Abstract
Mariner-family transposable elements are active in a wide variety of organisms and are becoming increasingly important genetic tools in species lacking sophisticated genetics. The Himar1 element, isolated from the horn fly, Haematobia irritans, is active in Escherichia coli when expressed appropriately. We used this fact to devise a genetic screen for hyperactive mutants of Himar1 transposase that enhance overall transposition from ≈4- to 50-fold as measured in an E. coli assay. Purified mutant transposases retain their hyperactivity, although to a lesser degree, in an in vitro transposition assay. Mutants like those described herein should enable sophisticated analysis of the biochemistry of mariner transposition and should improve the use of these elements as genetic tools, both in vivo and in vitro.
Mariner-family transposable elements are a diverse and taxonomically widespread group of transposons occurring throughout the Animal kingdom (1–4). They encode transposases that belong to an extended superfamily of transposases and retroviral integrases distinguished by a conserved D,D35E (or variants thereof: mariners = D,D34D) motif in the catalytic domain of the protein Fig. 1 (5). Transposition of these elements follows a conservative cut-and-paste mechanism (6).
Figure 1.
Schematic diagram of Himar1 transposase showing notable landmarks, functional domains, and the positions of the hyperactive mutations isolated in this study. The putative DNA-binding domains are based on comparisons to the Tc1 and Tc3 transposases of C. elegans and to computer predictions of Tc1/mariner transposase structures (3, 37, 40). “D” indicates the positions of the catalytic residues of the putative D, D34D catalytic domain. Specific amino acids noted are hyperactive mutations isolated in this study. C9 and A7 refer to clones for those specific mutants as described in the text. NLS refers to the positions of two putative nuclear localization signals (beginning at positions 184 and 243, respectively) as predicted by the program psort (55).
Most mariners are known only from their sequences obtained either through homology-based PCR screens or by the examination of sequenced genes or ESTs (1, 7). Hundreds of different mariners have been detected in this way. Of these, only two are known to be active. The first is the canonical mariner element from Drosophila mauritiana discovered by its activity in that fly (8). The most active copy of this particular element is known as Mos1 (9). The second is the Himar1 element discovered by using homology-based PCR in the horn fly, Haematobia irritans, and reconstructed as a consensus sequence (7, 10). Both Mos1 and Himar1 require no host-specific factors for transposition and so have been advanced as generalized genetic tools (11–13). Indeed, Mos1 has been used as a transformation vector for chicken (14), zebrafish (15), the yellow fever mosquito, Aedes aegypti (16), Drosophila melanogaster (17), Drosophila virilis (11), and Leishmania major (12), with varying degrees of success. Himar1 has been used as a prokaryotic genetic tool, via in vitro transposition and subsequent homologous recombination in Haemophilus influenzae and Streptococcus pneumoniae, and in vivo in Escherichia coli and Mycobacterial spp. (18, 19). It is also active in human cells (20).
Whereas mariner elements are becoming increasingly important tools for eukaryotic genetics, neither Mos1 nor Himar1 appear to be as active as would be desired to make them efficient tools, particularly for whole metazoa (13, 15). In fact, these transposases may have evolved to be less active in their hosts and, therefore, be less deleterious (13, 21, 22). Such low transposition activity makes the use of these elements for genetic manipulations less practical. Identifying mutant transposases with higher activity might help to solve this problem but is difficult to carry out in metazoan systems (23). We have exploited the ability of Himar1 to transpose in prokaryotes to create a genetic system for isolation of transposase mutants with altered activity in vitro and in vivo. We have isolated highly active mutants that significantly improve the efficiency of Himar1-derived elements as genetic tools. Analysis of these mutants suggests the locations of functional domains and amino acids within the Himar1 transposase and mechanisms that may have led to the evolution of decreased transposase activity.
METHODS
Media and Antibiotics.
Strains were grown at the temperatures indicated in LB broth or on agar plates prepared as described (24). Papillation assays were performed on thick MacConkey lactose agar plates. Antibiotics concentrations were ampicillin (Amp), 100 μg; gentamicin (Gen), 10 μg; kanamycin (Kan), 40 μg; tetracycline (Tet), 15 μg; naladixic acid (Nal), 20 μg; chloramphenicol (Cam), 34 μg, per ml, respectively, except where otherwise noted.
Plasmids and Bacterial Strains.
pMarNco was constructed via PCR using the primers 5′-CCCCTCGAGCCATGGAAAAAAAGGAATTTCGTG-3′ and 5′-CCGCTCAGAATCATCAACACGTT-3′ and pMarNde18 (10) as a template. The resulting PCR fragment and pMarNde18 were cut with XhoI and EcoRV and were ligated to created pMarNco, which contains the Himar1 transposase coding sequence with an NcoI site at the start codon. pBCMar was constructed by cleaving pMarNde18 with PstI, creating a blunt end using T4 DNA polymerase, then digesting with XhoI. The fragment that encodes the transposase was gel purified and cloned into pBCKS+ that had been digested with XhoI and Ecl136I.
A miniHimar1 transposon containing an ORF through the 3′ inverted terminal repeat (ITR) was constructed by PCR using a PCR-ligation-PCR method (25). The 5′ ITR was amplified with primers 5′ TACCCGGGAATCATTTGAAGGTTGGTAC (76rSma) and 5′ TAATACGACTCACTATAGGG (T7), and the 3′ ITR was amplified with the primers 5′AACGAATTTTAACAAAAAAATGTG (Mar3′r) and 5′CGATTTAGGTGACACTATAG (SP6), both using pMinimariner (13) as a template and Pfu polymerase. The two separate PCR reactions were treated with T4 polynucleotide kinase and ATP. Five microliters of each kinase reaction were ligated with T4 DNA ligase for 15 min at room temperature. Another PCR reaction then was performed by using the SP6 and T7 primers and 1 μl of the ligation as a template, using Taq DNA polymerase. The resulting product was cloned as a t-tailed fragment into pTAdv1 (CLONETECH), producing pTAdvMMOrf. This clone was cut with BamHI, and the fragment containing the minimariner was isolated and ligated to the BamHI site of pCDNAII (Invitrogen) to produced pMMOrf, which contains a unique BglII site in the middle of the element.
A papillation construct was produced by cleaving pMMOrf with BglII and ligating it to the BamHI/BglII fragment of pRZ1495 (26) to produce pMMLacTet. The lacZ gene of this insert has no transcriptional or translational controls (27).
An F-plasmid containing the papillation transposon from pMMLacTet was produced by transforming E. coli β2155 (28) by electroporation with pBCMar and pMMLacTet with selection on Amp, Cam (20 μg/ml), and diaminopimelic acid. Diaminopimelic acid is required for growth of β2155, which is a dapA mutant. Approximately 5,000 colonies were pooled and mixed with HB101 for a 6 h of mating on LB agar. Exconjugants in which the F′ from β2155 was mated out into HB101 were selected on Tet and Xgal (20 μg/ml) in the absence of diaminopimelic acid, resulting in colonies that were dark blue, light blue, white, or mosaic. White colonies were picked and colony purified on a second LB-Tet-Xgal plate. Resulting clones were patched to plates containing either Amp, Cam, Kan, or Tet, and clones confirmed to be Amps, Cams, Kans, and TetR were named HBFlac.
To verify that the HBFlac strains contain functional Himar1 ′lacZ elements, they were transformed with pBCMar and were selected on Cam and Xgal. The resulting colonies were dark blue, light blue, white and mixed (i.e., exhibited blue papillae on white colonies). The F′ from HBFlac3 (the lightest blue strain) was transferred to DH5α by conjugation with selection on Nal and Tet, yielding the strain DL1.
A Himar1 transposon for use in the mating-out assay was constructed by ligating the BamHI/EagI fragment of pMarKan (10) containing a KanR-marked Himar1 transposon to the BamHI/EagI fragment of pACYC184, resulting in the plasmid pACMarKan.
E. coli protein expression plasmids were made for each of the hyperactive transposase mutants by cutting the pBAD24 vectors containing the hyperactive inserts with NcoI, making this site blunt with Klenow, and cutting again with KpnI. The coding sequence fragments were purified from a 0.5% 1× TAE (40 mM Tris-acetate/1 mM EDTA) gel and were ligated to the NdeI (made blunt with Klenow as above)/KpnI sites of pET29b+ (Novagen), yielding pET29A7 and pET29C9 for the pBADA7- and pBADC9-containing transposase mutants, respectively.
Transposase Mutagenesis.
Mutations were introduced into the coding region of Himar1 transposase by error-prone PCR using pMarNco as a template (29). The reactions contained ≈2 ng of template DNA, 50 mM KCl, 10 mM Tris⋅HCl (pH 9.0 at 25°C), 0.1% Triton X-100, 1.5 mM MgCl2, and either 200 or 100 μM MnCl2 in a volume of 25 μl and were run for 30 cycles at 95°C for 1 min, 52°C for 1 min, and 75°C for 1.5 min. PCR products were cut with NcoI and PstI at 37°C for 45 min. The cleaved products were isolated from a 0.5% agarose gel in 1× TAE buffer by using a Qiagen (Chatsworth, CA) gel purification kit. Purified products were ligated into the NcoI/PstI sites of pBAD24 (30). These ligation reactions were used as the source of transposase mutants in the papillation assay.
Papillation Assay.
A papillation assay to detect mutants of Himar1 was performed by transforming 1 μl of a ligation of mutated Himar1 coding sequences in pBAD24 into electrocompetent DL1 cells (see Fig. 2A). Screens of similar design have been used for bacterial transposons, including in Tn5 and Tn10 transposases (31–33). Cells also were transformed with pBADMar1 as a wild-type control. Transformed cells were resuspended in 1 ml of cold LB medium and were shaken at 37°C for 1 h. Dilutions of these cells were plated onto thick (50 ml in 100- × 15-mm dishes) MacConkey lactose agar plates containing Amp and Tet so that there were ≈100–150 colonies per plate, and the plates were incubated at 32°C for 2–3 days. Typically, papillae could be detected by using the wild-type transposase source at ≈50 h after plating. Potential hypertransposers were picked and grown overnight in LB with Amp, and the mutant transpose source DNA was purified. Putative hypertransposers were examined again by using the papillation assay to confirm hyperactivity.
Figure 2.
Overview of the papillation and mating-out assays used for detection and measurement of transposition frequency of mutant Himar1 transposases. (A) Papillation assay (after ref. 43). The papillation screen was used to examine a pool of mutant transposase sequences to recover hyperactive transposases. The Himar1 coding sequence was mutagenized via error-prone PCR and was cloned into pBAD24. Transposase from this construct can mobilize a nonautonomous Himar1 transposon carrying an in-frame fusion of the 3′ ITR and a defective lacZ gene from the F plasmid to the E. coli chromosome. If the transposon fuses in the correct orientation and frame to an E. coli gene, a fusion protein can be produced with β-galactosidase activity. The subpopulation of E. coli in the colony in which this occurs are thus transformed from Lac(−) to Lac(+). When plated on MacConkey lactose agar, transposition can be scored by the formation of papillae, or red bumps, that form on the colonies. The greater the number of papillae after a given period of time, the higher the frequency of transposition. (B) Mating-out assay. The mating-out assay was used to quantify the relative frequency of individual transposase constructs. A plasmid expressing transposase isolated from the papillation screen was transformed into a strain of E. coli that contains a GenR F plasmid and a plasmid (p15 ori) carrying a KanR Himar1 transposon. Cells were plated on media to select for each plasmid. Individual colonies were grown overnight under the same selection in rich broth. The next day, these cells were mated to a NalR strain of E. coli. The total number of exconjugates were detected by selecting recipient cells with Nal-Gen. Transpositions into the target F plasmid were detected by plating the mated cells on Nal-Kan. The ratio of NalR-KanR exconjugates to NalR-GenR exconjugates is a measure of the frequency of transposition.
Mating-Out Assay.
A mating-out assay, which measures the frequency of transposition of a KanR Himar1 minitransposon from a plasmid to an F factor, was carried out to quantify the activity of the putative hyperactive mutants (see Fig. 2B). RZ212(MK) cells were transformed with individual mutant transpose sources isolated in the papillation assay, and the cells were grown as above. Cells were plated on LB agar containing Amp, Gen, and Cam and were grown overnight at 37°C. Five colonies were picked the following day and were grown for 16 h in LB containing Amp, Gen, and Kan at 37°C. These cells were mated to RZ221 cells by mixing 10 μl of the donor strain and 30 μl of recipient cells from overnight cultures in 1 ml of LB medium. The mating mixture was shaken gently at 37°C for 6–10 h. Mating cultures were vortexed vigorously, and suitable dilutions were plated on LB agar plates containing Nal and Gen to detect total numbers of exconjugates and on Nal and Kan to measure the number of exconjugates that contained a Himar1 insertion.
Transposase Purification.
Transposases were purified as described in ref. 10. Protein purity was determined by Coomassie blue-stained 10–20% polyacrylamide gradient gels. Protein concentrations were determined spectrophotometrically as described in ref. 13 and were confirmed visually on Coomassie blue-stained 4–20% SDS/PAGE gels.
In Vitro Transposition Assay.
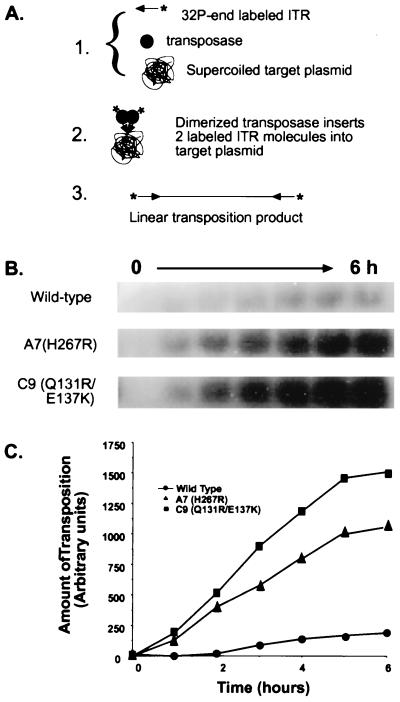
Comparative rates of transposition were determined by measuring the relative ability of the transposases to incorporate a radiolabeled DNA fragment containing the left ITR of Himar1 into an unlabeled supercoiled plasmid target in a reaction similar to that for Tn10 in vitro (Fig. 4) (34). The fragment containing the ITR was labeled by cutting p27fH5′ (10) with EcoRI and isolating the 111-bp fragment on a 1.5%, 1× TAE agarose gel. The DNA was purified from the agarose as described above and then was radiolabeled by filling in the overhanging ends with 32P-α-dATP using Klenow under standard conditions. The reaction was stopped by heating to 70°C for 20 min, and the labeled DNA was purified by passing the reaction over a G50 spin column.
Figure 4.
Comparison of the activity of purified wild-type and hyperactive Himar1 transposases in vitro. (A) Assay overview. Purified transposase is mixed with a short 32P-end-labeled DNA fragment containing the 5′ ITR of Himar1 and cold supercoiled plasmid target DNA. The reaction is incubated at 28°C, and aliquots of the reaction are withdrawn every hour for 6 h. The aliquots are run on an agarose gel, and the gel is dried and analyzed. Transposition using two labeled ITR fragments is equivalent to a normal transposition event by Himar1. This event linearizes the target DNA and labels it with 32P, which is easily measured by autoradiography and phosphorimaging. The rate at which the product accumulates is a measure of the transposition frequency. (B) Typical results of the in vitro assay. Autoradiograph showing the accumulation of the radiolabeled linear transposition product for wild-type, A7 (H267R), and C9 (Q131R/E137K) transposases, respectively, over a period of 6 h. (C) Graphical representation of the data in B. The gel was analyzed by using a Molecular Dynamics PhosphorImager. The values on the y axis are density units based on the numbers of pixels per unit area as measured by imagequant software.
Transposition reactions contained 10% glycerol (vol/vol), 25 mM Hepes (pH 7.9 at room temperature), 250 of μg acetylated BSA, 2 mM DTT, 100 mM NaCl, 5 mM MgCl2, 450 ng of target plasmid DNA, ≈10,000 cpm labeled ITR DNA, and a 10 nM concentration of one of the purified transposases. Reactions were performed at 28°C, the optimal temperature for wild-type Himar1 transposase (13). Ten-microliter aliquots were removed at 1-h intervals for 6 h, and the reaction was stopped by adding 2 μl of stop solution (60 mM EDTA/0.25% bromophenol blue/0.25% xylene cyanol/15% ficoll). Reaction products were separated on a 0.5% 1× TAE agarose gel. The gel was photographed, was placed on a piece of exposed x-ray film as a support, and then was dried until completely flat in a forced-air oven set at 55°C for 5–6 h. Reaction products in dried gels were analyzed by using a Molecular Dynamics PhosphorImager and imagequant software.
Mutagenesis of Haemophilus influenzae.
In vitro reactions for H. influenzae mutagenesis were conducted as above except that 100 nM transposase was added to reactions containing 500 ng of target PCR product and 200 ng of transposon donor plasmid pENT3 carrying Tn-magellan1. Independent reactions and transformations were performed in triplicate. Repair of transposon junctions and transformation of H. influenzae was as described (18).
RESULTS
Transposase Mutation and Papillation Screen.
A papillation screen was used to detect altered levels of transposition frequency for a pool of transposase mutants constructed by error-prone PCR (Fig. 2A). The Himar1 papillation screen is based on the ability of Himar1 to mobilize a nonautonomous Himar1 transposon carrying an in-frame fusion of a lacZ gene off an F plasmid and into the E. coli chromosome. If the transposon insertion fuses in frame with an expressed E. coli gene, a protein fusion can be produced that contains β-gal activity. The Lac(+) subpopulation of cells in an otherwise Lac(−) E. coli colony can metabolize lactose if plated on MacConkey agar. The Lac(+) cells grow faster than the surrounding Lac(−) cells and thus will produce bumps, or papillae, on the colony. Moreover, these papillae turn red on MacConkey lactose agar because of the production of lactic acid and its detection by the neutral red in the media. The greater the number of papillae produced after a given period of time, the greater is the frequency of transposition.
Transposition of Himar1 in E. coli was readily detected by using the papillation screen. Papillation only occurred when using Himar1 transposase constructs. The proportion of colonies showing any transpositional activity depended strongly on the concentration of MnCl2 used in the error-prone PCR. Using 250 μM MnCl2, we recovered only one hypertransposer (A7) of 2,500 colonies screened, the vast majority of which were nulls or hypomorphs, presumably through introduction of multiple mutations. Using 100 μM MnCl2, we were able to increase the number of colonies showing papillation of some degree to ≈30% and recovered 10 potential hypertransposers from 2,300 colonies screened.
Mating-Out Assays.
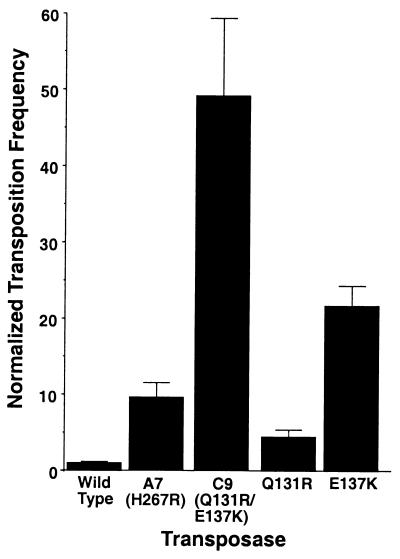
A mating-out assay is used to measure quantitatively the relative frequency of transposition produced by individual transposases (31, 35). This assay measures the frequency with which an F plasmid is used as a target by a nonautonomous Himar1 transposon mobilized by a given transposase source. After mating the target F plasmids to a recipient strain, the transposition frequency is determined by measuring the ratio of F plasmids carrying KanR (the marker in Himar1) to all exconjugates (Fig. 2B).
Mutants A7 and C9 were particularly active in the papillation screen and so were chosen for further analysis in the mating-out assay. The A7 mutant was ≈10-fold more active in E. coli than the wild type whereas the C9 mutant was ≈50-fold more active (Fig. 3). Sequencing showed that both mutants contained multiple amino acid changes (two each in A7 and C9). By substituting wild-type sequences for mutated ones and testing the isolated mutant amino acid changes again in the papillation assay, it was possible to determine which amino acid changes actually conferred the hyperactivity. Doing this, we found that a H267R change in mutant A7 and both the Q131R and E137K changes in mutant C9 conferred hyperactivity (Fig. 1).Testing the individual C9 mutations in the mating-out assay showed that the Q131R mutation alone was ≈4-fold more hyperactive whereas the E137K alone was ≈20-fold more hyperactive. The combination of these mutations is ≈50-fold more active, and, thus, it would appear that these mutations act synergistically, and not simply additively.
Figure 3.
Relative activity of wild-type and hyperactive Himar1 transposases as measured in the mating-out assay. The frequency of transposition is expressed relative to that of wild-type Himar1 transposase, which has been normalized to a value of 1.0. A typical transposition frequency for wild-type transposase under the conditions described here is ≈4 × 10−6. Relative frequencies are calculated by dividing each of the absolute frequencies by the average absolute transposition frequency of wild-type Himar1 transposase. The errors are SEM. Relative errors are computed by dividing the absolute errors by the mean absolute frequency for the wild-type transposase.
Most of the seemingly hyperactive mutants isolated in the papillation assay were not hyperactive in the mating-out assay (data not shown). The reason for this is unclear, but false positive results have been reported when using similar assays with both Tn10 and Tn5 (31, 32). Wild-type Himar1 is most active at 30°C (13), so the fact that we isolated the mutants at 32°C and quantified their activity at 37°C may mean that A7 and C9 are more stable at the higher temperature used for the mating-out assay.
In Vitro Transposition.
It is possible that the hyperactive mutations we recovered were attributable to some novel interaction with the E. coli host and not to some property in the transposase itself. To address this question, we examined purified A7 and C9 transposases in an in vitro transposition assay. This assay measures the relative ability of purified transposase to process and insert two 32P end-labeled ITR DNA fragments into an unlabeled supercoiled DNA target, thus producing a labeled linear transposition product that is easily quantified (Fig. 4A). The rate at which this labeled product accumulates is a measure of transposition frequency. Fig. B and C shows that the A7 and C9 transposases were both hyperactive compared with the wild type. By measuring the slope of the linear portions of the curves in Fig. 4C (between 1 and 5 h), it is possible to compare the rate of product accumulation. Doing this, we found that the A7 transposase was 4.8-fold more active than the wild type whereas C9 was 7-fold more active. Thus, neither of the purified transposases was as active in the in vitro assay as in the mating-out assay. These results, however, do confirm that hyperactivity is intrinsic to the transposase protein and not wholly the result of some novel interaction with E. coli.
Activity in Mutagenesis Applications.
In vitro mutagenesis is rapidly becoming an important tool for studies of gene function. We have developed a technique using the Himar1 mariner system to mutagenize targeted genomic regions of the chromosome of a human respiratory pathogen, Haemophilus influenzae. (18). Analysis of such regions is enhanced by using large pools of mutants, which requires high efficiency of transposition. We examined whether the hyperactive transposases could improve mutagenesis frequencies in this system. Wild-type, A7, and C9 transposases were tested for the ability to move the minimariner elementTn-magellan1 carrying the gene for kanamycin resistance into a PCR-amplified chromosomal segment of H. influenzae. After repair of single-strand gaps introduced into the target DNA by the transposition reaction, DNA was transformed into competent H. influenzae cultures as described (18). The number of Kan-resistant H. influenzae colonies obtained with the wild-type, A7, and C9 transposases were 217 ± 122, 633 ± 50, and 733 ± 49, respectively. These results indicate that mutagenesis of H. influenzae with Himar1 could be significantly improved by the use of hyperactive transposases.
DISCUSSION
Mariner transposons are well known for their wide distribution in animals, which suggests that they do not rely on any host-specific factors for transposition. Indeed, members of the Tc1-mariner superfamily are active in a wide range of organisms, and both Tc1 and Himar1 transposases are capable of catalyzing transposition in the absence of any host proteins (10, 36). Our recent finding that Himar1 is active in E. coli (19) has provided the opportunity to utilize bacterial genetic methods to create and study transposase mutants of this eukaryotic transposon in a manner that would be very difficult in a metazoan system. Mutants of Mos1 mariner have been isolated by ethane methylsulfonate mutagenesis in Drosophila melanogaster, but this system is laborious and only mutations that negatively affected transposition were detected (23). The combination of the papillation screen and mating-out assay in E. coli described above is a simple method to produce mutants of mariners and related transposases and ascertain their level of activity.
Hyperactive Transposase Mutants of Himar1.
We were able to isolate two hyperactive mutants in an E. coli papillation screen that produced significant levels of hyperactivity in a quantitative mating-out assay. Both were double mutants, and, in one case (C9), both amino acid changes contributed synergistically to the overall hyperactivity, suggesting that additional combinations of mutants constructed directly, or by shuffling during the mutagenesis, might be even more hyperactive.
Although there is no structural data available for the Himar1 transposase, analysis of the mutant sequence along with comparison to other known transposases suggests the locations of functional domains. The only structural information for any of the Tc1-mariner superfamily of transposons is that for the Caenorhabditis elegans transposase Tc3, and then only for the specific DNA binding domain, a region that does not include the Himar1 mutations (37). Functional studies have been performed for both Tc1 and Tc3 transposases that demonstrated the existence of a separate, nonspecific DNA binding domain in each (38, 39). By comparing Tc1 and Tc3 transposases and computer models (40) with Himar1 transposase, we can assume that specific DNA binding is encoded by the first ≈113 amino acids of Himar1, nonspecific DNA binding by approximately amino acids 114–173, and catalysis by at least amino acids 158 and 287, the first and last amino acids of the D, D34D catalytic triad (Fig. 1). The C-terminal-most region is of unknown function. Given the fact that the Q131R and E137K mutations occur in a region of the transposase implicated in nonspecific DNA binding, the enhanced activity in these mutants may be attributable to increased affinity for DNA in general (Fig. 1). Similarly, the H267R mutation, which occurs in the putative catalytic domain, may be attributable to increased or altered catalysis. In Tn5 transposase, these various regions are known to overlap extensively, so a mutation in one region may effect a completely different property of the transposase (41). Indeed, the ways in which a transposase can become hyperactive are diverse. For example, at least three different classes of hyperactive mutations have been uncovered for Tn5. These affect the production of cotranslated inhibitor protein (42), an increase in the affinity of Tn5 transposase for ITR DNA (29), and a decrease in the self-inhibitory activity of intact Tn5 transposase (43). The combination of these three classes of hyperactive mutants are synergistic, leading to an extraordinarily active transposase (44).
Comparison of Himar1 Hyperactive Changes to Amino Acid Sequences in Other Tc1-mariners.
Abundant sequence data is available for members of the Tc1-mariner family of transposons. Alignment of the available transposase sequences (3) allowed us to determine whether any of the amino acid replacements we uncovered are present in related transposases. Interestingly, one of the amino acid changes is present in a homologous position in the highly active Mos1 mariner. This transposase contains an arginine residue at the position of the Q131R mutation we isolated. This is not a highly conserved position in mariner transposases generally, although Tc1-like elements are biased toward basic residues at this position. The E137R mutation is not present in most other mariner family elements because this region is a unique small insertion in the irritans subfamily of mariner transposases to which Himar1 belongs. Finally, the H267R replacement of A7 is shared with one other member of the irritans subfamily (Hsmar2) and two members of the mellifera subfamily (Gpmar1 and Demar1), but, again, this is not a widely conserved position in mariner transposases.
Evolutionary Implications of Hyperactivity.
Reznikoff and coworkers have stressed that the ability to isolate hyperactive transposases of Tn5 strongly suggests it has not evolved for maximal activity (41). Tn5 transposase mutants have been isolated that can increase the intrinsic activity of the transposase or eliminate regulatory mechanisms (29, 45). Low intrinsic activity and self-regulation appear to allow Tn5 to persist in E. coli without producing serious levels of genetic damage. Our ability to isolate hyperactive mutants for Himar1 may be attributable to similar evolutionary forces at work on mariner−family transposons. Horizontal transfer is a major feature in the evolutionary history of these mobile genes (4, 7, 46, 47). Clearly, the elements must be active enough to make copies of themselves when they transfer to a new host to persist. If not, they will be eliminated because of stochastic mechanisms (47). Their activity, however, cannot be so high as to significantly reduce the fitness of the host. Unregulated transposition can be highly deleterious to a host organism (48). Thus, the mariners that persist are not likely to be present in their most active forms. From the standpoint of copy number in the host organism, Himar1 is very successful, being present in 17,000 copies in the H. irritans genome (7). It may be that this success is attributable to a fairly benign level of activity in that genome because of either a comparatively low intrinsic activity level, some self-regulatory mechanism (13), or both. Our ability to isolate hyperactive forms of this transposase lends credence to this view, assuming that such mutants retain their hyperactivity in the natural host as well as in E. coli.
Practical Uses.
The mariner-family transposons Himar1 and Mos1 are being developed in a number of organisms for a variety of uses and hold great promise to extend the techniques of transposon-based genetic analyses available for E. coli and Drosophila to other organisms (12, 15, 18, 19, 49). For these elements to be used easily and efficiently, especially Himar1, their activity needs to be improved (13, 15). One way to do this is to establish optimum conditions under which transposition can take place. This method is most suitable for in vitro work by using purified transposases (13).
Alternatively, hyperactive transposase mutants such as those described here can be used and are well suited for both in vivo and in vitro work. The two mutants we recovered appear to be somewhat tolerant of high temperatures, which would be particularly useful in E. coli and human cells. Functional genomic applications of these transposons, like gambit (18), are improved by using these enhanced transposases. Clearly, labor-intensive methods such as germline transformation could be eased by more active transposases. A better understanding of the biochemistry of mariner transposases provided by mutagenesis methods described herein can be expected to provide transposases that have differing target sites, improved thermal stability, and enhanced activity in specific hosts. Finally, mutants may be uncovered that are amenable to crystallization and structural study, a prospect that is not currently possible because of the poor solubility of mariner transposases.
Table 1.
Bacterial strains and plasmids used in this study
| Strains, plasmids | Description | Reference or Supplier |
|---|---|---|
| Strains | ||
| β2155 | thrB1004 pro thi strA hsdS lacZΔM15 | 28 |
| (F′ lacZΔM15 lacI9 traD36 proA+ proB+) | ||
| ΔdapA∷erm (=EmR) pir∷RP4 [∷kan (KmR) from SM10] | ||
| HB101 | supE44 hsdS20 (rB−mB−) recA13 ara-14 | 50 |
| proA2 acY1 galK2 rpsL20 xyl-5 mtl-1 | ||
| HBfLac | HB101 F∷miniHimar1LacTet from pMMLacTet | This study |
| DH5α | Δ(lac)U169 endA1 gyrA96 hsdR17 recA1 | 51 |
| relA1 supE44 thi-1 φ80lacZΔM15 | ||
| DL1 | DH5α F∷miniHimar1LacTet from pMMLacTet | This study |
| RZ212/pOX38-Gen | D(lac-pro), ara, str, recA56, srl, thi, | 52 |
| RZ212MK | RZ212/ pACMarKan (mating-out strain) | This study |
| RZ221 | polA, Δ(lac-pro), ara, str, nal | 53 |
| BL21(DE3) | F−, omp T, rB− mB− l DE3 | 54 |
| Plasmids | ||
| pMMOrf | Like pMMar but with ORF through 3′ ITR | This study |
| pMMLacTet | pMMOrf containing lacZYA and Tetr gene | This study |
| pBCMAR | Himar1 coding sequence under Plac control | This study |
| pMarNco | Himar1 coding sequence with NcoI at start site | This study |
| pRZ1495 | Tn5 papillation factor | 26 |
| p27fH-5′ | pK19 containing left (5′) ITR of Himar1 | 10 |
| pACMarKan | pACYC184 carrying KanrHimar1 | This study |
| pMarNde18 | Himar1 lacking ITR sequences | 10 |
| pMinimariner | Himar1 of only the first and last 100 bp | 13 |
| pBAD24 | Expression plasmid with araBAD promoter | 30 |
| pBADMar1 | Himar1 tpase under araBAD promoter | This study |
| pBADH267R | As pBADMar1 but with H267R mutation | This study |
| pBADC9 | As pBADMar1 but with Q131R and E137K mutations in Himar1 tpase | This study |
| pET29A7 | pET29b+ carrying H267R mutation in Himar1 tpase | This study |
| pET29C9 | pET29b+ carrying Q131R and E137K mutations in Himar1 tpase | This study |
| pCDNAII | Target plasmid for in vitro reactions | Invitrogen |
Acknowledgments
We acknowledge W. S. Reznikoff and W. Metcalf for the gift of plasmids and helpful discussions. This work was supported by a U.S. Department of Agriculture/National Research Initiative grant to H.M.R. (95-37302-1796), a Public Health Service grant to H.M.R. and D.J.L. (2R01AI33586-04), a Damon-Runyon grant (DRG1371) to B.J.A., and National Institutes of Health Grants AI-02137 to E.J.R. and AI-26289 to J.J.M.
ABBREVIATIONS
- Amp
ampicillin
- Gen
gentamicin
- Kan
kanamycin
- Tet
tetracycline
- Nal
naladixic acid
- Cam
chloramphenicol
- ITR
inverted terminal repeat
References
- 1.Robertson H M. Nature. 1993;362:241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 2.Robertson H M, MacLeod E G. Insect Mol Biol. 1993;2:125–139. doi: 10.1111/j.1365-2583.1993.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 3.Robertson H M, Asplund M L. Insect Biochem Mol Biol. 1996;26:945–954. doi: 10.1016/s0965-1748(96)00061-6. [DOI] [PubMed] [Google Scholar]
- 4.Robertson H M, Soto-Adames F N, Walden K K O, Avancini R M P, Lampe D J. In: Horizontal Gene Transfer. Syvanen M, Kado C, editors. London: Chapman & Hall; 1998. [Google Scholar]
- 5.Doak T G, Doerder F P, Jahn C L, Herrick G. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig N L. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 7.Robertson H M, Lampe D J. Mol Biol Evol. 1995;12:850–862. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson J W, Medhora M M, Hartl D L. Proc Natl Acad Sci USA. 1986;83:8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medhora M M, MacPeek A H, Hartl D L. EMBO J. 1988;7:2185–2189. doi: 10.1002/j.1460-2075.1988.tb03057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lampe D J, Churchill M E, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 11.Lohe A R, Hartl D L. Genetics. 1996;143:365–374. doi: 10.1093/genetics/143.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gueiros-Filho F J, Beverley S M. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 13.Lampe D J, Grant T E, Robertson H M. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D, Sang H. Nat Biotechnol. 1998;16:1050–1053. doi: 10.1038/3497. [DOI] [PubMed] [Google Scholar]
- 15.Fadool J M, Hartl D L, Dowling J E. Proc Natl Acad Sci USA. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coates C J, Jasinskiene N, Miyashiro L, James A A. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lidholm D A, Lohe A R, Hartl D L. Genetics. 1993;134:859–868. doi: 10.1093/genetics/134.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin E J, Akerley B J, Novik V N, Lampe D J, Husson R N, Mekalanos J J. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Sankar U, Lampe D J, Robertson H M, Graham F L. Nucleic Acids Res. 1998;26:3687–3693. doi: 10.1093/nar/26.16.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohe A R, Hartl D L. Mol Biol Evol. 1996;13:549–555. doi: 10.1093/oxfordjournals.molbev.a025615. [DOI] [PubMed] [Google Scholar]
- 22.Hartl D L, Lohe A R, Lozovskaya E R. Genetica. 1997;100:177–184. [PubMed] [Google Scholar]
- 23.Lohe A R, De Aguiar D, Hartl D L. Proc Natl Acad Sci USA. 1997;94:1293–1297. doi: 10.1073/pnas.94.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Ali S A, Steinkasserer A. BioTechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 26.Makris J C, Nordmann P L, Reznikoff W S. Proc Natl Acad Sci USA. 1988;85:2224–2228. doi: 10.1073/pnas.85.7.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hediger M A, Johnson D F, Nierlich D P, Zabin I. Proc Natl Acad Sci USA. 1985;82:6414–6418. doi: 10.1073/pnas.82.19.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehio C, Meyer M. J Bacteriol. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Reznikoff W S. J Mol Biol. 1997;271:362–373. doi: 10.1006/jmbi.1997.1188. [DOI] [PubMed] [Google Scholar]
- 30.Guzman L M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huisman O, Kleckner N. Genetics. 1987;116:185–189. doi: 10.1093/genetics/116.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krebs M P, Reznikoff W S. Gene. 1988;63:277–285. doi: 10.1016/0378-1119(88)90531-8. [DOI] [PubMed] [Google Scholar]
- 33.Reznikoff W S, Jilk R, Krebs M P, Makris J C, Nordmann P L, Weinreich M, Wiegand T. Methods Enzymol. 1993;217:312–322. doi: 10.1016/0076-6879(93)17072-d. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy A K, Haniford D B. J Mol Biol. 1996;256:533–547. doi: 10.1006/jmbi.1996.0106. [DOI] [PubMed] [Google Scholar]
- 35.Johnson R C, Reznikoff W S. J Mol Biol. 1984;177:645–661. doi: 10.1016/0022-2836(84)90042-1. [DOI] [PubMed] [Google Scholar]
- 36.Vos J C, De Baere I, Plasterk R H. Genes Dev. 1996;10:755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- 37.van Pouderoyen G, Ketting R F, Perrakis A, Plasterk R H, Sixma T K. EMBO J. 1997;16:6044–6054. doi: 10.1093/emboj/16.19.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vos J C, van Luenen H G, Plasterk R H. Genes Dev. 1993;7:1244–1253. doi: 10.1101/gad.7.7a.1244. [DOI] [PubMed] [Google Scholar]
- 39.Colloms S D, van Luenen H G, Plasterk R H. Nucleic Acids Res. 1994;22:5548–5554. doi: 10.1093/nar/22.25.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietrokovski S, Henikoff S. Mol Gen Genet. 1997;254:689–695. doi: 10.1007/s004380050467. [DOI] [PubMed] [Google Scholar]
- 41.Braam L A M, Goryshin I Y, Reznikoff W S. J Biol Chem. 1999;274:86–92. doi: 10.1074/jbc.274.1.86. [DOI] [PubMed] [Google Scholar]
- 42.Wiegand T W, Reznikoff W S. J Bacteriol. 1992;174:1229–1239. doi: 10.1128/jb.174.4.1229-1239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinreich M D, Gasch A, Reznikoff W S. Genes Dev. 1994;8:2363–2374. doi: 10.1101/gad.8.19.2363. [DOI] [PubMed] [Google Scholar]
- 44.Goryshin I Y, Reznikoff W S. J Biol Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 45.Weinreich M D, Mahnke-Braam L, Reznikoff W S. J Mol Biol. 1994;241:166–177. doi: 10.1006/jmbi.1994.1486. [DOI] [PubMed] [Google Scholar]
- 46.Hartl D L, Lohe A R, Lozovskaya E R. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 47.Lohe A R, Moriyama E N, Lidholm D A, Hartl D L. Mol Biol Evol. 1995;12:62–72. doi: 10.1093/oxfordjournals.molbev.a040191. [DOI] [PubMed] [Google Scholar]
- 48.Engels W R, Benz W K, Preston C R, Graham P L, Phillis R W, Robertson H M. Genetics. 1987;117:745–757. doi: 10.1093/genetics/117.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozovskaya E R, Nurminsky D I, Hartl D L, Sullivan D T. Genetics. 1996;142:173–177. doi: 10.1093/genetics/142.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyer H W, Roulland-Dussoix D. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 51.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 52.Johnson R C, Reznikoff W S. Genetics. 1984;107:9–18. doi: 10.1093/genetics/107.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson R C, Yin J C, Reznikoff W S. Cell. 1982;30:873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- 54.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 55.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]