly affect the occupancy of FLAG-Af9 at the R0–R3 subregions compared with the vector control, indicating that Sgk1-mediated Af9 phosphorylation has little impact on FLAG-Af9 DNA binding. On the contrary, ChIP with anti-Dot1a antibody confirmed our reported basal level of Dot1a association at the Ra subregion (8), which likely occurs because of the non–sequence-specific DNA binding activity of Dot1a and is likely responsible for the residual level of histone H3 Lys79 methylation in the absence of FLAG-Af9 binding to this subregion (Figure 4, C and E). Overexpression of WT Sgk1 variably reduced Dot1a abundance at the R0, R1, and R3, but not the Ra or R2, subregions of the ENaCα promoter compared with the vector control (Figure 4D). Transfection of the mutant Sgk1 significantly decreased the association of Dot1a only in the R3 subregion (Figure 4D). Decreased association of Dot1a with the ENaCα promoter was consistently accompanied by histone H3 Lys79 hypomethylation at the corresponding subregions (Figure 4E). As stated above, a lower abundance of Dot1a at the ENaCα promoter could be the result of impaired Dot1a DNA binding, its interaction with Af9, or both. Dot1a–FLAG-Af9 sequential ChIP revealed that overexpression of WT Sgk1 markedly decreased the abundance of the Dot1a-Af9 complex associated with the R0–R3 subregions of the ENaCα promoter compared with the vector control (Figure 4F). Overexpression of the kinase-dead Sgk1 mutant, however, only had a significant impact on the Dot1a-Af9 complex associated with subregion R1. No detectable sequential ChIP signals were obtained in any reactions for the Ra subregion, in accord with the lack of FLAG-Af9 binding in Ra. Taken together, these data confirm our previous findings that Af9 binds directly or indirectly with each subregion except Ra, but also indicate that Sgk1 impairs the ability of Af9 to interact with Dot1a at these subregions without affecting Af9 DNA-binding capacity, resulting in histone H3 Lys79 hypomethylation. In most — but not all — subregions, Sgk1 exercised these effects in a kinase-dependent manner and presumably by phosphorylating Af9 (see Discussion).
Mutation of the Sgk1 phosphorylation site Ser435 in Af9 impairs Af9 overexpression–dependent histone H3 Lys79 hypermethylation, but not Af9 association with the ENaCα promoter.
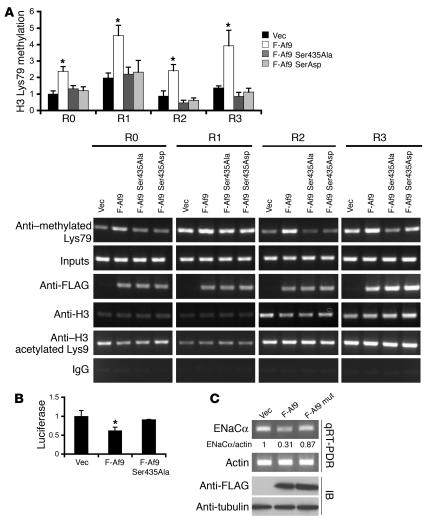
To test further whether Af9 Ser435 plays an important role in regulating Af9-mediated histone H3 Lys79 hypermethylation at the ENaCα promoter, we constructed phosphorylation-deficient and phosphorylation-mimetic Af9 mutants by converting Ser435 to Ala or Asp, respectively, and repeated ChIP assays with mIMCD3 cells transfected with vector, pFLAG-Af9, pFLAG-Af9 Ser435Ala, or pFLAG-Af9 Ser435Asp, focusing on the R0–R3 subregions. As we previously reported (8), FLAG-Af9 overexpression increased histone H3 Lys79 methylation 2- to 3-fold in each of the R0–R3 subregions of the ENaCα promoter compared with vector controls, but these increases were not evident in cells transfected with pFLAG-Af9 Ser435Ala or pFLAG-Af9 Ser435Asp (Figure 5A). Consistent with our previous results, ChIP assays with antibodies specific for histone H3 acetylated Lys9 or the N-terminal tail of histone H3 performed in parallel experiments detected no significant alterations in any subregion analyzed (Figure 5A), suggesting that the changes in histone H3 Lys79 methylation were specific and not the result of a generalized effect on histones. Moreover, ChIP with IgG yielded barely detectable background signals by agarose gel analysis.
Figure 5. Af9 Ser435Ala and Ser435Asp mutations impair Af9 overexpression–dependent histone H3 Lys79 hypermethylation and ENaCα promoter repression.
(A) ChIP assay showing that Af9 Ser435Ala and Ser435Asp mutations decreased Dot1a-Af9–mediated histone H3 Lys79 methylation, but not Af9 association with the ENaCα promoter, in mIMCD3 cells. pFLAG-Af9, pFLAG-Af9 Ser435Ala, and pFLAG-Af9 Ser435Asp were used, and ChIP assays were performed with the indicated antibodies (n = 3–4). The relative histone H3 Lys79 methylation of R0 from the vector-transfected cells was assigned as 1. Representative agarose gel analyses of the final quantitative PCR products are shown to verify the quantification determined by quantitative real-time PCR for each sample. (B) Luciferase assay indicating that the Ser435Ala mutation impaired Af9-dependent repression of ENaCα promoter activity. mIMCD3 cells stably carrying pGL3Zeocin-1.3ENaCα (12) were transiently transfected with pCMV500, pFLAG-Af9, or pFLAG-Af9 Ser435Ala, followed by luciferase assay (n = 4). *P < 0.05 versus vector control. (C) Quantitative RT-PCR (qRT-PCR) demonstrating that the Ser435Ala mutation partially relieves Af9-dependent repression of endogenous ENaCα transcription. As in B, except that total RNA was analyzed by quantitative real-time RT-PCR with primers specific for ENaCα or β-actin as control and that whole-cell lysates were examined by IP with the indicated antibodies. The relative level of ENaCα mRNA was normalized to actin in the same sample, and that of vector-transfected cells was assigned as 1. The means of 3 independent measurements with SE less than 20% are shown.
The failure of Af9 Ser435Ala or Af9 Ser435Asp to increase histone H3 Lys79 methylation at the ENaCα promoter could result from a weak or failed association of the mutated Af9 with Dot1a, the ENaCα promoter, or both. To distinguish among these possibilities, ChIP with the anti-FLAG antibody was performed. Consistent with and extending our previous observations (8), all WT and mutant Af9 fusions were efficiently immunoprecipitated by the anti-FLAG antibody and found in the chromatin associated with all 4 subregions (R0–R3) of the ENaCα promoter. Only background signals were detected in the vector-transfected cells (Figure 5A). Therefore, the Ser435Ala or phosphomimetic Ser435Asp mutations, while impairing the Dot1a-Af9 interaction, did not significantly alter the ability of Af9 to interact with the ENaCα promoter. These data are consistent with our GST pulldown assay results (Figure 2) as well as the conclusions that Ser435 is one of the key residues in mediating the Dot1a-Af9 association and that its perturbation by phosphorylation or Ala or Asp mutations probably induces conformational changes in Af9 that weakens Af9 binding to Dot1a (see Discussion).
Mutation of the Sgk1 phosphorylation site Ser435 in Af9 partially relieves Dot1a-Af9-mediated ENaCα transcriptional repression.
In order to determine the importance of Af9 Ser435 in Dot1a-Af9 mediated ENaCα repression, mIMCD3 cells stably expressing an ENaCα promoter–luciferase construct (12) were transiently transfected with empty vector, pFLAG-Af9, or pFLAG-Af9 Ser435Ala. We performed luciferase assays to assess ENaCα promoter–luciferase activity, quantitative real-time RT-PCR to examine endogenous ENaCα mRNA levels (with β-actin as control), and IB with the anti-FLAG antibody to monitor FLAG-Af9 expression. Overexpression of the WT, but not the mutant, Af9 fusion significantly decreased the ENaCα promoter–luciferase activity and endogenous ENaCα mRNA levels (Figure 5, B and C). The abundance of ENaCα protein in mIMCD3 cells was below the level needed to consistently discriminate differences on IB with the anti-ENaCα antibody (data not shown). The Af9 Ser435Ala mutation also impaired Af9-mediated repression of period homolog, connective tissue growth factor, and preproendothelin (Supplemental Figure 2), 3 genes known to be upregulated by aldosterone (10) and to be repressed by Af9 and/or Dot1a overexpression in mIMCD3 cells (8).
Af9 phosphorylation and Sgk1 mRNA expression are coordinately regulated in kidneys of mice with different salt intakes.
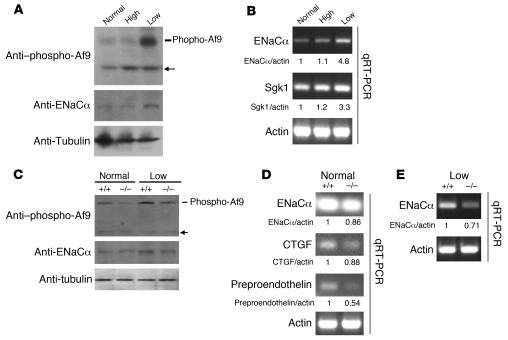
Aldosterone and low salt intake strongly induce Sgk1 expression in the ASDN (reviewed in ref. 13). If Af9 is a bona fide substrate of Sgk1 for phosphorylation in vivo, salt restriction should coordinately induce Sgk1 expression and Af9 phosphorylation in the kidney, which in turn should relieve Dot1a-Af9–mediated repression of ENaCα. Accordingly, whole kidney lysates from mice fed normal-, high-, or low-salt diets were analyzed by IB with anti–phospho-Af9, anti-ENaCα, or control anti–α-tubulin. Mice with normal or high salt intakes exhibited low levels of Af9 phosphorylation and ENaCα protein in their kidneys, but Af9 phosphorylation and ENaCα protein levels were coordinately increased about 10-fold and 3-fold, respectively, in the kidneys of mice fed the salt-restricted diet (Figure 6A). Under these conditions, the abundance of α-tubulin was not significantly changed (Figure 6A). Quantitative real-time RT-PCR of total RNAs isolated from these same kidneys revealed that ENaCα and Sgk1 mRNA levels were similar to those of controls (normal salt) in the mice fed the high-salt diet, but 3- to 4-fold greater than controls in mice fed the low-salt diet (Figure 6B). These findings are consistent with previous studies showing that a low-salt diet induces ENaCα mRNA expression in renal cortex and outer and inner medulla (14) as well as Sgk1 mRNA levels in renal cortex (15).
Figure 6. Sgk1 phosphorylates Af9 and regulates transcription of ENaCα and other aldosterone target genes in vivo in mouse kidneys.
(A and B) Low-salt diet coordinately increases Af9 phosphorylation and mRNA expression of Sgk1 and ENaCα in mouse kidneys. Whole lysates from the right kidneys of WT mice (Sgk1+/+) fed the indicated diet were pooled within the group (n = 3 in each group, similar for B–E) and analyzed by IB probed with the antibodies shown. Similarly, total RNA from the left kidneys was pooled within the group (n = 3) and analyzed with primers specific for the indicated genes. Representative agarose gel analyses of the final quantitative RT-PCR with the mean of 3 different measurements are shown. SEM was less than 20% in all cases. (C–E) Af9 phosphorylation is regulated by salt intake, is achieved in an Sgk1-dependent or -independent manner, and correlates with expression of several aldosterone target genes. As in A, except that Sgk1+/+ mice or their age- and sex-matched Sgk1–/– littermates fed the indicated diets were used and that additional aldosterone target genes were examined (n = 3 for each group and condition). Arrow denotes unknown protein. CTGF, connecting tissue growth factor.
Sgk1–/– mice exhibit markedly lower levels of Af9 Ser435 phosphorylation in the kidney.
We previously generated and characterized Sgk1–/– mice and demonstrated that Sgk1 plays an important role in regulating ENaC activity in the ASDN (13, 16–18). Here, we used these mice to test the hypothesis that Sgk1 phosphorylates Af9 and regulates ENaCα transcription in vivo in the kidney. Whole kidney lysates isolated from Sgk1+/+ and Sgk1–/– littermates fed a normal- or low-salt diet were analyzed by IB with the anti–phospho-Af9 antibody. Under a normal-salt diet, Sgk1–/– mice exhibited renal Af9 phosphorylation levels that were significantly lower than those of controls, if not completely abolished (Figure 6C). Lower aldosterone levels cannot explain the difference in Af9 phosphorylation in Sgk1–/– mice under these conditions, because these mice exhibit modestly (approximately 2-fold) greater compensatory plasma aldosterone concentrations compared with Sgk1+/+ mice (17, 19). Moreover, the Sgk1–/– mice on a normal-salt diet displayed subtle changes in ENaCα protein abundance and modestly lower mRNA levels of ENaCα, connecting tissue growth factor, and preproendothelin compared with WT littermates (86%, 88%, and 54% of WT, respectively; Figure 6D), suggesting that Sgk1 likely regulates transcription of these genes, which we have previously shown to be regulated by the aldosterone-Dot1a-Af9 signaling network (8). In contrast, with a low salt intake, Sgk1–/– mice exhibited renal Af9 phosphorylation levels that were substantially higher than for mice fed the normal-salt diet, yet still about 3 times lower than those of the WT littermates (Figure 6C), indicating that Af9 can be phosphorylated in a Sgk1-dependent or -independent manner in vivo. In keeping with our hypothesis, the reduced Af9 phosphorylation in Sgk1–/– mice compared with WT littermates was accompanied by reduced levels of ENaCα mRNA (71% of control; Figure 6E) and protein (approximately 60% of control; Figure 6C) despite approximately 4-fold higher plasma aldosterone concentrations in plasma of Sgk1–/– mice compared with WT littermates (17).
Discussion
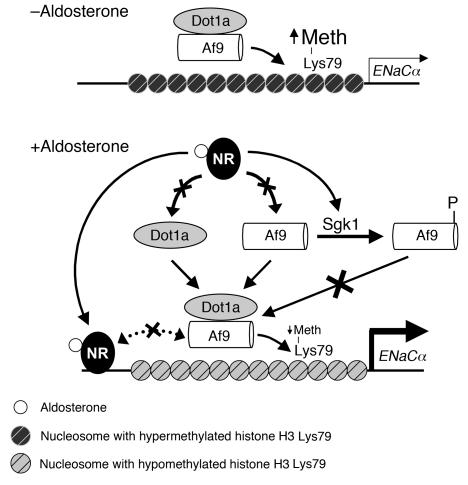
Sgk1 is rapidly induced by a spectrum of stimuli and phosphorylates a variety of proteins, including transcription factors (reviewed in ref. 20), to serve as a point of convergence for multiple signaling pathways. Recently, Sgk1 was shown to regulate the transcription of the ENaCα and ENaCβ genes in the mouse renal cortical collecting duct M1 cell line (5) and connective tissue growth factor in the heart by a mechanism involving NF-κB activation and translocation into the nucleus (21), but the effector mechanisms were not fully elucidated. Moreover, the molecular mechanisms by which Sgk1 regulates transcription in the context of chromatin has not to our knowledge been addressed until now. Here, we identified Af9 as a physiologic target for Sgk1 phosphorylation and Sgk1 as what we believe to be a novel component and negative regulator of the Dot1a-Af9 complex. These data cast light on the mechanisms governing Sgk1-mediated transcriptional activation of genes such as ENaCα and offer additional insights into mechanisms by which aldosterone alters the local chromatin structure of its target genes to activate transcription. Accordingly, we propose a refined model of this alternative aldosterone signaling network that now includes the abilities of aldosterone, not only to downregulate the abundance of the components of the Dot1a-Af9 complex, but also (via Sgk1-mediated phosphorylation of Af9) to weaken their interaction and relieve the basal repression on ENaCα transcription (Figure 7).
Figure 7. Model for aldosterone-sensitive repression of ENaCα expression by targeted histone H3 Lys79 hypermethylation mediated by the Dot1a-Af9-Sgk1 complex.
Under basal conditions, Dot1a and Af9 form a nuclear complex that directly or indirectly binds specific sites of the ENaCα promoter, leading to hypermethylation of histone H3 Lys79 and repression of ENaCα transcription. Aldosterone stimulates ENaCα transcription by regulating 2 different pathways. In the classical pathway, aldosterone binds and activates the nuclear hormone receptors (NR) that are either glucocorticoid receptor and/or mineralocorticoid receptor homo- or heterodimers to bind the glucocorticoid response element in the ENaCα promoter and transactivate ENaCα. In a parallel pathway, aldosterone releases the repression of ENaCα by (a) downregulating Dot1a (12) and Af9 expression (8), and thus the amount of Dot1a-Af9 complex, presumably via nuclear receptor–dependent or –independent (not shown) mechanisms and/or (b) decreasing the Dot1a-Af9 interaction via Sgk1-mediated phosphorylation of Af9 to limit their availability, leading to histone H3 Lys79 hypomethylation at specific subregions of the ENaCα promoter. Sgk1 might join and downregulate the Dot1a-Af9 complex associated with the ENaCα promoter (not shown for simplicity). In all cases, Af9-free Dot1a binds DNA nonspecifically and catalyzes histone H3 Lys79 methylation throughout the genome under basal conditions (not shown). A hypothetical opposing interaction between the nuclear receptor–aldosterone complex and the Dot1a-Af9 complex (dotted line) may tune the ultimate level of ENaCα transcription. Meth, methylation.
Af9 was discovered as one of the most common fusion partners of the mixed-lineage leukemia (MLL) protein in humans (22), and it is abundantly expressed in the kidney and in other tissues. Specific functions of Af9 protein are largely unknown, but — as shown in our work and that of others — it regulates transcription in promoter-reporter gene assays and its structural domains, which include a YEATS domain and a nuclear targeting sequence, are consistent with a role as a transcription factor. Its close structural relative in yeast, ANC1/TFG3, is a component of the SWI/SNF chromatin-remodeling complex (23). In addition to Dot1a, Af9 can interact with AF4 (11), specific isoforms of the BCL-6 corepressor (24), and the polycomb group member MPc3 (25). Ultimately the actions of Af9 as activator or repressor of transcription for ENaCα or other genes may depend on the interplay and relative proportions of these Af9-binding proteins in the cell. Whether interactions with Dot1a and/or Sgk1 influence the role of Af9 as an MLL fusion partner in leukemogenesis remains to be tested.
Both Af9 and Sgk1 specifically associate with the R0–R3, but not the Ra, subregions of the ENaCα promoter. It remains to be defined whether Sgk1, Af9, or one or more other factors contribute to the DNA-binding activity. The ultimate level of histone H3 Lys79 methylation at each specific region of the genome is determined by the abundance of Dot1a that directly binds DNA in a nonspecific manner (26), the DNA-binding factor(s) interacting with Dot1a targeting it to specific loci, and the factors regulating their interaction. ChIP with the anti-Dot1a antibody should theoretically immunoprecipitate all ENaCα promoter–associated Dot1a molecules, regardless of whether they are associated with endogenous Af9 or FLAG-Af9 or are free from any Af9 molecules. Conversely, only those Dot1a molecules that interact with FLAG-Af9 would be potentially reimmunoprecipitated by the anti-Dot1a antibody in the second round of immunoprecipitation of the Dot1a–FLAG-Af9 sequential ChIP experiments (Figure 4F). These assumptions likely explain why, in ChIP assays, anti-Dot1a antibody (Figure 4D) and anti–methylated H3 Lys79 antibody (Figure 4E) yielded consistent results that were generally, but not completely, in accord with the data generated by the Dot1a–FLAG-Af9 sequential ChIP (Figure 4F). At the R2 subregion, the FLAG-Af9–Dot1a complex was reduced to 10% of control levels in Sgk1-transfected cells (Figure 4F), yet the corresponding difference in histone H3 Lys79 methylation in these cells was not significant (Figure 4E), indicating that histone H3 Lys79 methylation in this region might be primarily mediated by Dot1a itself through its nonspecific DNA-binding activity. This scenario became even more complicated in the case of R1, in which the association of Dot1a with FLAG-Af9 was decreased by both WT and kinase-dead mutant Sgk1 overexpression (Figure 4F), while histone H3 Lys79 methylation was only affected by WT Sgk1 (Figure 4E). We do not know the exact mechanism to account for these observations. One possibility is that additional factors such as glucocorticoid receptor and/or mineralocorticoid receptor (Figure 7) exist to regulate the Dot1a-Af9 complex and/or Af9-free Dot1a at the ENaCα promoter. Sgk1 might directly or indirectly regulate these factors in a kinase-dependent and/or -independent manner, leading to the ultimate output of the kinase-insensitive Dot1a-Af9 association and kinase-sensitive histone H3 Lys79 hypermethylation at this particular subregion. Targeted histone H3 Lys79 methylation, as we observed here, has previously been observed in yeast and mammalian cells (27, 28), yet Dot1 proteins are generally thought to catalyze histone H3 Lys79 methylation in a nontargeted manner (27, 29). The present work offers clues as to how targeted histone H3 Lys79 methylation is achieved and regulated. Such targeting may occur only under conditions and in cell types in which Af9 and Sgk1 are coexpressed.
Results of previous studies of Sgk1–/– mice indicate that ENaC expression and function and mineralocorticoid action in the kidney are partially, but not completely, dependent on Sgk1 (16, 17). Therefore, other critical mechanisms of aldosterone signaling must be operative and might work in parallel with those described here. Moreover, it remains possible that the greater concentrations of plasma aldosterone in Sgk1–/– mice (see above) may partially compensate for impaired Sgk1-dependent Af9 phosphorylation by activation of Sgk1-independent control of Af9 phosphorylation and gene expression. Further studies will be needed to determine the potential involvement of these components and/or the mineralocorticoid receptor in the network described here. An integrative consideration of the following results, however, is consistent with a dominant role of Sgk1 in aldosterone-induced Af9 phosphorylation under physiologic conditions: (a) plasma aldosterone concentrations in Sgk1+/+ mice fed a low-salt diet are of similar magnitude to those of Sgk1–/– mice fed a normal-salt diet (17); (b) Af9 phosphorylation was significantly lower, if not completely abolished, in Sgk1–/– versus Sgk1+/+ mice on a normal-salt diet (Figure 6C); and (c) Sgk1+/+ mice exhibited further enhancement of Af9 phosphorylation when dietary salt intake was reduced (Figure 6A).
Mechanistically, our data suggest that Sgk1-mediated phosphorylation of Af9 impairs, but does not eliminate, the ability of Af9 to associate with Dot1a. Therefore, other factors controlling the Dot1a-Af9 complex may be operative. Moreover, Af9 phosphorylation by Sgk1 did not significantly alter the ability of Af9 to interact with the ENaCα promoter (Figure 4C). We predicted that the phosphorylation site–deficient Ser435Ala and phosphomimetic Ser435Asp mutants of Af9 would yield opposite results when tested for their ability to modulate histone H3 Lys79 methylation associated with the ENaCα promoter. However, these mutants were equally competent in their ability to associate with the ENaCα promoter but incapable of increasing histone H3 Lys79 methylation, possibly due to their impaired ability to interact with Dot1a in ChIP assays. These results suggest that Ser435 plays an important role in mediating the Af9-Dot1a interaction, and this role is sensitive to changes in structure and/or charge induced by phosphorylation or mutation into Ala or Asp. Analogous effects of phosphorylation on protein-protein interactions have been reported for other proteins (30–32). Furthermore, identical behavior of Ala and Asp mutations of putative Ser phosphorylation sites has also been reported (33, 34). In addition, the kinase activity of Sgk1, while necessary for disrupting the Dot1a-Af9 interaction, did not appear to be required for its association with the R0–R3 subregions of the ENaCα promoter in mIMCD3 cells. Elucidation of the factors controlling the ability of Sgk1 to associate with the Dot1a-Af9 in the native chromatin environment of specific regions of the ENaCα promoter will be critical to understanding this repression-derepression cycle of ENaCα transcription. It will also be important to determine whether stimuli known to activate Sgk1 (other than aldosterone) play similar roles in activating ENaCα transcription. In the aggregate, the present results indicate that, in addition to reversing the inhibition of tonic posttranslational repression of ENaC by Nedd4-2 (2), aldosterone-stimulated Sgk1 activity also signals the Dot1a-Af9 complex to relieve the basal repression of ENaCα transcription and augment ENaC-mediated Na+ transport. Moreover, the impaired, but robust, 32P labeling of FLAG-Af9 Ser435Ala in mIMCD3 cells (Figure 1E) suggests that Ser435 is a major site for phosphorylation, but not the sole site. Similarly, Sgk1 is not the sole kinase responsible for Af9 Ser435 phosphorylation, because significant levels of Af9 Ser435 phosphorylation were detected in Sgk1–/– mice fed a low-salt diet (Figure 6C). Identification of these additional phosphorylation sites and corresponding kinases should reveal the function and regulation of these proteins.
Methods
Reagents.
Aldosterone (Sigma-Aldrich); GST purification kit (Amersham Biosciences); Lipofectamine 2000 reagent and Geneticin (Invitrogen); Dual Luciferase Assay kit (Promega); antibodies against phosphorylated serine, dimethyl histone H3 Lys79, acetylated H3 Lys9, H3 N-terminal tail, and Sgk1 (Upstate USA Inc.); antibody against α-tubulin (Santa Cruz Biotechnology Inc.); antibody against EGFP (BD Biosciences — Clontech); and antibody against FLAG (Sigma-Aldrich) were purchased. Rabbit polyclonal antibody against ENaCα was a gift from M.A. Knepper (NIH, Bethesda, Maryland, USA; ref. 35), and pcDNA3.1 Sgk1 and pcDNA3.1 Sgk1 Lys127Met were kindly provided by P.A. Welling (University of Maryland School of Medicine, Baltimore, Maryland, USA; ref. 36). pEGFP-Dot1a, pEGFP-Dot1a 479–659, pGST-Dot1a, pGST-Af9 397–557, pFLAG-Af9, antibodies specific for Dot1a, and mIMCD3 cells stably transfected with pGL3Zeocin-1.3 ENaCα have been described previously (8, 12, 37). Anti–phospho-Af9 antibody was generated by immunizing rabbits with a peptide corresponding to mouse Af9 aa 421–441 harboring phosphorylated Ser435 and affinity purified using the corresponding nonphosphorylated peptide (New England Peptide Inc.). Its specificity was characterized (Supplemental Figure 1). pGST-Af9 397–557 Ser435Ala, pFLAG-Af9 Ser435Ala, and pFLAG-Af9 Ser435Asp were generated by overlapping PCR using the corresponding constructs (8) as template. For RNAi, 2 Sgk1-specific target sequences (RNAi7, 5′-TGGTAGCGATTCTCATCGC-3′, encoding aa 19–26; and RNAi9, 5′-CCACGGGCTCGATTCTACG-3′, encoding aa 196–203) were annealed and cloned into pSilencer-4.1–CMV–Neo (Ambion) at BamHI-HindIII sites according to the manufacturer’s instructions. The resulting constructs were called pSgk1-RNAi7 and pSgk1-RNAi9. Both RNAi target sequences perfectly matched with only their own sequences in the Sgk1 gene in BLAST searches against the mouse genome. The Sgk1 coding region was PCR amplified with pcDNA3.1-Sgk1 or pCDNA3.1-Sgk1 Lys127Met as template and cloned into pFLAG-Af9 at EcoRI-XhoI to replace the Af9 coding region for expression of the corresponding FLAG-Sgk1 fusions. DNA sequencing verified the authenticity of all inserts in the constructs.
Cell culture, aldosterone treatment, transient and stable transfections, RNAi and luciferase assays, 32P metabolic labeling, and IB assays.
mIMCD3 cells were routinely cultured and maintained with DMEM/F12 plus 10% FBS. For aldosterone time-course experiments, cells were cultured in DMEM/F12 plus 10% charcoal-stripped FBS for a minimum of 50 hours, followed by treatment with 1 μM aldosterone or its vehicle 0.01% ethanol at different time points as we have previously described (8). To knock down Sgk1 expression, mIMCD3 cells were transfected with Sgk1-specific RNAi constructs pSgk1-RNAi7 and pSgk1-RNAi9 and selected with Geneticin (50 μg/ml) for 15–20 days. Stably transfected colonies with the lowest Sgk1 expression were identified by IB with the anti-Sgk1 antibody and chosen for further study. The pSilencer-4.1–CMV–Neo negative control construct was transfected similarly and used as a negative control. For 32P metabolic labeling, 16 hours after transfection with the corresponding plasmids, cells were briefly washed with and cultured in phosphate-free DMEM plus 10% dialyzed FBS for 2 hours, labeled with 0.25 mCi/ml [32P]orthophosphate for 3 hours, and then harvested. Whole cell lysates of mIMCD3 cells or mouse kidneys were prepared with the cell lysis buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 1 mM EDTA, pH 8.0; 1% Triton X-100; and a cocktail of protease inhibitors) and used for IB, IP, or GST pulldown assays according to our previously published protocols (8). To immunoprecipitate FLAG-Af9 fusions without coimmnoprecipitating Af9-interacting partners, immunoprecipitation was performed in a manner similar to coimmunoprecipitation (8), except that whole lysates were made with the cell lysis buffer containing 0.1% SDS, anti-FLAG M2 affinity gel was used, and more stringent washing conditions with the cell lysis buffer were applied. IB with various antibodies was performed as we described previously (8, 12, 37). Protein bands on IB or Coomassie-stained gels were quantified using VersaDoc Model 4000 Imaging System (Bio-Rad).
ChIP, sequential ChIP, quantitative real-time PCR, and quantitative RT-PCR.
ChIP-coupled quantitative real-time PCR of DNA isolated in ChIP assays or quantitative RT-PCR for mRNA expression were performed with SYBR dye as we previously detailed (8, 12, 37). Briefly, chromatin was first sonicated using a Sonifier Cell Disruptor 350 (3 × 10 s; duty cycle, 50%; output control, 6; Branson) and immunoprecipitated with various antibodies as indicated in the text or figure legends. Sequential ChIP assays were conducted in a manner similar to that previously reported (38). In brief, chromatin from FLAG-Af9–expressing mIMCD3 cells was first immunoprecipitated with anti-FLAG antibody (8). Immunoprecipitated protein-DNA complexes were then eluted with 10 mM DTT, diluted with the cell lysis buffer 20 times, and subjected again to the ChIP procedure with anti-Dot1a antibody.
Animal experiments.
All animal experiments were conducted according to protocols reviewed and approved by the Institutional Animal Care and Use Committee of the VA San Diego Healthcare System. Female Sgk1+/+ mice (3 months of age; Sv129 genetic background) were placed for 10 days either on normal-salt diet (0.44% Na+), low-salt diet (0.05% Na+), or high-salt diet (normal-salt diet plus 0.9% NaCl in drinking water). Mice were then euthanized by decapitation, and their kidneys were rapidly placed in liquid nitrogen and stored at –80°C until use. Sgk1–/– mice were generated as previously described (17) and backcrossed to a Sv129 genetic background for 10 generations. Heterozygous offspring were mated to generate male Sgk1–/– and male Sgk1+/+ littermates, which were maintained on either a normal-salt or a low-salt diet as described above for 10 days, and at the age of 3 months, kidneys were harvested as described above.
Statistics.
Pooled data are presented as mean ± SEM for all figures. Unpaired 2-tailed Student’s t test was performed, and significance was set at P < 0.05. All experiments were repeated at least 3 times, with each independent observation representing a different set of cell cultures or assays (8, 12).
Supplementary Material
Acknowledgments
We thank Aniko Naray-Fejes-Toth for helpful discussions. W. Zhang was supported by NIH grant K01 DK70834; B. Kone was supported by NIH grants R01 DK075065 and R01 DK47981 and holds the James T. and Nancy B. Willerson Chair; V. Vallon was supported by American Heart Association Grant-in-Aid 655232Y; and T. Rieg, D. Kuhl, and F. Lang were supported by grants from Deutsche Forschungsgemeinschaft and the German Ministry for Science and Technology.
Footnotes
Nonstandard abbreviations used: Af9, ALL1-fused gene from chromosome 9; ASDN, aldosterone-sensitive distal nephron; ChIP, chromatin immunoprecipitation; Dot1a, disruptor of telomeric silencing alternative splice variant a; EGFP, enhanced GFP; ENaC, epithelial Na+ channel; GST, glutathione-S-transferase; RNAi, RNA interference; Sgk1, serum- and glucocorticoid-induced kinase–1.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:773–783 (2007). doi:10.1172/JCI29850
See the related Commentary beginning on page 592.
References
- 1.Thomas C.P., Itani O.A. New insights into epithelial sodium channel function in the kidney: site of action, regulation by ubiquitin ligases, serum- and glucocorticoid-inducible kinase and proteolysis. Curr. Opin. Nephrol. Hypertens. 2004;13:541–548. doi: 10.1097/00041552-200409000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Debonneville C., et al. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na+ channel cell surface expression. . EMBO J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asher C., Wald H., Rossier B.C., Garty H. Aldosterone-induced increase in the abundance of Na+ channel subunits. . Am. J. Physiol. 1996;271:C605–C611. doi: 10.1152/ajpcell.1996.271.2.C605. [DOI] [PubMed] [Google Scholar]
- 4.Sayegh R., et al. Glucocorticoid induction of epithelial sodium channel expression in lung and renal epithelia occurs via trans-activation of a hormone response element in the 5′-flanking region of the human epithelial sodium channel alpha subunit gene. J. Biol. Chem. 1999;274:12431–12437. doi: 10.1074/jbc.274.18.12431. [DOI] [PubMed] [Google Scholar]
- 5.Boyd C., Naray-Fejes-Toth A. Gene regulation of ENaC subunits by serum- and glucocorticoid-inducible kinase-1. Am. J. Physiol. Renal Physiol. 2005;288:F505–F512. doi: 10.1152/ajprenal.00242.2004. [DOI] [PubMed] [Google Scholar]
- 6.Escoubet B., Coureau C., Bonvalet J.P., Farman N. Noncoordinate regulation of epithelial Na channel and Na pump subunit mRNAs in kidney and colon by aldosterone. Am. J. Physiol. 1997;272:C1482–C1491. doi: 10.1152/ajpcell.1997.272.5.C1482. [DOI] [PubMed] [Google Scholar]
- 7.Mick V.E., et al. The alpha-subunit of the epithelial sodium channel is an aldosterone-induced transcript in mammalian collecting ducts, and this transcriptional response is mediated via distinct cis-elements in the 5′-flanking region of the gene. Mol. Endocrinol. 2001;15:575–588. doi: 10.1210/mend.15.4.0620. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Xia X., Reisenauer M.R., Hemenway C.S., Kone B.C. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCα in an aldosterone-sensitive manner. J. Biol. Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S.Y., et al. Epithelial sodium channel regulated by aldosterone-induced protein sgk. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2514–2519. doi: 10.1073/pnas.96.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumz M.L., Popp M.P., Wingo C.S., Cain B.D. Early transcriptional effects of aldosterone in a mouse inner medullary collecting duct cell line. Am. J. Physiol. Renal Physiol. 2003;285:F664–F673. doi: 10.1152/ajprenal.00353.2002. [DOI] [PubMed] [Google Scholar]
- 11.Erfurth F., Hemenway C.S., De Erkenez A.C., Domer P.H. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia. 2004;18:92–102. doi: 10.1038/sj.leu.2403200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W., et al. Aldosterone-sensitive repression of ENaCα transcription by a histone H3 lysine-79 methyltransferase. Am. J. Physiol. Cell Physiol. 2006;290:C936–C946. doi: 10.1152/ajpcell.00431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vallon V., et al. Role of Sgk1 in salt and potassium homeostasis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R4–R10. doi: 10.1152/ajpregu.00369.2004. [DOI] [PubMed] [Google Scholar]
- 14.Ono S., Kusano E., Muto S., Ando Y., Asano Y. A low-Na+ diet enhances expression of mRNA for epithelial Na+ channel in rat renal inner medulla. . Pflugers Arch. 1997;434:756–763. doi: 10.1007/s004240050462. [DOI] [PubMed] [Google Scholar]
- 15.Hou J., Speirs H.J., Seckl J.R., Brown R.W. Sgk1 gene expression in kidney and its regulation by aldosterone: spatio-temporal heterogeneity and quantitative analysis. J. Am. Soc. Nephrol. 2002;13:1190–1198. [PubMed] [Google Scholar]
- 16.Vallon V., et al. SGK1 as a determinant of kidney function and salt intake in response to mineralocorticoid excess. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;289:R395–R401. doi: 10.1152/ajpregu.00731.2004. [DOI] [PubMed] [Google Scholar]
- 17.Wulff P., et al. Impaired renal Na+ retention in the sgk1-knockout mouse. . J. Clin. Invest. 2002;110:1263–1268. doi: 10.1172/JCI200215696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D.Y., et al. Blunted hypertensive effect of combined fructose and high-salt diet in gene-targeted mice lacking functional serum- and glucocorticoid-inducible kinase SGK1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;290:R935–R944. doi: 10.1152/ajpregu.00382.2005. [DOI] [PubMed] [Google Scholar]
- 19.Huang D.Y., et al. Impaired regulation of renal K+ elimination in the sgk1-knockout mouse. . J. Am. Soc. Nephrol. 2004;15:885–891. doi: 10.1097/01.asn.0000120368.59693.a8. [DOI] [PubMed] [Google Scholar]
- 20.Lang F., et al. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol. Rev. 2006;86:1151–1178. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- 21.Vallon V., et al. SGK1-dependent cardiac CTGF formation and fibrosis following DOCA treatment. J. Mol. Med. 2006;84:396–404. doi: 10.1007/s00109-005-0027-z. [DOI] [PubMed] [Google Scholar]
- 22.Iida S., et al. Molecular cloning of 19p13 breakpoint region in infantile leukemia with t(11;19)(q23;p13) translocation. Jpn. J. Cancer Res. 1993;84:532–537. doi: 10.1111/j.1349-7006.1993.tb00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie Z., et al. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol. Cell. Biol. 2003;23:2942–2952. doi: 10.1128/MCB.23.8.2942-2952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan R.S., de Erkenez A.C., Hemenway C.S. The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene. 2003;22:3395–3406. doi: 10.1038/sj.onc.1206361. [DOI] [PubMed] [Google Scholar]
- 25.Hemenway C.S., de Erkenez A.C., Gould G.C. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene. 2001;20:3798–3805. doi: 10.1038/sj.onc.1204478. [DOI] [PubMed] [Google Scholar]
- 26.Min J., Feng Q., Li Z., Zhang Y., Xu R.M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 27.van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 28.Okada Y., et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Feng Q., et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 30.McAvoy T., et al. Regulation of neurabin I interaction with protein phosphatase 1 by phosphorylation. Biochemistry. 1999;38:12943–12949. doi: 10.1021/bi991227d. [DOI] [PubMed] [Google Scholar]
- 31.Weger S., Wendland M., Kleinschmidt J.A., Heilbronn R. The adeno-associated virus type 2 regulatory proteins rep78 and rep68 interact with the transcriptional coactivator PC4. J. Virol. 1999;73:260–269. doi: 10.1128/jvi.73.1.260-269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rong R., Surace E.I., Haipek C.A., Gutmann D.H., Ye K. Serine 518 phosphorylation modulates merlin intramolecular association and binding to critical effectors important for NF2 growth suppression. Oncogene. 2004;23:8447–8454. doi: 10.1038/sj.onc.1207794. [DOI] [PubMed] [Google Scholar]
- 33.Ku N.O., Liao J., Omary M.B. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 1998;17:1892–1906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seebohm G., et al. Regulation of KCNQ4 potassium channel prepulse dependence and current amplitude by SGK1 in Xenopus oocytes. Cell. Physiol. Biochem. 2005;16:255–262. doi: 10.1159/000089851. [DOI] [PubMed] [Google Scholar]
- 35.Masilamani S., Kim G.H., Mitchell C., Wade J.B., Knepper M.A. Aldosterone-mediated regulation of ENaC α, β, and γ subunit proteins in rat kidney. J. Clin. Invest. 1999;104:R19–R23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo D., et al. Cell surface expression of the ROMK (Kir 1.1) channel is regulated by the aldosterone-induced kinase, SGK-1, and protein kinase A. J. Biol. Chem. 2003;278:23066–23075. doi: 10.1074/jbc.M212301200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W., Hayashizaki Y., Kone B.C. Structure and regulation of the mDot1 gene, a mouse histone H3 methyltransferase. Biochem. J. 2004;377:641–651. doi: 10.1042/BJ20030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metivier R., et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763.. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.