Abstract
The promyelocytic leukemia zinc finger gene (PLZF) is involved in chromosomal translocation t(11;17) associated with acute promyelocytic leukemia. In this work, a 201-kilobase genomic DNA region containing the entire PLZF gene was sequenced. Repeated elements account for 19.83%, and no obvious coding information other than PLZF is present over this region. PLZF contains six exons and five introns, and the exon organization corresponds well with protein domains. There are at least four alternative splicings (AS-I, -II, -III, and -IV) within exon 1. AS-I could be detected in most tissues tested whereas AS-II, -III, and -IV were present in the stomach, testis, and heart, respectively. Although splicing donor and acceptor signals at exon–intron boundaries for AS-I and exons 1–6 were classical (gt–ag), AS-II, -III, and -IV had atypical splicing sites. These alternative splicings, nevertheless, maintained the ORF and may encode isoforms with absence of important functional domains. In mRNA species without AS-I, there is a relatively long 5′ UTR of 6.0 kilobases. A TATA box and several transcription factor binding sites were found in the putative promoter region upstream of the transcription start site. PLZF is a well conserved gene from Caenorhabditis elegans to human. PLZF paralogous sequences are found in human genome. The presence of two MLL/PLZF-like alignments on human chromosome 11q23 and 19 suggests a syntenic replication during evolution. The chromosomal breakpoints and joining sites in the index acute promyelocytic leukemia case with t(11;17) also were characterized, which suggests the involvement of DNA damage-repair mechanism.
Promyelocytic leukemia zinc finger (PLZF), first identified as a partner gene fused to retinoic acid receptor α (RARα) in a variant chromosomal translocation t(11;17) (q23;q21) in acute promyelocytic leukemia (APL), is a nuclear transcription factor of the BTB/POZ (for bric-a-brac/tramtrack/broad complex, poxvirus and zinc-finger) family (1–3). Recent studies showed that the PLZF-RARα fusion gene is able to induce leukemia in transgenic mice (4, 5), and PLZF also may be involved in the pathogenesis of APL with t(15;17) (6). Importantly, APL patients with PLZF-RARα had poor response to all-trans retinoic acid treatment, in contrast to those with promyelocytic leukemia (PML)-RARα (7–9). Recent studies revealed that the BTB/POZ domain of PLZF-RARα binds to the co-repressor complex even in the presence of high concentrations of all-trans retinoic acid (4, 10–16). The wild-type PLZF gene was expressed in the developing rhombomere boundaries and in CD34+ hematopoietic precursors (17–18). The PLZF knock-out mice showed striking hip devilment and impaired spermatogenesis in male animals (19). All of this indicates an important role of PLZF in both normal development and leukemogenesis.
In the present work, we cloned and sequenced the genomic region covering the entire PLZF gene by using the ordered shotgun sequencing (OSS) approach (20, 21). Comparison of genomic with cDNA sequences led to the characterization of the structural organization of this large gene. Homology search of PLZF-like sequences provided important information on the evolution of a multiple gene family. In addition, the chromosome breakpoints and joining regions in an APL case with t(11;17)(q23;q21) were cloned and analyzed to explore possible molecular mechanisms of aberrant gene rearrangement in APL.
MATERIALS AND METHODS
Library Screening.
Using a specific PLZF cDNA probe, we screened a human genomic DNA EMBL phage λ library (2, 22) and obtained seven positive clones. One cosmid clone (clone 45) was kindly provided by the National Center for Human Genome Research (Bethesda, MD). With the help of the Center for Genetics in Medicine, Washington University, we screened a bacterial artificial chromosome (BAC) library by using PLZF cDNA sequence-tagged site, a 163-bp PCR product on exon 6, and obtained three BAC clones. One clone that covers 95% of the PLZF gene was used for large-scale DNA sequencing.
BAC Subcloning, Long-Range PCR, and T3/T7 End Sequencing.
BAC DNA extracted with minipreparation (23) was partially digested with Sau3AI, and the resulting overhangs were partially filled in with the Klenow fragment of DNA polymerase I and dGTP and dATP. The DNA was fractionated, and fractions of 8–12 kilobases (kb) were recovered and cloned into BlueSTAR λ vector to yield a mini-library. Totally, 192 clones of 8–10 kb and 96 clones of 10–12 kb were obtained. To ensure efficient recovery of end sequences from inserts, long-range PCR amplification of subclones was conducted with GeneAmp XL-PCR kit (Perkin–Elmer). To prepare the sequencing template, each PCR product was treated with exonuclease I and shrimp alkaline phosphatase (EXoI/sAP) (24, 25). Dye-primer T3/T7 end sequencing was performed on a 377 automatic sequencer (26).
OSS Map Construction and Complete Sequencing of Amplified λ Inserts.
OSS strategy (20, 21) was adopted in this work. As outlined in Fig. 1, the building of contigs was initiated with an autoassembler (Perkin–Elmer) or phred/phrap (University of Washington) program to find overlaps among end sequences, and then a framework physical map was built by connecting groups of clones based on pairwise relationships of end sequences from individual BlueSTAR λ clones. Full sequencing was initiated for the clones constituting a minimal tiling path, including those anchored to known parts of the BAC vector arm. To do this, long-range PCR products from selected BlueSTAR λ clones were sonicated and size-fractionated for shotgun sequencing. The fractions of 1.6–2.0 kb were selected and cloned into M13 or pUC vectors. Templates were prepared by symmetric PCR, and Dye-terminator or Dye-primer cycle sequencing was performed. With a 10-fold coverage, sequences were assembled by using autoassembler and phred/phrap programs. Gap closure was performed by primer extension on BAC DNA by using dye-terminator reactions.
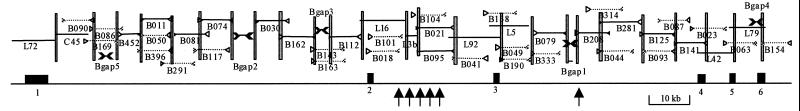
Figure 1.
PLZF genomic DNA sequence map shows coverage of BAC (BWXD 109) by λ clones (B), phage clones (L), and a cosmid clone (C). Sequenced ends are indicated by open (T3 end) or closed (T7 end) triangles. Selected subclones completely sequenced are shown as solid lines while clones with only ends sequenced are shown as dashed lines. Shaded vertical bars represent overlapping segments between subclones. “B gap 1–5” are not covered by the λ clones but are closed by amplification of BAC DNA. Exons are shown as black boxes, and arrows indicate the breakpoints identified in six patients previously described (7).
Computer Analysis of Sequence Data.
Several software tools were used to analyze the genomic sequence of PLZF (27, 28): (i) Repetitive sequences were determined by the program repeatmask (http://repeatmasker.genome.washington.edu/cgi-bin/RMZ-req.pl). (ii) GC content was sought by grail 1.3, which also was used to find latent exons with genscan, genie, genlang, genview, geneid, fgenes, fgenesh, and ebest (All above are from http://ares.ifrc.mcw.edu/MetaGene/). (iii) For analyzing regulatory sequences in the putative promoter region, nnpp (Promoter Predication by Neural Network), promoter scan, tssw, tess, and transfac were applied together to make a comprehensive prediction. (iv) cDNA was aligned with genomic sequence of PLZF by using sim4 and blast to give a view of the actual splicing sites. In addition, nnssp (Splice Site Prediction by Neural Network), netgene2 and spl (search for potential splice sites) were used to find possible alternative splicing sites in the genomic sequence while rnaspl was used to search for potential exon–exon junctions in cDNA (All above are from http://dot.imgen.bcm.tmc.edu:9331/seq-search/gene-search.html). (v) The position of PLZF with regard to adjacent loci, for example, MLL(ALL-1, HRX), was established by radiation hybrid (29) and by searching the Unigene cluster and genemap98 in the National Center for Biotechnology Information (30). (vi) Ortholog analysis was carried out for PLZF homologs from different organisms, including Caenorhabditis elegans, Gallus gallus, mouse, and human.
Rapid Amplification of cDNA Ends (RACE).
To get the 5′ and 3′ ends of PLZF transcript, the bone marrow and brain ready cDNA (CLONTECH) was used as template for PCR. AP1 and AP2 at each end of ready cDNA were used as anchor primers. PLZF-specific primers were designed according to cDNA sequence (Fig. 2). Successive rounds of nested PCR were carried out to get the 5′and 3′ ends of PLZF cDNA.
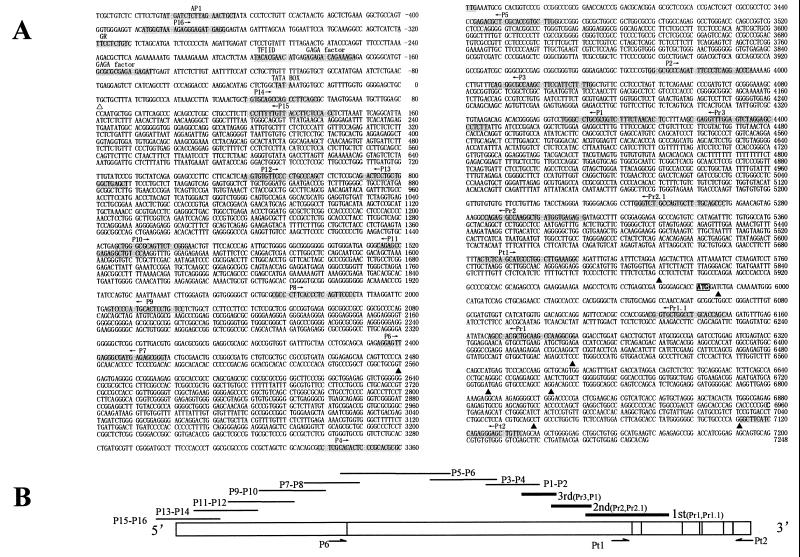
Figure 2.
(A) Sequence of the 5′ flanking region and exon 1 of PLZF. Open triangle points to the putative transcription start site. Possible binding sites of transcription factors are shadowed, and primers for PCR are indicated. Filled triangles indicate alternative splicing sites (AS-I through AS-IV). (B) Schematic representation of RACE and RT-PCR results of the extended exon 1. The upper solid lines indicate alignment of PCR fragments. The three thick lines at the right represent RACE results. Vertical lines within the exon 1 box denote alternative splicing sites. Arrows indicate primers for amplification of alternatively spliced fragments (see Fig. 3 A and B).
Reverse Transcription (RT)–PCR Analysis.
Total RNA was extracted from MDS cells treated with A23187 (31) and from different tissues by single-step acid guanidinium-thiocyanate-phenol method or by using TRIZOL reagent. RNA was treated with DNase I, and RT-PCR was performed with the Advantage cDNA amplification kit (CLONTECH) or the Taq thermal polymerase (Perkin–Elmer). To determine the alternative splicing patterns of PLZF, a series of PLZF-specific primers were designed according to the PLZF cDNA or genomic sequences (Fig. 2).
Molecular Analysis of Genomic Regions Encompassing PLZF-RARα and RARα-PLZF Reciprocal Translocations.
Based on the genomic sequences of PLZF and RARα, primers were designed to amplify both PLZF-RARα and RARα-PLZF fusion regions in a Chinese patient with t(11;17) (1, 2). A 5-kb fragment containing the PLZF-RARα genomic fusion was obtained and then sequenced with OSS method. By comparison of this 5-kb fragment with PLZF and RARα genomic sequences, the breakpoints in both PLZF and RARα genes were precisely determined, and the reciprocal joining region in RARα-PLZF also was amplified. The sequence of wide-type RARα encompassing the breakpoint also was determined.
RESULTS
Implementation of Sequence.
Using the OSS strategy, we obtained complete genomic DNA sequence of the PLZF gene in a series of clones, including one BAC, one cosmid, and seven EMBL λ phage clones. The BAC clone was partially digested into 8- to 12-kb fragments and was subcloned into the BlueSTAR λ vector. We were able to amplify 98% of DNA inserts from these subclones, and 90% of the amplified inserts yielded end sequences, a prerequisite for the success of an OSS approach. Sequence information from both ends of 288 subclones led to the construction of a contig with minimum overlap consisting of 23 λ clones. Total random shotgun sequencing of the 23 λ, 1 cosmid, and 7 EMBL λ phage clones produced 4,168 sequence tracts with autoassembler and phred/phrap programs, of which 3,983 (96.6%) with average usable length of 763 bp were assembled into 6 contigs. Internal redundancy levels were from 3- to 15-fold (10.5-fold coverage, on average). Most (98%) of the sequences were determined on both strands, but, at a few sites, the opposite strand was not recoverable. In these cases, consensus sequences were obtained by sequencing templates in the same orientation with two different sequencing chemistries (dye-terminator and dye-primer). The five gaps were closed by amplifying the intervening sequences from BAC DNA(32). The complete genomic DNA sequence of PLZF was 201,303 bp (submitted to the GenBank database under accession no. AF060568).
Computer-Assisted Sequence Analysis.
Comparison of genomic sequence with PLZF cDNA showed a good concordance (99.99% identical) and enabled us to assess the accuracy of the sequencing results. Minor discrepancies between cDNA and genomic sequences may result from individual variation or technical difficulties.
In repeat masker analysis, several frequently repeated elements in human DNA were detected. Total, there are 100 short interspersed elements (SINES) (47 Alu, 53 MIR), 46 long interspersed elements (LINES), 7 long terminal repeats, and 18 DNA elements, accounting for 10.28, 6.09, 1.41, and 2.05%, respectively, of the total sequence content. There are six GC-rich regions (Table 1). Further analysis suggested that, immediately upstream of the first GC-rich region, lies a possible promoter.
Table 1.
Results of computer analysis of PLZF genomic sequence
| GC-rich region | Percent of GC | Predicted exon | Tscr | Actual exon |
|---|---|---|---|---|
| 7,026–7,312 | 56.74 | |||
| 7,516–9,574 | 69.85 | 11,488–12,759+ | 152.52 | 6,849–12,755 |
| 18,337–18,624 | 54.57 | |||
| 104,557–104,654+ | 16.85 | 104,557–104,654 | ||
| 135,173–135,259+ | 9.64 | 135,173–135,259 | ||
| 147,370–147,624 | 73.06 | 147,664–147,248− | 16.73 | |
| 153,311–153,606 | 54.43 | 190,371–190,541+ | 24.31 | 190,371–190,541 |
| 197,693–197,949 | 55.97 | 195,403–195,570+ | 37.22 | 195,343–195,570 |
| 198,537–198,766+ | 38.59 | 198,537–198,860 |
+, forward strand; −, reverse strand.
We used the powerful metagene engine (http://ares.ifrc.mcw.edu/MetaGene/) to launch genescan, genie, geneid, geneview, genefinder, grail, genlang, nnpp, and ebest and used its feature annotation tools to analyze the result. genescan seems to give the best result. Among the seven predicted exons that have a Tscr (a score system for exon prediction depending on the information of length, splice sites, and coding region) >9, only one was located in the reverse strand. All of the predicted exons in the forward strand corresponded well to the actual exons. ebest revealed three clusters on the forward strand. Cluster 1 contains 26 expressed sequence tags (ESTs) dispersed along the genome sequence. Yet, assembly of these ESTs resulted in a sequence slightly similar to human β-globin region. Each of the other two clusters had one EST and was not analyzed because of a shortage of candidates.
After searching the whole 201-kb genomic sequence against the human EST database through the blast service of the National Center for Biotechnology Information, we picked the top 113 hits out of the GenBank database, with P/Z/E ranging from e-42 to 0.0. These ESTs fell into six clusters through Applied Biosystem’s autoassembler. However, except for one cluster that contains only two ESTs, all clusters showed high similarity to PLZF cDNA. Autoassembly analysis of these ESTs with PLZF cDNA sequences reduced the cluster number to two, one consisting of the PLZF cDNA and 73 ESTs, which extended the sequence from the 5′ end of published PLZF cDNA by 600 bp. The other contains 38 ESTs, which showed high similarity to Alu. Therefore, PLZF may be the only coding information over the 201-kb region.
Exon–Intron Organization of the PLZF Gene.
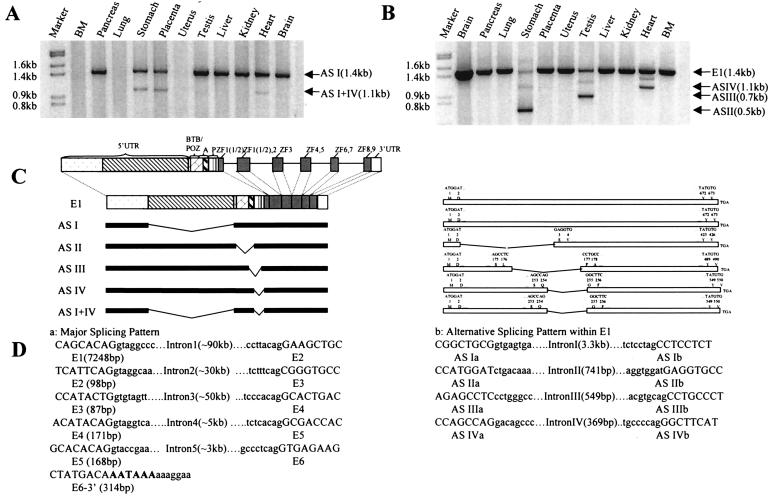
The reported PLZF cDNA was only 2.2 kb long, but Northern blot analysis showed an ≈8.5-kb transcript in most tissues investigated (33). To obtain full length PLZF cDNA, 3′ and 5′ RACE were performed by using bone marrow and brain ready cDNA. 3′ RACE showed that the real 3′ end could be already reached because three independently obtained clones all stopped near the previously reported polyadenylation signal AATAAA. Starting from the most 5′ PLZF cDNA sequence encoding the BTB/POZ domain, three successive rounds of 5′ RACE were performed and extended the 5′ end by 1.5 kb. However, the total cDNA length was still shorter than predicted by Northern blot analysis. Interestingly, using nnpp, tssw, tess, and promoter scan, we found a typical TATA box and a transcription start site ≈4.4 kb upstream of the most 5′ end of our cDNA contig. RT-PCR was performed to obtain the upstream sequence by using DNase I-treated RNA from MDS cells. Sequence analysis on the PCR products confirmed that they formed a contig that corresponded exactly to the genomic region (Fig. 2A). However, one pair of primers flanking the predicted TATA box and transcription start site did not produce any band in RT-PCR but primed amplification of a specific band of 500 bp from genomic DNA in which the transcription start site is located. Hence, the total length of the PLZF cDNA was extended to ≈8.1 kb, with a 5′UTR of 6.0 kb.
Alternative Splicing Within Exon 1 of PLZF.
As suggested by previous studies on human PLZF-RARα fusion transcript and mouse PLZF cDNA, possible alternative splicing patterns of the large exon 1 exist. As shown in Fig. 3A, when PCR primers 3.6 kb upstream of the initiation codon and at the 3′ end of exon 1 were used, a band of 1.4 kb was obtained in 5 of 11 tissues tested. In three tissues (heart, stomach, and placenta), in addition to the 1.4-kb band, there was an 1.1-kb band. Sequence analysis showed that the 1.4-kb band (designated here as AS-I) resulted from a splicing pattern similar to a mouse PLZF cDNA whereas the 1.1-kb band was generated by AS-I together with a previously reported alternative splicing (referred to AS-IV) to produce the short isoform of PLZF-RARα fusion transcript (Fig. 3). Using primers at the 3′ ends of AS-I intron and exon 1, however, we obtained shortened bands in the stomach, testis, placenta, and heart (Fig. 3B). These bands were reproducible in three independent experiments while a band of expected size was also present in these tissues, providing an internal control. When these PCR products of smaller sizes were subjected to sequence analysis, all turned out to be transcripts with intact ORFs. Southern blot analysis using specific oligonucleotide probes for putative new splicing joining sites also confirmed the above results. As shown in Fig. 3, the alternatively spliced products in stomach (AS-II), testis (AS-III), and heart, placenta, and stomach (AS-IV) were generated as a result of splicing between codons 3 and 249, 177 and 359, and 255 and 377, respectively. Of note, although AS-I occurs in the 5′UTR and therefore does not alter the protein sequence of PLZF, AS-II, -III, and -IV could delete the BTB/POZ domain, a domain rich in negatively charged amino acids and a proline rich domain. Our data thus suggest that the PLZF gene consists of six exons and five introns and that the huge exon 1 has at least four types of alternative splicing. The location of exon–intron junctions and flanking sequences are shown in Fig. 3D. Although splicing donor and acceptor signals at exon–intron boundaries for AS-I of exon 1 and other exons were classical (5′ donor-gt, 3′ acceptor-ag), AS-II, -III, and -IV within exon 1 were associated with atypical splicing sites.
Figure 3.
PCR analysis of alternative splicing. P6 and Pt2, Pt1 and Pt2 (Fig. 2) were used in A and B, respectively. (C Left) Relationship between functional domains in PLZF protein and exon–intron organization, as well as the position of exon 1 alternative splicing with regard to gene organization. (Right) Corresponding PLZF protein isoforms as a result of AS I–IV. (D Left) Splicing signals in exon–intron boundaries of exons 1–6. The sizes of exons and introns are indicated. (Right) Sequences used as splicing signals in ASI–IV.
Analysis of the Regulatory Region of PLZF Gene.
Analysis of the sequences upstream of transcription start site predicted presence of several potential transcription factor binding sites: a TATA box at −33 to −30 bp, two GAGA factor binding sites at −160 to −146 bp and −186 to −173 bp, a TFIID binding site at −199 to −189 bp, a GR site at −321 to −311 bp, and an AP1 binding site at −462 to −444 bp (Fig. 2).
Evolution of the PLZF Gene.
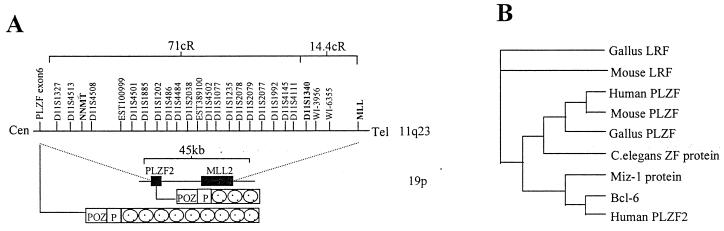
Search against public databases revealed sequences highly homologous to PLZF in several species, including C. elegans (CE19548), G. gallus PLZF (Z47402) (17) and leukemia/lymphoma-related factor (LRF) (AF086831) (34), mouse PLZF (Z47205) (17) and LRF (AF086830) (34), and human BCL-6 (U00115) and human Miz-1 (Y09723). Of note, a sequence highly similar to PLZF, designated as PLZF2, was found in a cosmid clone (AC003005) from human chromosome 19. Within this 45-kb cosmid lies another gene with high homology to MLL, referred to as MLL2. Interestingly, the MLL gene was located on 11q23, a region harboring the PLZF gene. Radiation hybrid analysis was thus carried out to determine the relative position of MLL and PLZF (35). As shown in Fig. 4A, the two genes are ≈2,587 kb apart, according to the current sequence-tagged site map. Using paupsearch and paupdisplay (GCG package), we constructed the phylogenetic tree of PLZF (Fig. 4B).
Figure 4.
(A) Comparison between the regions of PLZF/MLL on chromosome 11q23 and PLZF2/MLL2 on chromosome 19. 1 cR corresponds approximately to 30 kb. (B) Phylogenetic tree of PLZF homologous sequences through evolution. Note that PLZF ortholog is present in C. elegans.
Identification and Analysis of PLZF-RARα and RARα-PLZF Joining Regions.
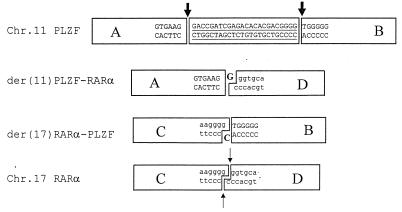
In a previous study on the index Chinese APL patient with t(11;17)(q23;q21), the breakpoint of PLZF gene was in an intron separating the exons encoding the second and third zinc fingers whereas that of RARα was at the 3′ end of the second intron. The alignment of the sequences of PLZF-RARα and RARα-PLZF joining regions obtained here by PCR against those of normal PLZF and RARα genes revealed a deletion of 24 nucleotides and an insertion of 1 nucleotide (Fig. 5). No recombination signals like heptamer and nanomer were found in regions flanking the breakpoints on both genes, eliminating the possible involvement of recombinase responsible for somatic rearrangement of antigen receptor genes. An intermediate repetitive sequence MIR was found adjacent to the breakpoint of the PLZF gene whereas an MER (one type of the DNA elements) was present on the RARα gene side. Because MIR and MER belong to two different categories of repetitive elements, it is unlikely that the translocation was mediated by homologous recombination. Trying to reconstruct a plausible translocation pathway, we realized that the sequence features could be best explained by a recently proposed “DNA damage-repair” model (38) if two double-strand and two single-strand cleavages are proposed for breaks on chromosomes 11 and 17, respectively (Fig. 5).
Figure 5.
Sequences around the breakpoints. One nucleotide (in bold) was inserted, and 24 nucleotides (underlined) were deleted in the joining regions. Thick arrows indicate double-strand DNA breaks, and thin arrows point to single-strand DNA breaks. The der(11) and der(17) could be generated by ligations of the blunt-ends of PLZF breaks with overhanging ends of RARα.
DISCUSSION
Features of the PLZF Gene.
To better understand the structure and function of PLZF, an important gene implicated in leukemogenesis, we cloned and sequenced the PLZF genomic DNA. Our study shows that PLZF is a large gene spanning a 201-kb genomic sequence. To determine its exon–intron organization, we also obtained the PLZF cDNA sequence. Several interesting features were noted. In spite of several big introns in the region, the PLZF gene does not seem to harbor other coding information. Exon 1 of PLZF contains a 5′ UTR of 6.0 kb, which is rare for known genes. The PLZF gene exhibits complex patterns of splicing. Although it has a basic structure of six exons and five introns, four alternative splicing patterns were detected in the first exon, one (AS-I) in the 5′UTR and the other three (AS-II, -III, and -IV) in the ORF. It is unclear whether alternative splicing of 5′UTR may result in different levels of transcriptional expression, but AS-II, -III, and -IV may generate PLZF protein isoforms lacking the BTB/POZ domain, the region rich in negatively charged amino acids, and the proline rich domain. Because these domains are seen typically in transcription factors and may possess important functions like transcriptional repression or dimerization, the three isoforms may show functional variations. That these alternative splicings occur in a tissue-specific manner adds more functional meanings. For example, AS-IV was mainly seen in the heart, stomach, and placenta but not in other tissues, such as bone marrow. Alternative splicing patterns AS-II, -III, and -IV use atypical splicing donor and/or acceptor signals, and the donor and acceptor signals in AS-II also were found in Ig genes. Special cellular mechanisms may exist to recognize these unique signals, which may be involved in crucial gene regulation events. The presence of a TATA box and many transcription factor binding sites in the putative promoter region indicates that PLZF gene expression may be accurately regulated by a complex network of regulators.
Orthologs and Paralogs of PLZF.
Computer-assisted homologous sequence search revealed that PLZF-related sequences are present as orthologs in several species at different stages of evolution, including C. elegans, G. gallus, and mouse. A gene highly similar to PLZF (PLZF2) was reported in a cosmid clone containing genomic DNA from human chromosome 19. PLZF2 also has a BTB/POZ domain at the N terminus and a proline-rich region, but there are only three zinc finger motifs on the C terminus. Little has been learned about its function. Interestingly, PLZF2 is closely linked to a gene highly homologous to MLL (MLL2), which was also found in the same cosmid. This is reminiscent of the fact that PLZF and MLL genes are both mapped on 11q23. Therefore, the two paralogous PLZF sequences may have arisen from a syntenic duplication in evolution, followed by a rearrangement event to place them onto different chromosomes (36).
Abnormal DNA Recombination in PLZF.
A subgroup of APL patients has t(11;17)(q23;q21) translocation resulting in the formation of both PLZF-RARα and RARα-PLZF chimeric genes (2, 7, 37). The presence of some unique morphological and immunophenotypical features, together with the poor response to all-trans retinoic acid in these patients, made some authors consider this leukemia a new, APL-like syndrome (7, 37). Most t(11;17) APL patients had chromosome 11 breakpoints mapped to intron 2, and rare cases to intron 3 of PLZF (7), both fusing to RARα in an “in frame” manner. The clustering of breakpoints in a limited region of the large intron 2 suggests that not only the translocations were biologically selected for cells to acquire growth advantage, but some sequence/structure features also could favor the breaking and fusing events. To understand the molecular mechanism of this translocation, we obtained and analyzed the sequences of joining regions of both PLZF-RARα and RARα-PLZF. The presence of a deletion of 24 nucleotides and an insertion of 1 nucleotide suggests that this translocation is reciprocal but not balanced at the fine structural level. It is unlikely that topoisomerase II is responsible for this translocation because it usually causes 4-bp duplication but no deletion. The absence of heptamer–nanomer signals and of homologous sequences on both chromosomal breaks implies that neither antigen receptor gene recombinase error nor homologous recombination could be the underlying mechanism. Of note, a recently proposed model implicating DNA damage-repair machinery could be more appropriate to explain this abnormal rearrangement event based on the sequence structure around the breakpoints of both fusion partners (38). Short deletion and duplication may thus indicate initial DNA damaging events in the form of two double-strand and two single-strand DNA breaks on chromosomes 11 and 17, respectively, followed by ligation of a blunt end with an overhanging end for both derivative chromosomes 11 and 17.
Acknowledgments
The authors thank Prof. Zhu CHEN and Prof. Zhen-Yi WANG for their support and constructive discussion. This work was supported in part by Chinese National Key Program for Basic Research (973), Chinese High Tech Program 863, National Natural Sciences Foundation of China, Shanghai Commission for Science and Technology, Samuel Waxman Cancer Research Foundation, and the Clyde Wu Foundation of Shanghai Institute of Hematology.
ABBREVIATIONS
- PLZF
promyelocytic leukemia zinc finger
- RACE
rapid amplification of cDNA ends
- BAC
bacterial artificial chromosome
- OSS
ordered shotgun sequencing
- RARα
retinoic acid receptor α
- APL
acute promyelocytic leukemia
- kb
kilobase
- RT
reverse transcription
- EST
expressed sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF060568).
References
- 1.Chen Z, Brand N J, Chen A, Chen S J, Tong J H, Wang Z Y, Waxman S, Zelent A. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S J, Zelent A, Tong J H, Yu H Q, Wang Z Y, Derre J, Berger R, Waxman S, Chen Z. J Clin Invest. 1993;91:2260–2267. doi: 10.1172/JCI116453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J Y, English M A, Ball H J, Yeyati P L, Waxman S, Licht J D. J Biol Chem. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 4.He L Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 5.Cheng G X, Meng X Q, Jin X L, Wang L, Zhu J, Xiong S M, Zhu X H, Waxman S, Licht J, Zelent A, et al. Blood. 1998;92, Suppl. 1:213. (abstr.). [Google Scholar]
- 6.Koken M H M, Reid A, Quignon F, Chelbi-Alix M K, Davies J M, Kabarowski J H S, Zhu J, Dong S, Chen S J, Chen Z, et al. Proc Natl Acad Sci USA. 1997;94:10225–10260. doi: 10.1073/pnas.94.19.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Licht J D, Chomienne C, Goy A, Chen A, Scott A A, Head D R, Michaux J L, Wu Y, DeBlasio A, Miller W H, Jr, et al. Blood. 1995;85:1083–1094. [PubMed] [Google Scholar]
- 8.Guidez F, Huang W, Tong J H, Dubois C, Balitrand N, Waxman S, Michaux J L, Martiat P, Degos L, Chen Z, Chomienne C. Leukemia. 1994;8:312–317. [PubMed] [Google Scholar]
- 9.Chen Z, Tong J H, Dong S, Zhu J, Wang Z Y, Chen S J. Genes Chromosomes Cancer. 1996;15:147–156. doi: 10.1002/(SICI)1098-2264(199603)15:3<147::AID-GCC1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Ruthardt M, Testa U, Nervi C, Ferrucci P F, Grignani F, Puccetti E, Grignani F, Peschle C, Pelicci P G. Mol Cell Biol. 1997;17:4859–4869. doi: 10.1128/mcb.17.8.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong S H, David G, Wong C W, Dejean A, Privalsky M. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 13.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, et al. Nature (London) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 14.Collins S J. Blood. 1998;91:2631–2633. [PubMed] [Google Scholar]
- 15.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Blood. 1998;91:2634–2642. [PubMed] [Google Scholar]
- 16.Cheng G X, Zhu X H, Men X Q, Wang L, Huang Q H, Chen J Q, Xu S F, So E, Chan L C, Waxman S, et al. Proc Natl Acad Sci USA. 1999;96:6318–6323. doi: 10.1073/pnas.96.11.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook M, Gould A, Brand N, Davies J, Strutt P, Shaknovich R, Licht J, Waxman S, Chen Z, Gluecksohn-Waelsch S, et al. Proc Natl Acad Sci USA. 1995;92:2249–2253. doi: 10.1073/pnas.92.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivins S, Zelent A. Blood. 1998;92, Suppl. 1:308. (abstr). [Google Scholar]
- 19.Hawe N, Soares V, Niswander L, Cattoretti G, Pandolfi P P. Blood. 1996;88,Suppl.:291. (abstr.). [Google Scholar]
- 20.Chen E Y, Schlessinger D, Kere J. Genomics. 1993;17:651–656. doi: 10.1006/geno.1993.1385. [DOI] [PubMed] [Google Scholar]
- 21.Chen C N, Su Y, Baybayan P, Siruno A, Najaraja R, Mazzarella R, Schlessinger D, Chen E. Nucleic Acids Res. 1996;24:4034–4041. doi: 10.1093/nar/24.20.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard O, Guglielmi P, Jonveaux P, Cherif D, Gisselbrecht S, Mauchauffe M, Berger R, Larsen C J, Mathieu-Mahul D. Genes Chromosomes Cancer. 1990;1:194–208. doi: 10.1002/gcc.2870010303. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritch E F, Mainiatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 459–463. [Google Scholar]
- 24.Werle E, Schneider C, Renner M, Volker M, Fiehn W. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanke M, Wink M. BioTechniques. 1994;17:858–860. [PubMed] [Google Scholar]
- 26.Chen E Y. In: Automated DNA Sequencing and Analysis Techniques. Adams M D, Fields C, Venter J C, editors. London: Academic; 1994. pp. 3–10. [Google Scholar]
- 27.Altschul S F, Gish W, Miller W, Myers E M, Lipman D J. J Mol Biol. 1990;15:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Chen E Y, Zollo M, Mazzarella R, Ciccodicola A, Chen C N, Zuo L, Heiner C, Burough F, Repetto M, Schlessinger D, D’Urso M. Hum Mol Genet. 1996;5:659–668. doi: 10.1093/hmg/5.5.659. [DOI] [PubMed] [Google Scholar]
- 29.Walter M A, Spillett D J, Thomas P, Weissenbach J, Goodfellow P N. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- 30.Collins F S, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- 31.Licht J D, Shaknovich R, English M A, Melnick A, Li J Y, Reddy J C, Dong S, Chen S J, Zelent A, Waxman S. Oncogene. 1996;12:323–336. [PubMed] [Google Scholar]
- 32.Zollo M, Mazzarella R, Bione S, Toniolo D, Schlessinger D, D’Urso M, Chen E Y. DNA Seq. 1995;6:1–11. doi: 10.3109/10425179509074693. [DOI] [PubMed] [Google Scholar]
- 33.Reid A, Gould A, Brand N, Cook M, Struff P, Li J, Licht J, Waxman S, Krumlauf R, Zelent A. Blood. 1995;86:4544–4552. [PubMed] [Google Scholar]
- 34.Davies J M, Hawe N, Kabarowski J, Huang Q H, Zhu J, Brand N J, Leprince D, Dhordain P, Cook M, Morriss K G, Zelent A. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 35.Baysal B E, van Schothorst E M, Farr J E, James M R, Devilee P, Richar C W., III Genomics. 1997;44:214–221. doi: 10.1006/geno.1997.4880. [DOI] [PubMed] [Google Scholar]
- 36.Henikoff S, Greene E A, Pietrokovski S, Bork P, Attwood T K, Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 37.Sainty D, Liso V, Cantu-Rajnoldi A, Head D, Mozziconacci M J, Benattar L, Arnoulet C, Grimwade D, Biondi A, Birg F, et al. Blood. 1998;92, Suppl. 1:603. (abstr.). [PubMed] [Google Scholar]
- 38.Reichel M, Gillert E, Nilson I, Siegler G, Greil J, Fey G H, Marschalek R. Oncogene. 1998;17:3035–3044. doi: 10.1038/sj.onc.1202229. [DOI] [PubMed] [Google Scholar]