Abstract
Mammalian target of rapamycin (mTOR) forms two complexes, mTORC1 and mTORC2, that play central roles in cell growth and functions. Only mTORC1 is directly inhibited by the immunosuppressive drug rapamycin. Despite recent progress in identifying new components and functions of the mTOR pathway, relatively little is known about the spatial arrangement of mTOR signaling and the underlying mechanisms. In a previous study, we showed that a large proportion of mTOR is localized to the endoplasmic reticulum (ER) and Golgi in many common cell lines. Here, we report the identification of an internal mTOR sequence that contains two HEAT (HT) repeats, HT18 and HT19, and two intervening interunit spacers (IUSs), IUS17 and IUS18, which is sufficient to target enhanced green fluorescent protein to the Golgi. Surprisingly, deletion of IUS17 from this Golgi localization sequence (GLS) converts it to an ER localization sequence (ELS). Deletion of HT19, a common element of both GLS and ELS from the full-length mTOR, causes delocalization of mTOR and inhibits the ability of mTOR to promote S6 phosphorylation. Moreover, overexpression of GLS and ELS inhibits both mTOR complexes. Together, our results reveal unusual ER- and Golgi-targeting sequences and suggest that anchoring to these organelles is important for the functions of mTOR complexes.
INTRODUCTION
Rapamycin and analogs are promising anticancer agents currently under clinical trials (Huang and Houghton, 2003). When complexed with FKBP12, rapamycin binds to and inhibits the target of rapamycin (TOR) protein, a 289-kDa phosphatidylinositide 3-kinase–related kinase (PIKK). TOR proteins are well conserved in all eukaryotes, with >45% amino acid sequence identity and 70% similarity between yeast and human TOR proteins. The C-terminal PIKK kinase domain has protein serine and threonine kinase activity (Hunter, 1995; Keith and Schreiber, 1995) essential for TOR functions (Brown et al., 1995; Zheng et al., 1995). PIKK and two other C-terminal domains, FAT and FATC, form the signature for the PIKK family that also includes ATM, ATR, and DNA-PK (Bossini et al., 2000). FKBP12-rapamycin binding (FRB) domain is a conserved 11-kDa region necessary and sufficient for FKBP12–rapamycin binding (Chen et al., 1995; Zheng et al., 1995). A conserved serine residue in FRB domain, Ser2035 in mTOR and Ser1972 in yeast Tor1, is crucial for the binding of FKBP12-rapamycin. When mutated to a bulkier residue such as threonine, it disrupts the binding of FKBP12-rapamycin and confers dominant rapamycin resistance (Stan et al., 1994; Brown et al., 1995; Lorenz and Heitman, 1995; Zheng et al., 1995). The N-terminal half of TOR contains 20 tandem repeats of 37–43 amino acids termed HEAT (HT) repeats (Andrade and Bork, 1995). A single HEAT repeat contains a pair of interacting antiparallel helices linked by a flexible intraunit loop. HEAT repeats occur in tandem clusters, linked by flexible interunit spacers (IUSs). Crystal structures of HEAT repeat proteins such as the A subunit of phosphatase 2A show that each HEAT repeat unit folds independently into helix-loop-helix. The tandem HEAT repeats form an extended thread-like structure with little interaction among different HEAT repeat units (Groves et al., 1999; Perry and Kleckner, 2003). HEAT repeats are thought to engage in interaction with other proteins. Indeed, the HEAT repeats of TOR proteins have been shown to mediate interaction with proteins such as Gln3 (Bertram et al., 2000) and Kog1/Raptor (Kim et al., 2002).
TOR forms two distinct protein complexes, TOR complexes 1 and -2 (TORC1 and TORC2, respectively). The major components of mammalian TORC1 (mTORC1) are mTOR, Raptor/mKog1, and GβL/mLst8 (Hara et al., 2002; Kim et al., 2002, 2003; Loewith et al., 2002), and those of mTORC2 include mTOR, mSin1/mAvo1, Rictor/mAvo3, and GβL/mLst8 (Jacinto et al., 2004, 2006; Sarbassov et al., 2004, 2006; Shiota et al., 2006; Yang et al., 2006). Although TORC1 is sensitive to rapamycin, TORC2 is not (Loewith et al., 2002), which is in agreement with two previously defined functions for TOR2 in yeast (Zheng et al., 1995). mTORC1 integrates signals from growth factors and nutrients, and it regulates diverse growth-related processes, including translational initiation, ribosome biogenesis, and autophagy. Translational initiation is the best understood TORC1-dependent process. mTORC1 phosphorylates several translational regulators, including ribosomal S6 kinases (S6Ks) and eIF4E-binding protein 1 (4E-BP1, also called PHAS-I) (Dennis et al., 1999; Kuruvilla and Schreiber, 1999; Schmelzle and Hall, 2000; Raught et al., 2001; Rohde et al., 2001; McDaniel et al., 2002). mTORC2 is required for actin organization (Jacinto et al., 2004; Sarbassov et al., 2004). More recently, mTORC2 has been shown to phosphorylate AKT/PKB at Ser473 (Sarbassov et al., 2005).
The endoplasmic reticulum (ER) and the Golgi apparatus are part of the secretory pathway engaging in synthesis, modification, and transport of secreted and plasma membrane proteins (Baumann and Walz, 2001). The ER and Golgi also participate in intracellular signaling and other aspects of cell regulation, including the unfolded protein response and the ER overload response (Pahl, 1999). More recently, several studies demonstrate that the ER is involved in signal transduction pathways traditionally thought to emanate from the plasma membrane. For example, the small GTPase Ras restricted to the Golgi apparatus can actively engage signal transduction to mitogen-activated protein kinases (Chiu et al., 2002). How the ER and Golgi anchor intracellular signaling is elegantly illustrated by the sterol-sensing and -signaling pathway involving sterol regulatory element-binding proteins (SREBPs) (Rawson, 2003). When cholesterol level is high, SCAP binds to cholesterol in the ER membrane and assumes a conformation that promotes binding to the ER-resident protein INSIG (Brown et al., 2002; Yang et al., 2002). This retains the SREBP–SCAP complex in the ER by preventing interaction of SCAP with COPII vesicle formation proteins Sar1, Sec23, and Sec24. When cholesterol level is low, SREBP-SCAP is dissociated from INSIG and transported by COPII-coated vesicles to the Golgi, where SREBP is sequentially cleaved by two proteases, S1P and S2P, leading to the release of the N-terminal transcriptional activation domain from the Golgi membrane to the nucleus, where it activates the target genes (Wang et al., 1994; Sakai et al., 1996).
Differential signaling output is fundamentally dependent on the spatial organization of the signaling molecules, their regulators, and effectors within the cell. How signaling molecules are targeted to different subcellular compartments is an important but still poorly understood question for signal transduction research. Despite the central role of mTOR in cell growth and functions, relatively little is known about its precise subcellular distribution, and to an even lesser degree the underlying mechanisms and functional significance. Previous studies indicate that mTOR is present in both the cytoplasm (Desai et al., 2002; Drenan et al., 2004) and nucleus (Kim and Chen, 2000; Zhang et al., 2002; Drenan et al., 2004). A large proportion of mTOR is localized to the ER and Golgi in several common cell lines (Drenan et al., 2004). In this study, we identified an internal region spanning HT18 and -19 that is sufficient to target the heterologous protein enhanced green fluorescent protein (EGFP) to the ER or Golgi. Interestingly, this region does not contain any known ER- or Golgi-targeting signal sequence. We find that mutation of this region causes mTOR delocalization and blocks normal mTOR signaling. Moreover, overexpression of ER and Golgi localization sequences inhibits the functions of both mTORC1 and mTORC2. These results demonstrate that the HT18-19 region form novel ER and Golgi localization signals and suggest that anchoring to the ER/Golgi is important for mTOR signal transduction.
MATERIALS AND METHODS
Cell Lines, Plasmids, Antibodies, and Other Reagents
Mammalian cell lines used in this study were purchased from American Type Culture Collection (Manassas, VA) and maintained as recommended by the supplier. To construct plasmids expressing EGFP– or FLAG–HA–mTOR fusion proteins, different mTOR regions were generated by polymerase chain reaction (PCR) and cloned into pEGFPN1 (Clontech, Mountain View, CA) and pFH-IRESneo (Teichmann et al., 2000). Further deletional analysis of different regions of Golgi localization sequence (GLS)-EGFP was carried out by the inverted PCR approach by using primers flanking the deleted sequence. FLAG-mTOR(S2035T, ΔHT19) was created by deletion of HEAT19 from FLAG-mTOR(S2035T)(Brown et al., 1995) by the inverted PCR approach. The N-terminal mTOR/FRAP antibody was described and characterized previously (Drenan et al., 2004). Other antibodies include FLAG antibodies (Sigma-Aldrich, St. Louis, MO); calnexin antibodies (BD Transduction Laboratories, Lexington, KY); green fluorescent protein (GFP) antibody (Affinity BioReagents, Golden, CO); Golgin-97 antibody and goat anti-mouse or rabbit secondary antibodies conjugated to Alexa Fluor 594 or 488 (Invitrogen, Carlsbad, CA); hemagglutinin (HA) monoclonal antibodies (Harlan Laboratories, Haslett, MI); AKT, P-AKT(Ser473), S6, P-S6(Ser235/236), and P-S6K1(Thr389) antibodies (Cell Signaling Technology, Beverly, MA); and horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit antibodies (Pierce Chemical, Rockford, IL). Proteinase K and phalloidin-tetramethylrhodamine B isothiocyanate (TRITC) were purchased from Sigma-Aldrich.
Immunofluorescence (IF), Western Blot, Immunoprecipitation, and In Vitro Kinase Assay
For immunofluorescence, cells were fixed in 3% paraformaldehyde, 2% sucrose in H2O for 10 min at 37°C; permeabilized with ice-cold HEPES–Triton X-100 buffer (0.5% Triton X-100 in 20 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose) for 5 min on ice; blocked with 0.1% bovine serum albumin in phosphate-buffered saline for 10 min on ice; and incubated with primary antibodies for 20 min (N-terminal mTOR antibody at 1:500, mouse anti-calnexin at 1:50, mouse anti-Golgin-97 at 1:100, and rabbit anti-FLAG at 1:1000) at 37°C in a moisture chamber. Unbound antibodies were removed by washing 10 times with Tris-buffered saline plus 0.1% Tween 20 (TBST). Fluorescence-labeled secondary antibodies (Invitrogen) were incubated for 15 min at room temperature and washed as with the primary antibodies. Glass cover slips carrying treated cells were mounted with Cytoseal mounting medium onto glass slides and analyzed using an Olympus BX51 fluorescence microscope equipped with a Qimaging Retiga EXi digital camera. Phalloidin-TRITC staining was carried according to the manufacturer's instruction (Sigma-Aldrich). Cell lysates for Western blot were prepared using ice-cold lysis buffer containing 50 mM HEPES-KOH, pH 7.4, 40 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 5% glycerol, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1.5 mM Na3VO4, 50 mM NaF, and 1× protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Protein samples were separated on SDS-polyacrylamide gels and transferred onto Immobilon-P membrane (Millipore, Billerica, MA). After blocking with 5% dry milk in TBST, the membrane was incubated with primary antibodies for 1 h to overnight, followed by incubation with HRP-conjugated secondary antibodies (1:10,000) for 30 min and with enhanced chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). For kinase assays, lysates of human embryonic kidney (HEK)293T cells transiently transfected with FLAG-mTOR(S2035T), FLAG-mTOR (S2035T, ΔHT19), or FLAG-mTOR(S2035T, D2357E) were immunoprecipitated with the FLAG M2 antibody and protein A-Sepharose beads. After extensive wash, the immunoprecipitated materials were assayed for mTOR kinase activity as described previously (Drenan et al., 2004).
Subcellular Fractionation and Related Biochemistry
HEK293T cells were washed with ice-cold HME buffer (10 mM HEPES, 250 mM mannitol, and 0.5 mM EDTA, pH 7.4), resuspended in 5 volumes of ice-cold HME buffer containing 0.1 mM phenylmethylsulfonyl fluoride, and Dounce-homogenized by 10 gentle strokes. Nuclei and unbroken cells were removed at 1500 × g. The supernatants were centrifuged at 10,000 × g (10 min at 4°C). The pellets (P10) were resuspended in HME buffer. The S10 supernatant (after saving an aliquot) was overlaid on a 20% sucrose cushion and further centrifuged at 100,000 × g (60 min at 4°C). The pellets (P100) were resuspended in HME buffer. The S100 was also saved for Western blot analysis. For protease protection assays, the P100 pellets were resuspended in HME plus 10 mM CaCl2 without or with 1% TritonX-100 and then incubated with different concentrations of trypsin-chymotrypsin (0, 5, and 50 μg/ml) for 40 min at 4°C. Reactions were stopped by addition of aprotinin and boiling in SDS protein sample buffer.
RESULTS
Identification of ER and Golgi Localization Sequences of mTOR
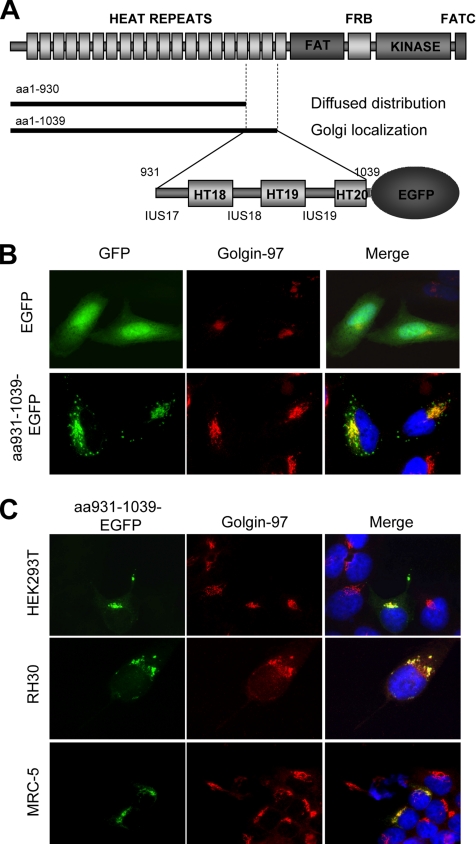
We have previously shown that both endogenous mTOR and transiently expressed full-length FLAG-tagged mTOR (FLAG-mTOR) are predominantly localized to the ER and Golgi in several common mammalian cell lines, including HeLa cells (Drenan et al., 2004). In the same study, we also found that the amino acids (aa)1-1039 region of FLAG-mTOR is exclusively localized to the Golgi, whereas the slightly shorter fragment (aa1-930) is diffusely distributed throughout the cytoplasm and nucleus. These observations suggest that aa931-1039 contains a Golgi localization sequence for mTOR. To verify this, we fused aa931-1039 to the N terminus of EGFP (aa931-1039-EGFP) (Figure 1A) and transiently expressed it in HeLa cells. We found that aa931-1039-EGFP is distributed in a unique pattern in the perinuclear area, reminiscent of the Golgi structure (Figure 1B). aa931-1039-EGFP is indeed colocalized with Golgi-97 as judged by indirect IF, confirming that aa931-1039-EGFP is localized to the Golgi (Figure 1B). Like in HeLa cells, aa931-1039-EGFP is exclusively localized in the Golgi of HEK293T, RH30, and MRC5 cells (Figure 1C), in which mTOR is localized to the ER and Golgi (Drenan et al., 2004). This result shows that Golgi localization is a common distribution pattern for aa931-1039.
Figure 1.
aa931-1039 of mTOR is sufficient to target EGFP to the Golgi apparatus. (A) Schematic presentation of mTOR and subcellular localization of two mTOR N-terminal fragments in HeLa cells. (B) aa931-1039-EGFP is predominantly localized to the Golgi in HeLa cells. EGFP or aa931-1039-EGFP is transiently expressed in HeLa cells (green). The Golgi is visualized by IF staining with a Golgin-97–specific antibody (red). The nuclei are stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (C) aa931-1039-EGFP is localized to the Golgi in several common mammalian cell lines. aa931-1039-EGFP is transiently expressed in HEK293T, RH30, and MRC-5 cells (green). The Golgi is visualized by IF with a Golgin-97–specific antibody (red). The nuclei are stained with DAPI (blue).
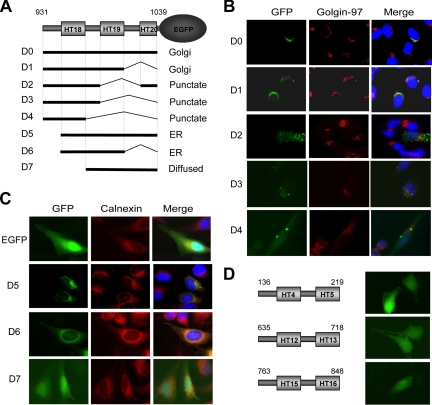
The aa931-1039 region contains HT18 and HT19, and part of HT20, as well as three IUSs—17, 18, and 19 (Figure 1A). To determine the minimal Golgi-localization sequence, we made systematic deletions from both the N and C termini of aa931-1039 of aa931-1039-EGFP. Each EGFP fusion was transiently expressed in HeLa cells and costained with Golgin-97 or the ER marker calnexin. The result is summarized in Figure 2A. Deletion of IUS19 and HT20 has no effect on Golgi localization (Figure 2B, D1). However, further deletions that remove HT19 lead to a punctate pattern that is distinct from that of Golgin-97 (Figure 2B, D2-4). Surprisingly, removal of IUS17 leads to an ER distribution pattern (Figure 2C, D5). Additional deletion of IUS19 and HT20 does not affect the ER distribution pattern of the EGFP fusion (D6). However, further deletion of HT18 causes the EGFP fusion to diffusely distribute throughout the cytoplasm and nucleus (D7), a pattern that is similar to that of EGFP alone. These results indicate that HT18, -19 and IUS17, -18 constitute a GLS (D1) and that HT18, -19 and IUS18 form an ELS (D6). As controls, we made EGFP-fusions with three randomly selected, tandem pairs of HEAT repeats together with an N-terminal IUS (HEAT4-5, 12-13, and 15-16). When expressed in HeLa cells, all three EGFP-fusions are diffusely distributed throughout the cytoplasm and nucleus, the same pattern as EGFP (Figure 2D), suggesting that the ER and Golgi localization property is intrinsic to the HT18-19 region.
Figure 2.
The aa931-1039 region contains a Golgi and an ER localization sequence. (A) Summary of the deletions of aa931-1039-EGFP and their subcellular localization. (B) Localization of D1-4 deletions. aa931-1039-EGFP (D0) and D1-4 deletions were transiently expressed in HeLa cells (green), and are analyzed for their localization to the Golgi. (C) Localization of D5-7 deletions. D5-7 deletions were transiently expressed in HeLa cells (green). The ER is visualized by IF with a calnexin-specific antibody (red). (D) ER/Golgi localization is not a general feature for tandem HEAT repeat pairs. EGFP-fusions with three randomly selected, tandem pairs of HEAT repeats and an N-terminal IUS (HT4-5, 12-13, and 15-16) were transiently expressed in HeLa cells and examined for their localization.
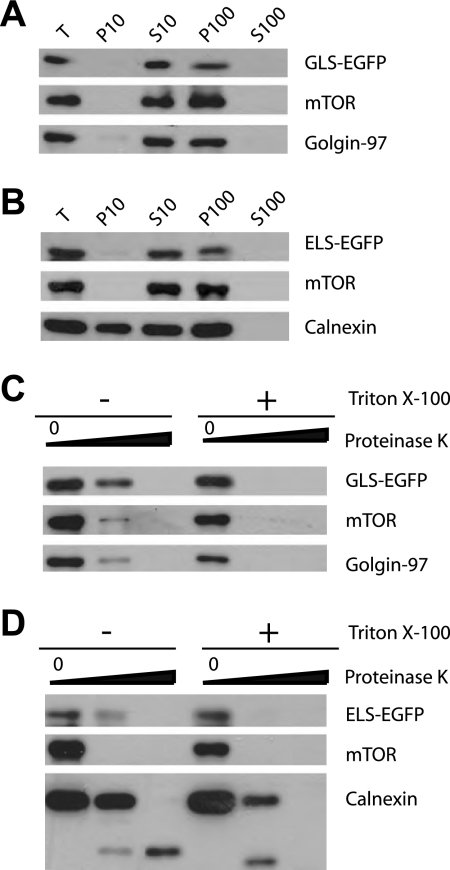
mTOR is tightly associated with the cytosolic side of ER and Golgi membranes (Drenan et al., 2004). We reasoned that if ELS and GLS mediate mTOR localization, their EGFP fusions should share the same topological characteristics. To test this, we transiently expressed ELS-EGFP and GLS-EGFP in HEK293T cells and then carried out subcellular fractionation by differential centrifugation. Like the full-length mTOR, GLS-EGFP and ELS-EGFP are copurified in fractions that are enriched with the Golgi and the ER (Figure 3, A and B, P10 and P100). We next performed proteinase K protection assay to determine the topology of the EGFP fusion proteins. Golgin-97 is associated with the cytosolic surface of the Golgi membrane. As expected, treatment with proteinase K caused complete degradation of Golgin-97 (Figure 3C). Calnexin is an integral ER membrane protein; proteinase K led to degradation of its cytosolic domain but not the ER lumen domain (Figure 3D). In the presence of the detergent Triton X-100, the ER lumen domain of calnexin was also digested by proteinase K (Figure 3D). These results show the quality of the ER/Golgi preparation and the specificity of the proteinase K assay. As shown previously (Drenan et al., 2004), mTOR is associated with the cytosolic face of the ER/Golgi, and proteinase K treatment of the purified ER and Golgi causes complete mTOR degradation (Figure 3, C and D). Under the same condition, proteinase K treatment also resulted in proteolysis of both GLS-EGFP and ELS-EGFP (Figure 3, C and D). Thus, like the full-length mTOR, ELS-EGFP and GLS-EGFP are localized on the cytosolic surface of the ER and Golgi.
Figure 3.
GLS and ELS share the same topology of ER/Golgi localization as mTOR. (A and B) Subcellular fractionation of GLS-EGFP and ELS-EGFP. GLS-EGFP and ELS-EGFP were transiently expressed in HEK293T cells. Lysates of these cells were fractionated by differential centrifugation. T, total cell lysates; P10, 10,000g pellet; S10, 10,000 × g supernatant; P100, 100,000 × g pellet; S100, 100,000 × g supernatant. (C and D) GLS-EGFP and ELS-EGFP are localized on the cytoplasmic face of Golgi and ER membranes. The P100 pellets from A and B were treated with different concentrations of proteinase K (0, 5, and 50 μg/ml) in the presence or absence of the detergent Triton X-100 and analyzed by Western blot with antibodies specific for GFP.
HT19 Is Required for Normal Subcellular Distribution and Function of mTOR
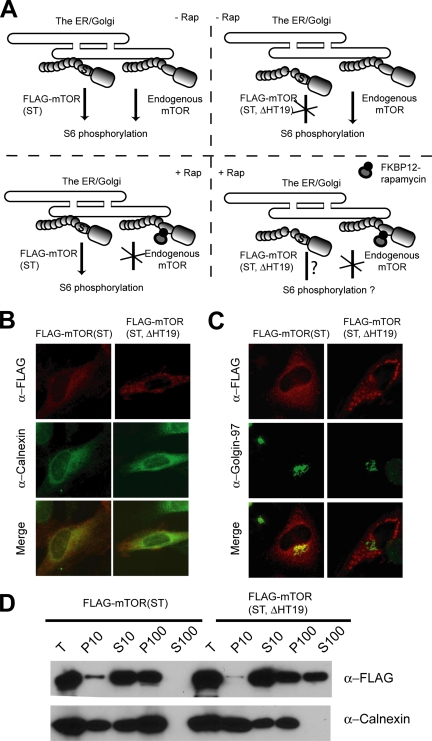
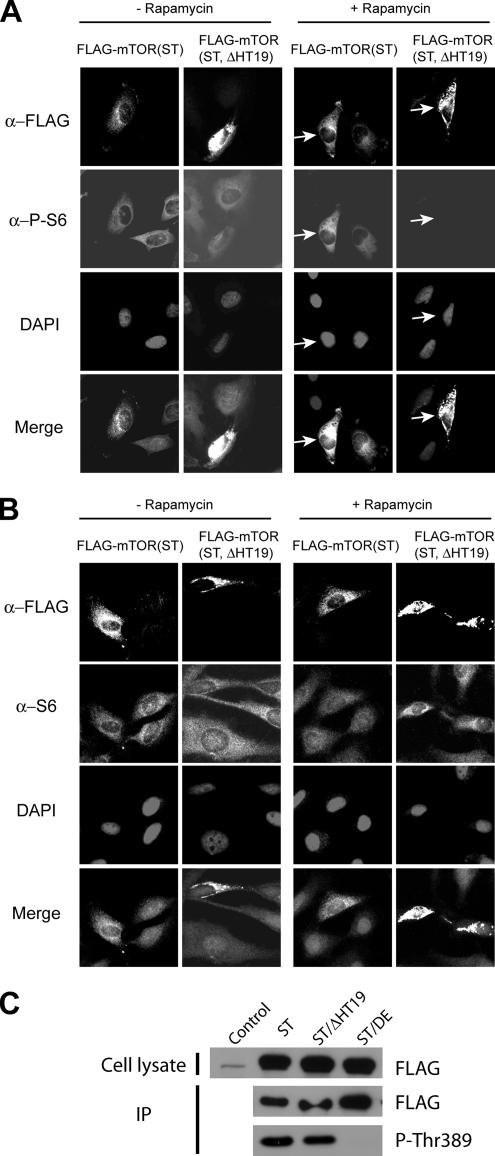
HT19 is a common element for GLS and ELS. To investigate whether GLS and ELS are important for mTOR localization, we deleted HT19 from FLAG-mTOR(S2035T). S2035 is a conserved residue located in FRB domain and is crucial for the binding of FKBP12-rapamycin (Chen et al., 1995; Zheng et al., 1995). The S2035→T mutation disrupts the binding of FKBP12-rapamycin and confers dominant rapamycin-resistant signaling for mTOR (Brown et al., 1995; Chen et al., 1995). Deletion of HT19 in this rapamycin-resistant mTOR allele is also useful to study the importance of ER and Golgi localizations for mTOR functions (Figure 4A; see below). Like endogenous mTOR and FLAG-mTOR (Drenan et al., 2004), transiently expressed FLAG-mTOR(S2035T) is localized to the ER and Golgi, as judged by the colocalization of FLAG-mTOR(S2035T) with calnexin and Golgin-97 by IF and subcellular fractionation (Figure 4, B, C and D). In contrast, IF shows that FLAG-mTOR(S2035T, HT19Δ) exhibits a highly aggregated pattern that is distinct from that of calnexin and Golgin-97 (Figure 4, B and C). Moreover, a significant proportion of FLAG-mTOR(S2035T, HT19Δ) is found in the cytosol (S100) (Figure 4D). Together, the above-mentioned results show that HT19 is important for proper mTOR localization.
Figure 4.
Deletion of HT19 causes delocalization of FLAG-mTOR from the ER and Golgi. (A) The strategy for analyzing the importance of HT19 in mTOR localization and function. FLAG-mTOR(S2035T) or FLAG-mTOR(S2035T, ΔHT19) is expressed in HeLa cells. IF and subcellular fractionation experiments are used to determine whether FLAG-mTOR(S2035T, ΔHT19) is localized to the ER/Golgi. The functional significance of mTOR localization is determined as follows. In the absence of rapamycin, both FLAG-mTOR(S2035T) and endogenous mTOR are localized to the ER and Golgi, and they are capable of promoting S6 phosphorylation. In the presence of rapamycin, FKBP12-rapamycin binds to and inhibits the endogenous mTOR but not FLAG-mTOR(S2035T) or FLAG-mTOR(S2035T, ΔHT19). This allows specific examination of the function of FLAG-mTOR(S2035T, ΔHT19) through S6 phosphorylation. (B) Deletion of HT19 inhibits mTOR localization to the ER. FLAG-mTOR(S2035T) and FLAG-mTOR(S2035T, ΔHT19) are transiently expressed in HeLa cells. Their localization was determined by IF staining. (C) Deletion of HT19 inhibits mTOR localization to the Golgi. The same as Figure 3B except Golgin-97 antibody was used instead. (D) Deletion of HT19 inhibits the ability of mTOR to associate with ER and Golgi membranes. FLAG-mTOR(S2035T) and FLAG-mTOR(S2035T, ΔHT19) were transiently expressed in HEK293T cells. The lysates of these cells were fractionated by differential centrifugation and analyzed by Western blot.
The strategy for studying the functional significance of mTOR localization is illustrated in Figure 4A. In FLAG-mTOR(S2035T)–expressing cells in the absence of rapamycin, both endogenous mTOR and FLAG-mTOR(S2035T) are localized to the ER and Golgi, and they are capable of normal signaling to downstream effectors such as S6, a ribosomal protein, and an mTOR downstream effector. Phosphorylation of S6, which is mTOR dependent, occurs at five residues at the C terminus (Ser235, Ser236, Ser240, Ser244, and Ser247) (Krieg et al., 1988). Rapamycin treatment results in a loss of S6 phosphorylation (Jefferies et al., 1994). In the presence of rapamycin, endogenous mTOR is inhibited, but FLAG-mTOR(S2035T) is not. FLAG-mTOR(S2035T) is still capable of normal signaling to S6 phosphorylation under this condition. This assay is useful for determining the signaling ability of FLAG-mTOR(S2035T, ΔHT19) to mTOR downstream effectors.
We transiently transfected HeLa cells with plasmids expressing FLAG-mTOR(S2035T) or FLAG-mTOR(S2035T, HT19Δ). We then treated these cells without or with rapamycin and performed IF staining with a FLAG-specific mouse monoclonal antibody to identify the cells expressing the FLAG-mTOR variant and with a rabbit polyclonal antibody specific for P-S6(Ser235/236) to observe S6 phosphorylation. As expected, rapamycin blocks S6 phosphorylation in nontransfected cells but not in cells expressing FLAG-mTOR(S2035T) (Figure 5A). In contrast, rapamycin potently inhibits S6 phosphorylation in FLAG-mTOR(S2035T, HT19Δ)– expressing cells (Figure 5A). In the absence of rapamycin, S6 and P-S6 levels are normal in cells expressing FLAG-mTOR(S2035T, HT19Δ), indicating that FLAG-mTOR(S2035T, HT19Δ) expression itself does not affect S6 phosphorylation or S6 protein level (Figure 5, A and B). Although HEAT repeats are known to fold independently (Groves et al., 1999; Perry and Kleckner, 2003), it is still possible that HT19 deletion somehow disturbs an important mTOR structure such as the kinase domain and affects mTOR signaling indirectly. To investigate this possibility, we assayed for the kinase activity of different mTOR variants. We found that the HT19Δ mutant, but not the D2357E kinase-dead mutant, retains the ability to phosphorylate Thr389 of S6K1 in vitro (Figure 5C). A functional mTOR kinase toward Thr389 requires assembly of a functional mTORC1 complex that includes Raptor bound to the N terminus and GβL associated with the C terminus (Hara et al., 2002; Kim et al., 2002). Our results indicate that mTOR maintains a relatively normal overall structure and enzymatic activity in the absence of HT19. Together, these observations suggest that ER and Golgi localization is crucial for normal mTOR signaling function.
Figure 5.
Deletion of HT19 inhibits the ability of mTOR to promote S6 phosphorylation. (A) FLAG-mTOR(S2035T) and FLAG-mTOR (S2035T, ΔHT19) were transiently expressed in HeLa cells. These cells were then treated with the drug carrier (−Rap) or with rapamycin (+Rap) for 2 h and analyzed for the expression of FLAG-mTOR variants and S6 phosphorylation by IF staining with a FLAG antibody and a P-S6 antibody, respectively. The nuclei were stained with DAPI. The arrowheads indicate FLAG-mTOR(S2035T)– and FLAG-mTOR(S2035T, ΔHT19)–expressing cells. (B) Expression of FLAG-mTOR proteins does not affect S6 expression. This experiment is performed the same way as Figure 5A except a S6-specific antibody instead of the P-S6 antibody was used. (C) Deletion of HT19 does not affect mTOR kinase activity. FLAG-mTOR(S2035T), FLAG-mTOR(S2035T, ΔHT19) or FLAG-mTOR(S2035T, D2357E)(kinase-dead) were transiently expressed in HEK293T cells. After immunoprecipitation, they were assayed for mTOR kinase activity toward GST-p70S6K1(aa333-412). Thr389 phosphorylation was detected by Western blot with a P-Thr389 antibody.
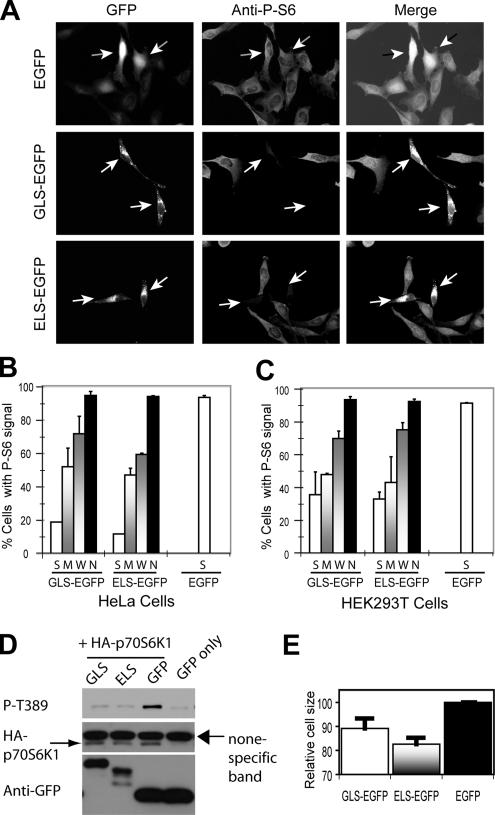
Overexpression of ELS and GLS Inhibits Both mTORC1 and mTORC2
One possible mechanism for ELS and GLS localization is that they interact with an ER and Golgi resident protein(s), as in the case of SREBP1 (Rawson, 2003). Conceptually, ELS and GLS overexpression may dominant negatively affect endogenous mTOR functions by interfering with one or both mTOR complexes. We therefore examined S6 phosphorylation in HeLa cells transiently expressing GLS-EGFP or ELS-EGFP. By IF with the P-S6–specific antibody, we found that S6 phosphorylation is strongly inhibited in cells overexpressing GLS-EGFP or ELS-EGFP but not in the untransfected cells or EGFP-expressing cells (Figure 6, A and B). The degree of inhibition of S6 phosphorylation is correlative with GLS and ELS expression levels: the lowest S6 phosphorylation is found in strong (S) GLS- and ELS-expressing cells; partial inhibition of S6 phosphorylation is seen in moderate (M) and weak (W) GLS- and ELS-expressing cells; and no inhibition of S6 phosphorylation is seen in the nontransfected cells (Figure 6B). Additionally, EGFP shows no discernible inhibition of S6 phosphorylation (Figure 6B). Essentially the same results were obtained with HEK293T cells (Figure 6C). To further demonstrate mTORC1 inhibition, we transiently expressed HA-p70S6K1 in HEK293T cells with ELS-EGFP, GLS-EGFP, or EGFP. We then assayed for HA-p70S6K1 phosphorylation by Western blot with a P-Thr389 antibody. Thr389 phosphorylation is indeed strongly inhibited by ELS-EGFP or GLS-EGFP but not EGFP alone (Figure 6D). Another established role of mTORC1 is cell size regulation. We found that ELS-EGFP– and GLS-EGFP–expressing cells are 10–20% smaller than EGFP cells (Figure 6E), which is consistent with the effect of rapamycin (Kim et al., 2002). Together, these observations show that overexpression of GLS and ELS strongly inhibits mTORC1 signaling.
Figure 6.
Overexpression of GLS-EGFP and ELS-EGFP inhibits mTORC1. (A) Overexpression of GLS-EGFP and ELS-EGFP inhibits S6 phosphorylation in HeLa cells. EGFP, ELS-EGFP, and GLS-EGFP were transiently expressed in HeLa cells. S6 phosphorylation was determined by IF using a P-S6–specific antibody. The arrowheads indicate transfected cells. (B) Quantification of cells with P-S6 signal from the experiment in B. The expression level of EGFP fusion proteins in individual cells was categorized into S, M, W, and no (N) signal according to the GFP signals. Virtually all the cells expressing EGFP alone show strong GFP signal. (C) Overexpression of GLS-EGFP and ELS-EGFP inhibits S6 phosphorylation in HEK293T cells. EGFP, ELS-EGFP, and GLS-EGFP were transiently expressed in HEK293T cells. P-S6 signal was quantified and the level of GFP signal in individual cells was categorized as in Figure 6B. (D) Overexpression of GLS-EGFP and ELS-EGFP inhibits Thr389 phosphorylation of p70S6K1 in HEK293T cells. HA-p70S6K1 was transiently expressed in HEK293T cells together with ELS-EGFP, GLS-EGFP, or EGFP. HA-p70S6K1 phosphorylation was assayed by Western blot using a P-Thr389–specific antibody. (E) ELS-EGFP, GLS-EGFP, or EGFP was transiently expressed in HEK293T cells. The size of EGFP-positive cells was measured by fluorescence-activated cell sorting.
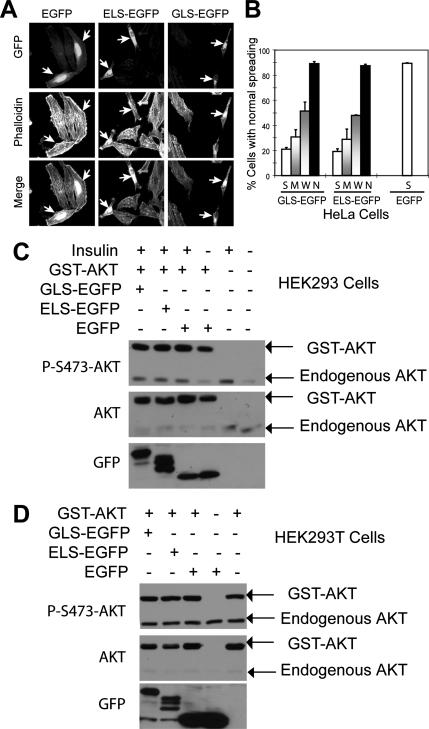
One established role of mTORC2 in cultured cells is related to cell spreading and organization of actin stress fibers (Jacinto et al., 2004; Sarbassov et al., 2004). When stained with TRITC-conjugated phalloidin, EGFP control cells and nontransfected cells are found to spread nicely, and their actin stress fibers are well organized (Figure 7A). In contrast, GLS-EGFP– and ELS-EGFP–expressing cells spread much less and show highly aggregated actin stress fibers (Figure 7, A and B). The morphology of ELS-EGFP– and GLS-EGFP–overexpressing cells is similar to that of mTOR and mAVO3/ Rictor small interfering RNA knockdown cells (Jacinto et al., 2004; Sarbassov et al., 2004), suggesting that mTORC2 is inhibited. Surprisingly, ELS-EGFP and GLS-EGFP do not seem to block AKT Ser473 phosphorylation in both HEK293 and HEK293T cells (Figure 7, C and D). Two recent studies show that deletion of Rictor in mice inhibits AKT/PKB phosphorylation but that it does not disturb actin organization (Guertin et al., 2006; Shiota et al., 2006). These observations suggest that mTORC2 is likely to control AKT/PKB phosphorylation and actin organization separately. One possible scenario is that different functions of mTORC2 require distinct subcellular localization and that only regulation of actin organization involves ER and Golgi localization.
Figure 7.
Overexpression of GLS-EGFP and ELS-EGFP inhibits mTORC2. (A) Overexpression of GLS-EGFP and ELS-EGFP disrupts normal actin organization and inhibits normal cell spreading. EGFP, GLS-EGFP, and ELS-EGFP were transiently expressed in HeLa cells. Actin cytoskeleton organization was determined by phalloidin-TRITC staining. The arrowheads indicate transfected cells. (B) Overexpression of GLS-EGFP and ELS-EGFP inhibits cell spreading. The level of GFP signal in individual cells was categorized as in Figure 6B. (C) ELS and GLS do not inhibit AKT/PKB phosphorylation in HEK293 cells. ELS-EGFP, GLS-EGFP, or EGFP was transiently expressed with GST-AKT in HEK293 cells. The cells were starved from fetal bovine serum for 4 h before incubated with insulin for 15 min. Phosphorylation of GST-AKT and endogenous AKT was analyzed by Western blot with a P-Ser473 antibody. (D) ELS and GLS do not inhibit AKT/PKB phosphorylation in HEK293T cells. ELS-EGFP, GLS-EGFP, or EGFP was transiently expressed with GST-AKT in HEK293T cells. Phosphorylation of GST-AKT and endogenous AKT was analyzed by Western blot with a P-Ser473 antibody.
DISCUSSION
Recent studies have shown that TOR proteins are associated with light intracellular membranes in mammals and yeast (Hara et al., 2002; Kim et al., 2002, 2003; Wedaman et al., 2003), likely to include the ER and Golgi. TCS2 and Rheb, two key upstream regulators of mTOR, have been found to be localized to the ER and Golgi (Wienecke et al., 1996; Jones et al., 2004; Buerger et al., 2006). Importantly, the small GTPase Rheb directly interacts with mTOR (Long et al., 2005a, b), and Golgi localization of Rheb is critical for its ability to stimulate mTOR activity (Buerger et al., 2006). These observations suggest that the ER and Golgi are common anchors for mTOR functions. Our data presented in this article show that these organelles are indeed crucial for signaling by both mTOR complexes. The ER and Golgi have been shown to anchor signaling by SREBPs into the nucleus (Brown et al., 2002; Yang et al., 2002; Rawson, 2003). In light of the findings that TOR proteins shuttle between the cytoplasm and the nucleus (Kim and Chen, 2000; Li et al., 2006), it is tempting to speculate that there is a similar mechanism for TOR proteins.
The ER and Golgi are part of the secretory pathway that actively engages transport of proteins to the plasma membrane through the vesicular trafficking. Each organelle also has to maintain a unique stable set of resident proteins that define its structural and functional properties. The ER residency is typically achieved by preventing resident proteins from entering the transport vesicles, or by retrieval of those being transported to the Golgi. The first mechanism is exemplified by SREBP, whose interaction with the COPII vesicle formation proteins Sar1, Sec23, and Sec24 is inhibited by high cholesterol concentration (Rawson, 2003). The second mechanism is directed by discrete retrieval motifs: soluble luminal proteins with the H/KDEL sequence at the carboxy terminus, or membrane proteins have a dibasic motif (KK or RR) located close to the cytosolic domain (Teasdale and Jackson, 1996). Recent work has identified the diphenylalanine (FF) in an Acidic Tract (FFAT) motif responsible for localizing cytosolic proteins to the cytoplasmic face of the ER (Loewen et al., 2003). The FFAT motif has the consensus amino acid sequence EFFDAxE. FFAT motifs bind to the highly conserved VAP proteins that are anchored to the cytoplasmic face of the ER (Loewen et al., 2003). Studies of several Golgi-resident proteins have revealed several mechanisms for Golgi targeting. For example, Imh1 is recruited to the Golgi membrane through the interaction of two GRIP domains with the Arf-like small GTPase Arl1. Arl1 is recruited to the Golgi by another member of the Arl family, Arl3, which requires an amino-terminal acetylated methionine residue to bind to a Golgi-localized, integral membrane protein called Sys1 (Graham, 2004). ArfGAP1 interaction with the Golgi is mediated by interaction of a hydrophobic motif with curved membrane lipid bilayer (Bigay et al., 2003; Parnis et al., 2006).
ELS and GLS do not bear any apparent sequence similarity to the known ER- or Golgi-targeting signals. Moreover, extensive homology search fails to reveal any significant similarity between ELS/GLS and known ER/Golgi surface proteins. How do ELS and GLS direct mTOR localization? Like SREBPs, their localization may be mediated by interaction with ER- and Golgi-resident proteins. This view is supported by the general role of HEAT repeats in mediating protein–protein interactions (Andrade and Bork, 1995). For example, the elongated HEAT repeats of importin β are involved in binding to various protein cargos for nuclear transport (Chook and Blobel, 1999; Vetter et al., 1999). Because ELS and GLS are overlapping with each other, it is possible that HT18,19 and IUS18 share a common receptor(s) on the ER and Golgi membranes, which allows its dynamic distribution on both the ER and Golgi. IUS17 may interact with a separate Golgi-resident protein, which allows its retention to the Golgi. Alternative, IUS17 could introduce a conformational constraint that blocks the interaction with an ER-resident receptor(s).
ACKNOWLEDGMENTS
We thank other members of the Zheng laboratory for discussions and technical assistance and Drs. Keqiang Ye and Kun-Liang Guan for GST-AKT plasmids. This work is supported by the National Institutes of Health.
Abbreviations used:
- EGFP
enhanced green fluorescence protein
- ER
endoplasmic reticulum
- ERS
endoplasmic reticulum localization sequence
- GLS
Golgi localization sequence
- mTOR
mammalian target of rapamycin
- mTORC
mammalian target of rapamycin complex.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0406 on January 10, 2007.
REFERENCES
- Andrade M. A., Bork P. HEAT repeats in the Huntington's disease protein. Nat. Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Baumann O., Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- Bertram P. G., Choi J., Carvalho J., Ai W. D., Zeng C. B., Chan T. F., Zheng X.F.S. Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J. Biol. Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Bigay J., Gounon P., Robineau S., Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. 2003;42:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- Bossini R., Isacchi A., Sonnhammer E. L. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- Brown A., Sun L., Feramisco J., Brown M., Goldstein J. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- Brown E. J., Beal P. A., Keith C. T., Chen J., Shin T. B., Schreiber S. L. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- Buerger C., DeVries B., Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem. Biophys. Res. Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Chen J., Zheng X. F., Brown E. J., Schreiber S. L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu V., Bivona T., Hach A., Sajous J., Silletti J., Wiener H., Johnson R., Cox A., Philips M. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- Chook Y. M., Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Dennis P. B., Fumagalli S., Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- Desai B. N., Myers B. R., Schreiber S. L. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA. 2002;99:4319–4324. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan R. M., Liu X., Bertram P. G., Zheng X.F.S. FKBP12-Rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- Graham T. Membrane targeting: getting Arl to the Golgi. Curr. Biol. 2004;14:R483–R485. doi: 10.1016/j.cub.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Groves M. R., Hanlon N., Turowski P., Hemmings B. A., Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals[no-break space]that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Huang S., Houghton P. Targeting mTOR signaling for cancer therapy. Curr. Opin. Pharmacol. 2003;3:371–377. doi: 10.1016/s1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- Hunter T. When a lipid kinase is not a lipid kinase? When a lipid kinase is a protein kinase? Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S., Huang Q., Qin J., Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Ruegg M., Hall A., Hall M. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jefferies H. B., Reinhard C., Kozma S. C., Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Jiang X., Yamamoto Y., Yeung R. S. Tuberin is a component of lipid rafts and mediates caveolin-1 localization: role of TSC2 in post-Golgi transport. Exp. Cell Res. 2004;295:512–524. doi: 10.1016/j.yexcr.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Keith C. T., Schreiber S. L. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Kim D., Sarbassov D. D., Ali S., King J., Latek R., Erdjument-Bromage H., Tempst P., Sabatini D. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Sarbassov D. D., Ali S., Latek R., Guntur K., Erdjument-Bromage H., Tempst P., Sabatini D. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kim J. E., Chen J. Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc. Natl. Acad. Sci. USA. 2000;97:14340–14345. doi: 10.1073/pnas.011511898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg J., Hofsteenge J., Thomas G. Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide [correction published in J. Biol. Chem. (1988). 263, 17887] J. Biol. Chem. 1988;263:11473–11477. [PubMed] [Google Scholar]
- Kuruvilla F., Schreiber S. L. The PIK-related kinases intercept conventional signaling pathways. Chem. Biol. 1999;6:R129–R136. doi: 10.1016/S1074-5521(99)80070-2. [DOI] [PubMed] [Google Scholar]
- Li H., Tsang C., Watkins M., Bertram P., Zheng X. F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- Loewen C.J.R., Roy A., Levine T. P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005a;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Long X., Ortiz-Vega S., Lin Y., Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J. Biol. Chem. 2005b;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- McDaniel M. L., Marshall C. A., Pappan K. L., Kwon G. Metabolic and Autocrine Regulation of the Mammalian Target of Rapamycin by Pancreatic [beta]-Cells Diabetes. 2002;51:2877–7438. doi: 10.2337/diabetes.51.10.2877. [DOI] [PubMed] [Google Scholar]
- Pahl H. L. Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol. Rev. 1999;79:683–701. doi: 10.1152/physrev.1999.79.3.683. [DOI] [PubMed] [Google Scholar]
- Parnis A., Rawet M., Regev L., Barkan B., Rotman M., Gaitner M., Cassel D. Golgi localization determinants in ArfGAP1 and in new tissue-specific ArfGAP1 isoforms. J. Biol. Chem. 2006;281:3785–3792. doi: 10.1074/jbc.M508959200. [DOI] [PubMed] [Google Scholar]
- Perry J., Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- Raught B., Gingras A.-C., Sonenberg N. The target of rapamycin (TOR) protein. Proc. Natl. Acad. Sci. USA. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson R. B. The SREBP pathway–insights from insigs and insects. Nat. Rev. Mol. Cell. Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- Rohde J., Heitman J., Cardenas M. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 2001;276:7027–7036. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- Sakai J., Duncan E., Rawson R., Hua X., Brown M., Goldstein J. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Sarbassov D., Ali S., Kim D., Guertin D., Latek R., Erdjument-Bromage H., Tempst P., Sabatini D. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Ali S. M., Sengupta S., Sheen J.-H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Molecular Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Hall M. N. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Shiota C., Woo J.-T., Lindner J., Shelton K. D., Magnuson M. A. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell. 2006;11:583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Stan R., McLaughlin M. M., Cafferkey R., Johnson R. K., Rosenberg M., Livi G. P. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J. Biol. Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- Teasdale R. D., Jackson M. R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- Teichmann M., Wang Z., Roeder R. G. A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl. Acad. Sci. USA. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I. R., Arndt A., Kutay U., Gorlich D., Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Wang X., Sato R., Brown M., Hua X., Goldstein J. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J., III, McCaffery J. M., Powers T. Tor Kinases Are in Distinct Membrane-associated Protein Complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienecke R., Maize J. C., Jr., Shoarinejad F., Vass W. C., Reed J., Bonifacino J. S., Resau J. H., de Gunzburg J., Yeung R. S., DeClue J. E. Co-localization of the TSC2 product tuberin with its target Rap1 in the Golgi apparatus. Oncogene. 1996;13:913–923. [PubMed] [Google Scholar]
- Yang Q., Inoki K., Ikenoue T., Guan K.-L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity 10.1101/gad. 1461206. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Espenshade P., Wright M., Yabe D., Gong Y., Aebersold R., Goldstein J., Brown M. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Zhang X., Shu L., Hosoi H., Murti K. G., Houghton P. J. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J. Biol. Chem. 2002;277:28127–23381. doi: 10.1074/jbc.M202625200. [DOI] [PubMed] [Google Scholar]
- Zheng X. F., Florentino D., Chen J., Crabtree G. R., Schreiber S. L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]