Abstract
Secreted proteins that fail to achieve their native conformations, such as cystic fibrosis transmembrane conductance regulator (CFTR) and particularly the ΔF508-CFTR variant can be selected for endoplasmic reticulum (ER)-associated degradation (ERAD) by molecular chaperones. Because the message corresponding to HSP26, which encodes a small heat-shock protein (sHsp) in yeast was up-regulated in response to CFTR expression, we examined the impact of sHsps on ERAD. First, we observed that CFTR was completely stabilized in cells lacking two partially redundant sHsps, Hsp26p and Hsp42p. Interestingly, the ERAD of a soluble and a related integral membrane protein were unaffected in yeast deleted for the genes encoding these sHsps, and CFTR polyubiquitination was also unaltered, suggesting that Hsp26p/Hsp42p are not essential for polyubiquitination. Next, we discovered that ΔF508-CFTR degradation was enhanced when a mammalian sHsp, αA-crystallin, was overexpressed in human embryonic kidney 293 cells, but wild-type CFTR biogenesis was unchanged. Because αA-crystallin interacted preferentially with ΔF508-CFTR and because purified αA-crystallin suppressed the aggregation of the first nucleotide-binding domain of CFTR, we suggest that sHsps maintain the solubility of ΔF508-CFTR during the ERAD of this polypeptide.
INTRODUCTION
The first committed step in the secretory pathway is the translocation of newly synthesized polypeptides into the endoplasmic reticulum (ER). Protein maturation and folding in the ER are prerequisites for subsequent transport through this pathway because soluble and integral membrane proteins are subject to ER protein quality control (ERQC) (Ellgaard and Helenius, 2003). Proteins that fail to pass the ERQC checkpoint may be degraded by the proteasome via a process referred to as ER-associated degradation (ERAD) (Tsai et al., 2002; Kostova and Wolf, 2003; McCracken and Brodsky, 2003). This frees the ER from housing aberrant proteins and eliminates the potential formation of toxic protein aggregates. The selection of ERAD substrates and in some cases their polyubiquitination are mediated by molecular chaperones, several of which were first identified as heat-shock proteins (Hsps). Hsps facilitate protein folding or degradation, depending on the conformation of the target protein (Fewell et al., 2001; Ellgaard and Helenius, 2003).
A prominent ERAD substrate is the cystic fibrosis transmembrane conductance regulator (CFTR), and many requirements for its degradation have been established. CFTR is a 1480-amino acid integral membrane glycoprotein that belongs to the ATP-binding cassette (ABC) transporter superfamily and functions as a cAMP-regulated chloride and bicarbonate channel in the apical membrane of epithelial cells, including those of airways, pancreas, intestine, and other fluid-secreting epithelia (Pilewski and Frizzell, 1999). CFTR is composed of two membrane-spanning domains (MSD1 and MSD2), each comprising six transmembrane segments, two nucleotide-binding domains (NBD1 and NBD2), and a central regulatory (R) domain. Mutations within the gene, particularly the deletion of the phenylalanine at position 508 (ΔF508-CFTR), may lead to cystic fibrosis (CF), one of the most common autosomal recessive disorders in individuals of European descent. Although perhaps ∼75% of wild-type CFTR is targeted for ERAD, essentially all of the ΔF508 variant is destroyed in the ER and thus fails to mature (Cheng et al., 1990).
From studies in mammalian cells and in yeast, it is clear that a large number of cytoplasmic and ER lumenal chaperones impact CFTR and ΔF508-CFTR biogenesis. For example, Hsp90, Hsp40s, and the Hsp70 nucleotide exchange factor (NEF) HspBP1 facilitate the Hsp70-dependent folding of CFTR (Strickland et al., 1997; Loo et al., 1998; Meacham et al., 1999; Rubenstein and Zeitlin, 2000; Choo-Kang and Zeitlin, 2001; Farinha et al., 2002; Alberti et al., 2004; Youker et al., 2004; Zhang et al., 2006). In addition, the ER chaperone calnexin interacts with the immature form of CFTR and ΔF508-CFTR and may also facilitate folding (Pind et al., 1994; Okiyoneda et al., 2004; Farinha and Amaral, 2005). However, when an Hsp70/Hsp90-interacting E3 ubiquitin ligase, known as CHIP, is recruited to chaperone-bound CFTR, there is a shift toward the degradation pathway (Connell et al., 2001; Meacham et al., 2001), and more recent data indicate that CHIP acts after the RMA1 ubiquitin ligase monitors CFTR biogenesis (Younger et al., 2006). Nevertheless, given the complexity of the CFTR-folding pathway (Du et al., 2005; Kleizen et al., 2005; Thibodeau et al., 2005) and the continued identification of factors that facilitate protein folding in the ER and ERAD, it is likely that additional chaperones and chaperone-like proteins that impact CFTR maturation remain to be discovered.
In this report, we show that small heat-shock proteins (sHsps) facilitate CFTR degradation in yeast and that overexpression of a mammalian sHsp selectively accelerates the degradation of ΔF508-CFTR. These data suggest that sHsps distinguish terminally misfolded forms of ΔF508-CFTR from the wild-type protein and add cystic fibrosis to the growing list of diseases and homeostatic processes that sHsps are thought to affect, a list that includes disorders associated with protein aggregation, ischemia/reperfusion injury, tumorigenesis, progression and metastasis, and apoptosis (Sun and MacRae, 2005).
MATERIALS AND METHODS
Yeast Strains, Plasmids, Molecular Methods, and Antisera
The yeast strains used in this study are listed in Table 1. The hsp26Δ strain in the RSY368 background was created using polymerase chain reaction (PCR)-based gene disruption (Brachmann et al., 1998). In brief, the RSY368 wild-type strain was transformed with an hsp26::KanMX4 disruption cassette generated by PCR from the template vector pRS400 (forward primer: GTGGTATTTCATAACAACGGTTCTTTTTCACCCTTATTCCCTGTGCGGTATTTCACACCG; reverse primer: TGACATATGTTTCAAGCCATATGCAAGCAACAATGGTCCTAGATTGTACTGAGAGTGCAC). Transformants were selected by growth on complete medium supplemented with 2% glucose and Geneticin (G-418) to a final concentration of 250 μg/ml (Invitrogen, Carlsbad, CA). Correct insertion of the disruption cassette and the absence of the HSP26 gene were confirmed by PCR by using primers up- and downstream of the HSP26 location in combination with primers within the sequence of the disruption cassette and HSP26 (KAN forward: ATTGATGTTGGACGAGTCGG; KAN reverse: ATCGCAGTGGTGAGTAACCA; HSP26 upstream, forward: GGATCGGAATAGTAACCGTC; HSP26 down-stream, reverse: GCCACATCCATAGAGATACC; HSP26 coding, forward: CACCAAGACGTCAGTTAGCA; and HSP26 coding, reverse: TGTTGTCTGCATCCACACCT).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JN516 (also referred to as wild type or SSA1) | MATα leu2-3, 112 his3-1 ura3-52 trp1Δ1 lys2 ssa2::LEU2 ssa3::TRP1 ssa4::LYS2 | Becker et al. (1996) |
| RSY368 (also referred to as wild type orHSP26) | Matα can1-100 leu2-3,112 his3-11, 15 trp1-1 ura3-1 ade2-1 | R. Schekman (University of California, Berkeley, Berkeley, CA) |
| hsp26Δ | Matα can1-100 leu2-3,112 his3-11, 15 trp1-1 ura3-1 ade2-1 hsp26::KanMX4 | This study |
| SEY6211 (also referred to as wild type or HSP26HSP42) | Mata ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 GAL10 | Robinson et al. (1988) |
| hsp26Δhsp42Δ | Mata ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 GAL10 hsp26::HIS3 hsp42::LEU2 | Haslbeck et al. (2004) |
| HRD1DOA10 (also referred to as wild type) | MATα, his3-Δ200, leu2-3, 112, ura3-52, lys2-801, trp1-1 | M. Hochstrasser (Yale University, New Haven, CT) |
| hrd1Δdoa10Δ | MATα, his3-Δ200, leu2-3, 112, ura3-52, lys2-801, trp1-1, doa10::HIS3, hrd1::LEU2 | M. Hochstrasser |
Cells were grown at the indicated temperatures with vigorous shaking in complete medium (YP) or in synthetic complete medium lacking uracil (Sc-ura) and containing glucose at a final concentration of 2%. Hemagglutinin (HA)-tagged CFTR, CPY*, and Ste6p* were expressed from a 2μ plasmid containing URA3 as the selectable marker under the control of the constitutive phosphoglycerate kinase promoter (CFTR and Ste6p*) or the endogenous promoter (CPY*) (Ng et al., 2000; Zhang et al., 2002b; Huyer et al., 2004); yeast transformed with the 2μ plasmid pRS426 (Christianson et al., 1992) without an insert served as controls. Plasmids were introduced into host strains by using lithium acetate-mediated transformation, and transformants were selected by growth on Sc-ura medium containing glucose. Protein extracts from yeast for immunoblot analysis were prepared as described previously (Brodsky et al., 1998).
The antibodies used in this study were raised against the C and N terminus of CFTR (M3A7 and MM13-4, respectively; Chemicon International, Temecula, CA/Upstate Biotechnology, Lake Placid, NY), the C terminus of CFTR (R&D Systems, Minneapolis, MN), αA-crystallin, Hsp70, and BiP (Stressgen Biotechnologies, San Diego, CA), Kar2p and Ssa1p (Brodsky and Schekman, 1993), Sec61p (Stirling et al., 1992), Hsp82p (a gift from A. Caplan, Mount Sinai School of Medicine, New York City, NY), Hsp26p (a gift from J. Buchner, Technical University of Munich, Munich, Germany), Hsp42p (a gift from S. Sandmeyer, University of California, Irvine, Irvine, CA), Hsp104p (a gift from J. Glover, University of Toronto, Toronto, Ontario, Canada), ubiquitin (a gift from C. Pickart and S. Michaelis, Johns Hopkins University School of Medicine, Baltimore, MD), or the HA-epitope (12CA5, Roche Molecular Biochemicals, Indianapolis, IN). To detect the primary antibody, horseradish peroxidase-conjugated secondary antibodies (anti-rabbit or anti-mouse; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were used.
Preparation of RNA and Northern Blot Analysis
A yeast colony was inoculated into 4 ml of Sc-ura supplemented with 2% glucose and incubated overnight at 30°C. The culture was diluted into 250 ml of the same medium and grown to an OD600 of ∼0.3 at 30°C. Cells were harvested by centrifugation at 4000 rpm for 10 min in a Sorvall GSA rotor at 20°C. The pellet was resuspended in 25 ml of sterile water and centrifuged at 4000 rpm for 5 min at 20°C in a Sorvall SS 34 rotor. Pellets were snap frozen in liquid nitrogen and stored at −80°C. Yeast RNA was extracted three times with hot acid-phenol (Sigma-Aldrich, St. Louis, MO) (Ausubel et al., 1988) and twice with chloroform and subjected to the RNeasy Midi kit (QIAGEN, Valencia, CA). Only samples with a ratio of A260/A280 higher than 1.8 were used. RNA integrity was confirmed on a formaldehyde-agarose gel.
For Northern blots, 20 μg of RNA was resolved on a formaldehyde-agarose gel, transferred to Gene Screen Plus membranes (PerkinElmer Life Science and Analytical Sciences, Boston, MA), and hybridized using 32P-labeled, randomly primed probes produced from PCR fragments by using primers against HSP26 (forward: CACCAAGACGTCAGTTAGCA, reverse: TGTTGTCTGCAT CCACACCT), FES1 (forward: CTACAGCAGTTATTCGGTGG, reverse: ACTCTCGT CAGACAAGCACT), HSP82 (forward: AGAGAGCACCATTCGACTTG, reverse: AATTGACCA GTTCTGATAGC), and SEC61 (forward: CCAACCGTGGTACTTT ACTG, reverse: GTCCAGAAGAGCTTCGGATA). Filters were processed as described previously (Ausubel et al., 1988), except that the high-temperature wash was performed at 55°C for 10 min. Bound probe was removed from the filters by incubation in 0.1X SSC, supplemented with an additional 15 mM NaCl, and containing 1% SDS for 20 min at 100°C.
ERAD Assays
The ERAD of HA-tagged CFTR in yeast was performed as described previously (Zhang et al., 2002b) but by using only fresh transformants, and data were quantified with a Kodak 440CF Image Station and Kodak 1D (version 3.6; Eastman Kodak, Rochester, NY) software. The degradation of HA-tagged CPY* and HA-tagged Ste6p* were determined by pulse-chase immunoprecipitations from 35S-metabolically labeled yeast as published previously (Zhang et al., 2001). Wild-type CFTR/ΔF508-CFTR degradation and maturation in human embryonic kidney (HEK)293 cells were assessed by pulse-chase immunoprecipitation as reported previously (Zhang et al., 2002a). The immunoprecipitates were resolved by SDS-PAGE and analyzed by PhosphorImager analysis by using Image Gauge software (version 3.45; FujiFilm Science Lab, Stamford, CT). Statistical analyses were performed via the program available at http://faculty.vassar.edu/lowry/t_ind_stats.html.
Detection of CFTR Polyubiquitination
Cells expressing CFTR-HA and containing a vector for the copper-inducible expression of a myc-tagged form of ubiquitin (provided by M. Hochstrasser, Yale University, New Haven, CT) were grown to log phase in 20 ml of selective medium at 30°C before 100 μM CuSO4 was added, and the cells were incubated for 3 h. Pelleted cells were washed with ice-cold 10 mM NaN3 and disrupted with glass beads in 200 μl of 20 mM HEPES, pH 7.4, 50 mM KOAc, 2 mM EDTA, 0.1 M sorbitol, 1 mM dithiothreitol and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 0.5 μg/ml pepstatin A) by agitation on a Vortex mixer five times for 1 min with 1-min intervals on ice between each cycle. The homogenate was collected and pooled with two 500-μl rinses of the beads with buffer 88 (20 mM HEPES, pH 6.8, 150 mM KOAc, 250 mM sorbitol, and 5 mM MgOAc). After unbroken cells were removed by centrifugation, the supernatant was centrifuged at 18,000 × g for 20 min at 4°C. The crude membrane pellet was washed and resuspended in buffer 88 so that the absorbance at 280 nm in 2% SDS was ∼40. Membranes (10 μl) were mixed with 10 μl of buffer 88 and solubilized in 80 μl of 50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1.25% SDS, 10 mM N-ethylmaleimide, and protease inhibitors at 37°C for 20 min. Samples were added to 400 μl of 50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 2% Triton X-100, 10 mM N-ethylmaleimide, and protease inhibitors and insoluble material was removed by centrifugation at 18,000 × g for 2 min. CFTR was immunoprecipitated from the supernatants with anti-HA antibody and resolved on a 6% SDS-polyacrylamide gel. CFTR and polyubiquitinated CFTR were decorated on immunoblots with anti-HA antibody and anti-ubiquitin antibody, respectively, after the nitrocellulose filter was incubated in boiling water for 20 min. The bound antibodies were visualized using enhanced chemiluminescence, and the data were quantified as described above.
Mammalian Cell Culture, Plasmids, and Molecular/Cellular Methods
HEK293 cells were cultured in DMEM (Sigma-Aldrich) with 10% fetal bovine serum (Invitrogen), 4 mM l-glutamine (Sigma-Aldrich), and penicillin-streptomycin (Invitrogen) at 37°C in a humidified environment containing 5% CO2. The αA-crystallin gene was removed from a pCIneo vector (provided by U. Andley, Washington University, St. Louis, MO) with SalI (3′) and XbaI (5′), and the gene was subcloned into pcDNA3.1 (the SalI site was lost in the cloning process). Construction of CFTR and ΔF508-CFTR expression plasmids (pcDNA3.1) and the CFTR C-terminal deletion constructs terminating at residue 588 were described previously (Zhang et al., 2002a; Sun et al., 2006).
HEK293 cells were transiently transfected with the indicated pcDNA3.1 expression plasmids aided by Lipofectamine 2000 (Invitrogen). After 24 h, the cells were subjected to a pulse-chase analysis as described previously (Zhang et al., 2002a) or were lysed in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, and 0.5% NP-40. For coimmunoprecipitation experiments, the cultures were treated with 50 μM MG-132 after transfection for 12 h before cell lysis.
Small Heat-Shock Protein–CFTR Interaction Assays
In total, 500 μg of HEK293 cell lysate was added to 13.3 mM HEPES, pH 7.0, 85 mM potassium-glutamate, 3.3 mM NaCl, 4.7 mM MgSO4, 13.3 mM EGTA, and 2.2 mM CaCl2 (vol/vol, 2:1) and incubated with 1 μg of anti-CFTR antibody M3A7 (Upstate Biotechnology) and anti-CFTR C-terminal antibody (R&D Systems) or with anti-αA-crystallin antibody (Stressgen Biotechnologies) for 2 h. The immunocomplex was isolated by incubation with protein A and G agarose beads for 2 h. Precipitates were isolated and washed three times with 13.3 mM HEPES, pH 7.0, 85 mM potassium-glutamate, 3.3 mM NaCl, 4.7 mM MgSO4, 13.3 mM EGTA, 2.2 mM CaCl2, and 0.25% NP-40, and the proteins were resolved on 4–15% SDS-polyacrylamide gels and subjected to Western blot analysis. Data were quantified using ImageJ software (http://rsb.info.nih.gov/ij/) and analyzed as described above.
Purification of αA-Crystallin and the NBD1 Aggregation Assay
αA-crystallin was expressed in Escherichia coli BL21 DE from the pAED4 vector (provided by K. P. Das, Bose Institute, Kolkata, India), and the protein was purified as described previously (Biswas and Das, 2004). The molar ratio stated assumes that the sHsp is a monomer. The ability of αA-crystallin to suppress NBD1 aggregation (provided by R. Youker, University of Pittsburgh, Pittsburgh, PA) was determined as described previously (Strickland et al., 1997; Youker et al., 2004).
RESULTS
Induction of sHsps in Yeast Expressing CFTR
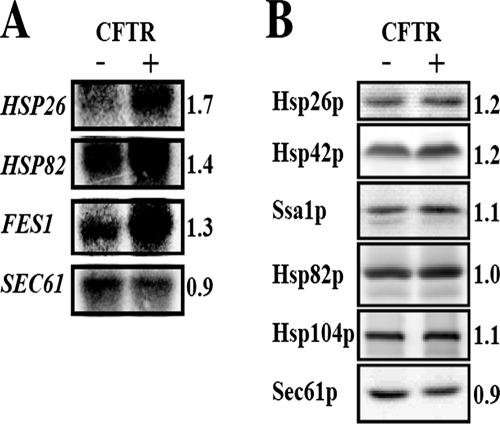
Because yeast expressing CFTR and ΔF508-CFTR grow as well as cells lacking the CFTR-expression vector, and because CFTR fails to elicit an unfolded protein response (UPR) in yeast (Zhang et al., 2001), we surmised that the expression of specific factors required for the ERAD of CFTR might be induced. To identify these factors, we examined the transcriptional profiles of yeast expressing wild-type CFTR and control strains by microarray analysis (Ahner and Brodsky, our unpublished data; details on the protocol and statistical analyses of the profiles will be reported elsewhere). Among the ∼150 transcripts up-regulated in strains expressing CFTR were HSP82, which encodes the yeast Hsp90 homologue, and FES1, which is a NEF for a cytosolic Hsp70; both of the corresponding proteins are known to play a role in CFTR biogenesis in yeast and/or mammalian cells (Loo et al., 1998; Zhang et al., 2001; Kabani et al., 2002; Alberti et al., 2004). In addition, we found that the message encoding Hsp26p increased significantly. These results were confirmed by Northern blot analysis (Figure 1A). We also examined the levels of Hsp26p and Hsp42p, two sHsps in yeast (also see below) as well as other chaperones (Figure 1B), some of which were unaffected by CFTR expression as determined by transcriptional profiling. We first noted a reproducible, ∼20% increase in the sHsps, whereas little or no change was observed for Ssa1p (a yeast Hsp70) or Hsp104p. Although the HSP82 message increased, there was no increase in protein levels, suggesting posttranscriptional control. Interestingly, we also noted that the message and protein levels corresponding to Sec61p, a component of the translocon and a UPR target (Casagrande et al., 2000) mildly decreased. These data indicate that CFTR expression does not induce a general heat-shock/stress response in yeast, although the levels of two sHsps rise when CFTR is produced.
Figure 1.
HSP26, HSP82, and FES1 mRNA levels are elevated in yeast expressing CFTR. (A) The indicated mRNAs were detected by Northern blot analysis from yeast strain JN516 transformed with a plasmid expressing CFTR and yeast transformed with a vector control (–; pRS426). The -fold up-regulation upon CFTR expression as determined by Northern blot analysis is indicated, and when standardized to the levels of SEC61 message, they are HSP26, 2.0; HSP82, 1.6; and FES1, 1.5. By comparison, the average -fold up-regulation from the microarray analyses (n = 6) are HSP26, 1.4; HSP82, 1.3; and FES1, 1.4. (B) Total cell extracts were prepared from yeast either lacking or containing the CFTR expression vector, as in A, and proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed with the indicated antisera. The ratios of protein in each sample from two independent sets of experiments were quantified as described in Materials and Methods.
sHsps Facilitate the Degradation of CFTR in Yeast
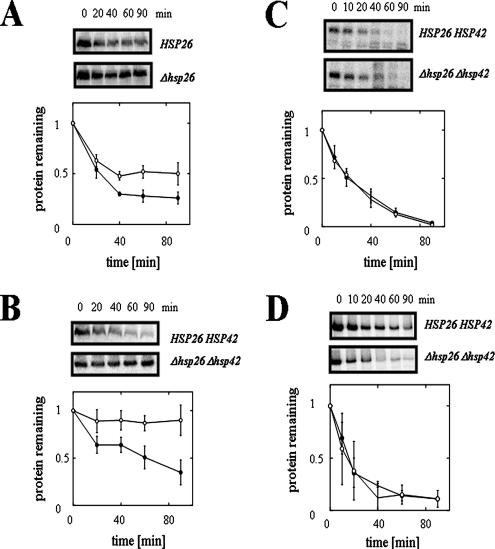
The expression of Hsp26p is induced by elevated temperature and Hsp26p prevents protein aggregation (Haslbeck et al., 2005). To assess whether Hsp26p depletion affects CFTR turnover, we expressed an HA-epitope–tagged form of wild-type CFTR under the control of a constitutive promoter in wild type yeast and in yeast deleted for HSP26. The rate of CFTR degradation was determined by cycloheximide chase analysis and levels of Sec61p were measured to provide a loading control (data not shown). As shown in Figure 2A, we found that the extent of CFTR proteolysis at 90 min was reduced in hsp26Δ yeast compared with wild-type cells by ∼50%, indicating that Hsp26p function is important for maximal CFTR degradation.
Figure 2.
Deletion of the small heat-shock protein-encoding genes slows CFTR degradation in yeast. CFTR degradation was assessed in wild-type and hsp26Δ yeast (A) and in wild type and hsp26Δhsp42Δ yeast (B). CFTR protein levels were quantified and averaged from at least three independent sets of experiments and were standardized to the levels detected directly after cycloheximide addition (0 min). Closed circles represent the CFTR protein levels in wild-type yeast, and open circles represent the CFTR protein levels in the mutant strains. Vertical bars indicate the SEs of the mean. p values in A at the 40, 60, and 90-min time points are 0.011, 0.022, and 0.062, respectively, and in part B are <0.03. CPY* (C) and Ste6p* (D) degradation rates were determined by pulse-chase immunoprecipitation experiments in HSP26HSP42 wild-type yeast and in the hsp26Δhsp42Δ double mutant strain after addition of cycloheximide. Protein levels were quantified from two independent sets of experiments and averaged. Standardization of the data and symbols are the same as described above. Vertical bars indicate the range of the data.
sHsps are distinguished by the presence of an α-crystallin domain (van Montfort et al., 2001), and Saccharomyces cerevisiae encodes two α-crystallin domain-containing proteins, Hsp26p and Hsp42p. Although they recognize ∼90% of the same client proteins, only Hsp42p is present constitutively at high levels (Haslbeck et al., 2004). This suggested that the continued expression of Hsp42p might have ameliorated the ERAD defect when HSP26 was deleted. To examine this hypothesis, CFTR was expressed in a different strain deleted for both HSP26 and HSP42, and indeed a much stronger degree of CFTR stabilization was observed (Figure 2B). For comparison, the deletion of either HSP26 or HSP42 in this strain background led to only subtle effects on CFTR degradation (Supplemental Figure 1S). These results suggest that the sHsps function redundantly during ERAD.
Different ERAD substrates require distinct factors for their degradation, depending on their solubility, membrane association, and location of structural alterations (see Discussion). Thus, it was important to establish whether the proteolysis of other substrates was altered by the loss of Hsp26p and Hsp42p. To this end, wild-type and hsp26Δhsp42Δ strains were transformed with a vector for the constitutive expression of CPY*, a soluble ERAD substrate (Hiller et al., 1996), and Ste6p*, a truncated form of the yeast a-mating factor pheromone transporter that, like CFTR, is an ABC transporter and is degraded via ERAD (Loayza et al., 1998). As shown in Figure 2, C and D, we found that the hsp26Δ hsp42Δ mutant proficiently degraded both CPY* and Ste6p*, demonstrating that sHsps are important for the proteolysis of only a select class of ER substrates. It is also important to note that these experiments were performed at a permissive temperature for the sHsp mutant strain (30°C) (Petko and Lindquist, 1986). Moreover, Kar2p, a UPR-regulated luminal Hsp70, was not induced in hsp26Δ hsp42Δ cells (our unpublished data). These observations indicate that the observed ERAD defect is due to deletion of the sHsps rather than to a nonspecific up-regulation of stress-inducible genes.
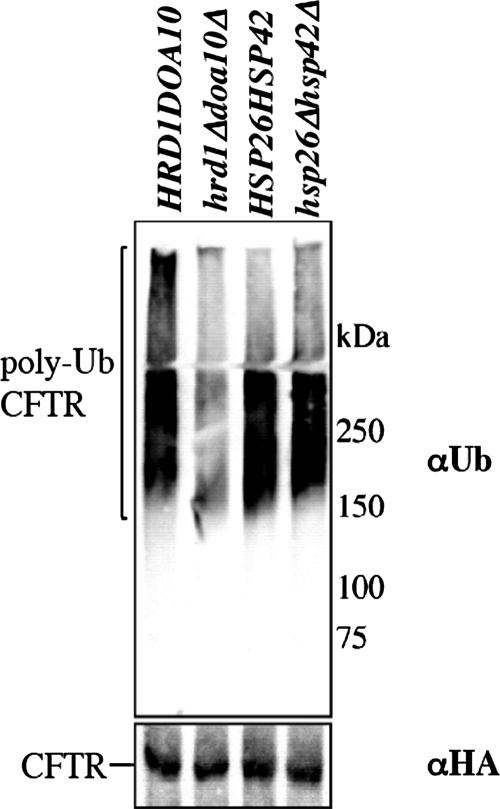
To identify the stage at which the yeast sHsps impact CFTR turnover, CFTR was immunoprecipitated from wild-type and hsp26Δ hsp42Δ cells, and the degree of polyubiquitination was assessed by immunoblot analysis. As a control, CFTR polyubiquitination was also measured in wild-type yeast and in cells lacking DOA10 and HRD1, two ubiquitin ligases that catalyze CFTR degradation in this organism (Gnann et al., 2004). Surprisingly, we found that CFTR polyubiquitination was unaffected in cells lacking the sHsps, although the degree of modification when the ubiquitin ligases were disabled was <50% of the control (Figure 3). We also noted that the steady-state level of total polyubiquitinated proteins in yeast deleted for both HSP26 and HSP42 was unaffected, but was modestly higher when ubiquitin was overexpressed (Supplemental Figure 2S).
Figure 3.
Deletion of the small heat-shock protein-encoding genes does not impact CFTR polyubiquitination in yeast. CFTR polyubiquitination was assessed in wild-type and hsp26Δhsp42Δ yeast and in wild-type and hrd1Δdoa10Δ yeast as described in Materials and Methods.
Overexpression of the Human sHsp αA-Crystallin Accelerates ΔF508-CFTR Degradation in HEK293 Cells
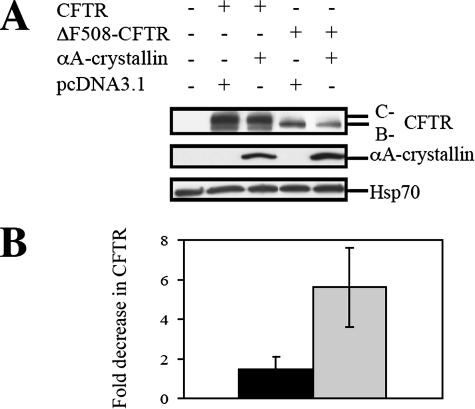
To determine whether sHsps impact CFTR biogenesis in mammals, HEK293 cells were transfected with vectors for the overexpression of wild-type CFTR or ΔF508-CFTR and either a vector control or a vector engineered for the overexpression of αA-crystallin. αA-crystallin is one of 10 “α-crystallin-like” proteins in humans (Kappe et al., 2003), and as observed previously for Hsp26p and Hsp42p, αA-crystallin associates with unfolded proteins and prevents their aggregation (Horwitz, 1992; Jakob et al., 1993; Biswas and Das, 2004). In addition, αA-crystallin is expressed in human cells that normally synthesize CFTR (our unpublished data). When CFTR expression was examined in extracts from each cell type by Western blot analysis, we noted that αA-crystallin overexpression did not alter the amount of wild-type CFTR but that ΔF508-CFTR steady-state levels decreased (Figure 4). Because Hsp70 levels were unchanged, the observed effect was likely not a result of secondary consequences from enhanced αA-crystallin expression.
Figure 4.
αA-crystallin overexpression decreases the steady state levels of ΔF508-CFTR. HEK293 cells were transfected with 1.5 μg of pcDNA3.1-CFTR or pcDNA3.1-ΔF508-CFTR and 0.5 μg of pcDNA3.1 or pcDNA3.1-αA-crystallin per 60-mm dish. (A) A representative immunoblot of the indicated proteins is shown after protein concentrations were normalized in all samples. Because Hsp70 levels remained constant, the amount of this chaperone served as an additional loading control. (B) CFTR protein levels were quantified from three independent sets of experiments and averaged. All values were obtained after standardization to the levels of Hsp70. The black bar represents the -fold decrease of wild-type CFTR total levels upon αA-crystallin overexpression, and the gray bar depicts the -fold decrease of ΔF508-CFTR levels upon αA-crystallin overexpression. Vertical bars indicate the SEs of the means (p = 0.013; t test).
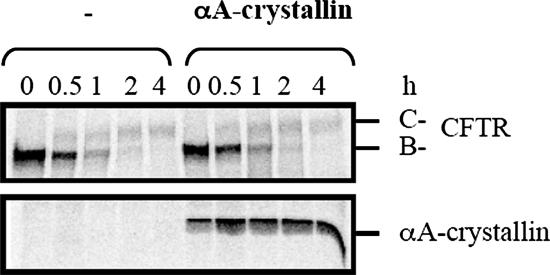
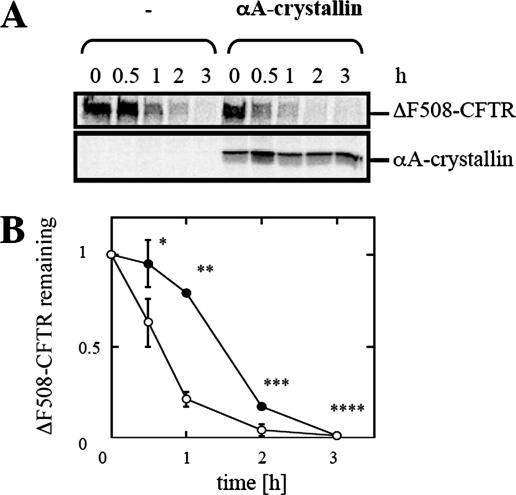
To investigate directly whether αA-crystallin accelerated CFTR and/or ΔF508-CFTR degradation, HEK293 cells were again transfected with the CFTR or ΔF508-CFTR expression vector and either the vector control or the αA-crystallin expression vector. Pulse-chase immunoprecipitation experiments were then performed to follow the stability of core-glycosylated immature CFTR (B-band) and its maturation to the complex-glycosylated form (C-band) (Figure 5); ΔF508-CFTR fails to mature and thus only the B-band is evident (Figure 6). We observed that for wild-type CFTR the disappearance of CFTR B-band and appearance of the C-band proceeded at equal rates in cultures transfected with the empty vector and in those overexpressing αA-crystallin. In contrast, the overexpression of αA-crystallin enhanced ΔF508-CFTR proteolysis by diminishing a reproducible lag in the disappearance of the B-band (Figure 6). This result indicates that αA-crystallin impacts only the ERAD of ΔF508-CFTR and thus might be able to distinguish between the wild type and mutant channels in mammals.
Figure 5.
αA-crystallin overexpression has no effect on the biogenesis of wild-type CFTR. HEK293 cells were transfected with 1.5 μg of pcDNA3.1-CFTR and 0.5 μg of pcDNA3.1 or pcDNA3.1-αA-crystallin per 60-mm dish. Rates of CFTR maturation (C-band) and CFTR B-band degradation as well as αA-crystallin expression levels were determined by pulse-chase immunoprecipitation in cells transfected with CFTR and containing a vector control or the αA-crystallin expressing vector.
Figure 6.
αA-crystallin overexpression accelerates the degradation of ΔF508-CFTR. (A) HEK293 cells were transfected with 1.5 μg of pcDNA3.1-ΔF508-CFTR and 0.5 μg of pcDNA3.1 or pcDNA3.1-αA-crystallin per 60-mm dish. Rates of ΔF508-CFTR degradation as well as αA-crystallin expression levels were determined by pulse-chase immunoprecipitation. (B) ΔF508-CFTR degradation was determined from four independent sets of experiments and averaged. All values were obtained after standardization to the levels detected at the beginning of the chase period (0 h). Closed circles represent ΔF508-CFTR protein levels in HEK293 cells with the vector control, and open circles represent ΔF508-CFTR protein levels in HEK293 cells overexpressing αA-crystallin. Vertical bars indicate the SEs of the mean. *p = 0.076, **p = 0.0001, ***p = 0.003, and ****p = 0.5.
αA-Crystallin Interacts Preferentially with ΔF508-CFTR and Prevents the Aggregation of NBD1
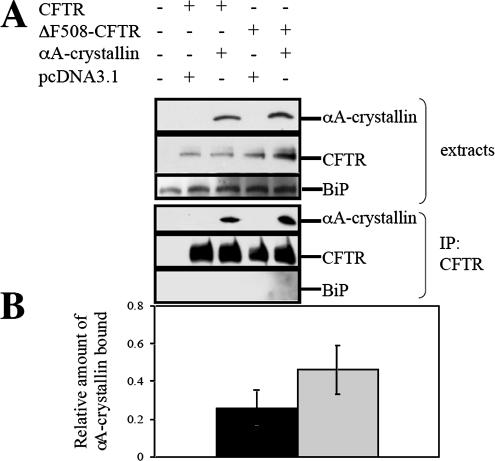
The interaction between αA-crystallin and CFTR and ΔF508-CFTR was assessed by immunoprecipitation after cotransfection of HEK293 cells with the αA-crystallin expression vector and expression vectors for either the wild-type or mutant protein. As displayed in Figure 7, the sHsp was isolated in a complex with CFTR or ΔF508-CFTR and by quantifying the amount of CFTR/ΔF508-CFTR relative to the chaperone, we estimated that approximately twofold more αA-crystallin bound to ΔF508-CFTR than to the wild-type protein.
Figure 7.
αA-crystallin coprecipitates preferentially with ΔF508-CFTR. (A) HEK293 cells were transfected with 6 μg of pcDNA3.1-CFTR or pcDNA3.1-ΔF508-CFTR and 2 μg of pcDNA3.1 or pcDNA3. 1-αA-crystallin per 100-mm dish. The interaction between CFTR or ΔF508-CFTR and αA-crystallin was assessed by immunoblot analysis for αA-crystallin after CFTR immunoprecipitation. The ER luminal chaperone BiP served as a negative control. (B) Relative binding efficiency was determined from 3 independent sets of experiments and averaged. All values were obtained after standardization to the amount of CFTR immunoprecipitated. The black bar represents αA-crystallin protein levels coprecipitated with wild-type CFTR, and the gray bar represents αA-crystallin protein levels coprecipitated with ΔF508-CFTR. Vertical bars indicate the SEs of the mean. *p = 0.012 (one-way analysis of variance for 2 correlated samples).
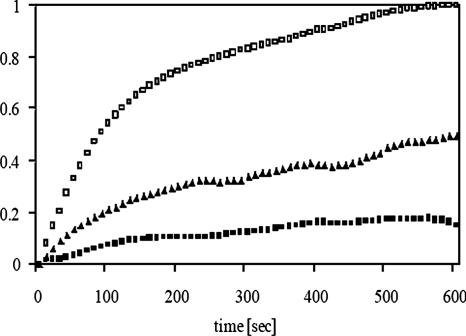
Eukaryotic sHsps associate with unfolded proteins and prevent their aggregation, which may be important for the proteasome-mediated degradation of some substrates (Horwitz, 1992; Jakob et al., 1993; Ehrnsperger et al., 1997; Lee et al., 1997; Haslbeck et al., 1999, 2004; Biswas and Das, 2004). Because NBD1 in CFTR is aggregation prone and interacts with other chaperones that prevent its aggregation (Qu and Thomas, 1996; Strickland et al., 1997; Meacham et al., 1999; Youker et al., 2004), we examined whether αA-crystallin also inhibits the formation of high-molecular-weight NBD1 oligomers. Toward this goal, αA-crystallin was expressed in and purified from E. coli and then incubated with NBD1 upon its dilution from denaturant. At a chaperone-to-substrate molar ratio of 0.5:1, αA-crystallin suppressed NBD1 aggregation by ∼50%, and at a molar ratio of 1:1 aggregation was suppressed by ∼80% (Figure 8). Although most chaperones suppress NBD1 aggregation to a similar extent only when used at higher molar ratios (e.g., Hdj2:NBD1 at 5:1, Hsc70-Hdj2:NBD1 at 2:1, and Hsp90:NBD1 at 5:1; Meacham et al., 1999; Youker et al., 2004), the near complete prevention of aggregation at a 1:1 molar ratio of sHsps to other substrates is common (Horwitz, 1992; Jakob et al., 1993; Chang et al., 1996; Ehrnsperger et al., 1997; Haslbeck et al., 1999; Studer and Narberhaus, 2000). Overall, and combined with the results presented above, we propose that αA-crystallin plays an important and previously unappreciated role in chaperoning the degradation of ΔF508-CFTR.
Figure 8.
αA-crystallin inhibits NBD1 aggregation. NBD1 aggregation was measured as a change in absorbance at 400 nm after its dilution from guanidinium hydrochloride in the absence or presence of αA-crystallin. Data were standardized to the amount of aggregation in the absence of chaperone at the 600-s time point. Open squares represent endogenous NBD1 aggregation, and closed symbols represent NBD1 aggregation in the presence of αA-crystallin. Triangles, αA-crystallin:NBD1 molar ratio of 0.5:1; squares, αA-crystallin:NBD1 molar ratio of 1:1.
DISCUSSION
By profiling which yeast genes are induced upon CFTR expression, we discovered that sHsps catalyze the degradation of CFTR in yeast and ΔF508-CFTR in mammalian cells. We also found that one sHsp, αA-crystallin, suppressed NBD1 aggregation and coprecipitated preferentially with ΔF508-CFTR. Because CFTR polyubiquitination was robust in yeast deleted for α-crystallin domain-containing proteins, we suggest that sHsps increase the accessibility of CFTR to the proteasome after polyubiquitination. If this is true, one might have expected an increase in the steady-state level of polyubiquitinated CFTR, but we cannot exclude the possibility that there was a concomitant increase in the activity of a deubiquitination enzyme or in the amount of aggregated protein. It is also formally possible that sHsps increase the accessibility of CFTR to the ubiquitin-conjugating machinery. The simplest model, though, is that sHsps maintain the solubility of a select group of ERAD substrates after ubiquitin modification.
An increasing number of papers implicate sHsps in the ubiquitin-proteasome pathway. Consistent with our findings αB-crystallin and Hsp27 are up-regulated by and are recruited to aggresomes (Ito et al., 2002), which are large, cytoplasmic inclusions of mis-folded proteins (Johnston et al., 1998). In addition, Hsp27 binds to the 26S proteasome and to a polyubiquitinated substrate (Parcellier et al., 2003). sHsps might directly catalyze the proteolysis of ERAD substrates because α-crystallins regulate proteasome activity and interact with a subunit in the proteasome core (Wagner and Margolis, 1995; Conconi et al., 1998, 1999; Boelens et al., 2001; Ito et al., 2002). However, other papers suggest a role for sHsps during substrate ubiquitination: αB-crystallin has been proposed to recruit FBX4, an adaptor molecule for the SCF ubiquitin ligase and promote substrate ubiquitination (den Engelsman et al., 2003; den Engelsman et al., 2004), and in vitro Hsp27 seems to facilitate the ubiquitination reaction (Parcellier et al., 2006). Overall, understanding the precise mechanism of sHsp action during ERAD represents an important, ongoing goal.
The observation that sHsps regulate the half-life of CFTR but not the turnover of a soluble ERAD substrate is in agreement with previous reports demonstrating distinct requirements for the degradation of different classes of ERAD clients (Plemper et al., 1997; Brodsky et al., 1999; Hill and Cooper, 2000; Fewell et al., 2001; Nishikawa et al., 2001; Vashist et al., 2001; Zhang et al., 2001; Kabani et al., 2003; Taxis et al., 2003; Ahner and Brodsky, 2004; Huyer et al., 2004; Vashist and Ng, 2004; Sayeed and Ng, 2005). However, a recent study reported similar requirements for the ERAD of CFTR and Ste6p* in yeast, which is in contrast to the results presented in Figure 2D (Huyer et al., 2004). Nevertheless, these newer data are consistent with the exquisite ability of sHsps to recognize subtle conformational differences between substrates (Basha et al., 2006, and see below) and with the fact that CFTR and Ste6p* are structurally distinct members of the same transporter family. Specifically, the sequence similarity between Ste6p* and CFTR is low and restricted to the NBDs (Kuchler et al., 1989) and only CFTR contains an R domain. Therefore, it will be important in the future to assess whether other aberrant ABC transporters in yeast require Hsp26p/Hsp42p for their degradation, and combined with the continued structural analysis of ABC transporters (see, for example, Lewis et al., 2004), it is hoped that the determinant(s) for their degradation can be ascertained.
Our observation that αA-crystallin enhances exclusively the degradation of ΔF508-CFTR in mammals implies that this sHsp distinguishes the wild type and the mutant variants of the channel. Although immature CFTR and ΔF508-CFTR are often assumed to adopt the same aberrant conformation—one that directs the proteins for ERAD—other evidence indicates discernible differences between immature wild-type and ΔF508-CFTR. First, Meacham et al. (1999) observed an approximately twofold higher abundance of Hsc70–ΔF508-CFTR complexes than Hsc70–CFTR complexes. Second, cotransfection of Hsc70 and Hdj-1 stabilized exclusively the immature form of wild-type CFTR (Farinha et al., 2002). Third, coexpression of Csp1 slowed the degradation of immature wild-type CFTR but increased the turnover of ΔF508-CFTR (Zhang et al., 2006; our unpublished data). Fourth, complexes between calnexin and ΔF508-CFTR were more stable than those between calnexin and wild-type CFTR (Pind et al., 1994). Fifth, the overexpression of RMA1, which may monitor folding defects in ΔF508-CFTR, has a greater effect on ΔF508-CFTR than on wild-type CFTR degradation (Younger et al., 2006). And finally, ΔF508-CFTR seems to be degraded independently of glycan modification, whereas proteolysis of the wild-type protein requires the glycan moiety and calnexin function (Farinha and Amaral, 2005). Moreover, we note that sHsps can distinguish between wild-type protein conformations and structurally identical mutant variants that differ only in their free energies of unfolding; thus, sHsps may be able to recognize transient nonnative states (McHaourab et al., 2002; Koteiche and McHaourab, 2003; Sathish et al., 2004; Shashidharamurthy et al., 2005), and indeed the cytosolic domain of ΔF508-CFTR exhibits a distinct conformation as assessed by limited proteolysis (Zhang et al., 1998). Interestingly, however, we found that αA-crystallin bound with equal efficiencies to an N-terminal, NBD1-containing fragment of wild-type and ΔF508-CFTR (Supplemental Figure 3S), suggesting that the chaperone recognizes the ΔF508-folding defect only after the full-length protein has been synthesized.
A hint that sHsps might also respond to the long-term expression of ΔF508-CFTR in humans was recently provided (Roxo-Rosa et al., 2006). A proteomic analysis of nasal cells from healthy and ΔF508-CFTR–expressing individuals uncovered 65 differentially expressed “spots,” and among these polypeptides was Hsp27. Interestingly, Hsp27 levels decreased approximately twofold in the samples from CF patients. Although the nature of this effect is unclear, one hypothesis—consistent with previous data—is that the chaperone negatively regulates the immune response (De et al., 2000). Another hypothesis is that the synthesis of this sHsp responds to the concentration of CFTR: Because the steady-state level of ΔF508-CFTR is low, Hsp27 levels may also drop. However, it is not clear whether the reduction of this sHsp impacts ΔF508-CFTR turnover.
If sHsps only affect the degradation of ΔF508-CFTR in mammals, why is the wild-type protein stabilized in yeast lacking Hsp26p and Hsp42p? One answer to this question is that CFTR might be unable to assume the native conformation in this organism, and support for this hypothesis derives from the observation that CFTR fails to exit the ER and is quantitatively degraded in yeast (Kiser et al., 2001; Zhang et al., 2001; Fu and Sztul, 2003). We therefore propose that the yeast CFTR expression system better reflects ΔF508-CFTR biogenesis than wild-type CFTR biogenesis. Nevertheless, our data suggest that yeast can be used to identify factors that affect the maturation of any mammalian protein, assuming that an expression system has been developed and validated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Fei Sun, Johannes Buchner, Cecile Pickart, Susan Michaelis, Mark Hochstrasser, Robert Youker, Usha Andley, Kali Das, Avrom Caplan, Suzanne Sandmeyer, John Glover, and Rebecca Hughey for providing reagents. This work was supported by grants from the Cystic Fibrosis Foundation to J.L.B. and R.A.F. and by National Institutes of Health Grants DK-075061 and DK-68196 (to J.L.B. and R.A.F., respectively). K.N. acknowledges support from the Uehara Memorial Foundation and a postdoctoral fellowship from the American Heart Association.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0458) on December 20, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ahner A., Brodsky J. L. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. doi: 10.1016/j.tcb.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Alberti S., Bohse K., Arndt V., Schmitz A., Hohfeld J. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell. 2004;15:4003–4010. doi: 10.1091/mbc.E04-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidmann J. G., Smith J. G., Struhl K. New York: Greene Publishing Associates and Wiley Interscience; 1988. Current protocols in Molecular Biology. [Google Scholar]
- Basha E., Friedrich K. L., Vierling E. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 2006;281:39943–39952. doi: 10.1074/jbc.M607677200. [DOI] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E. A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A., Das K. P. Role of ATP on the interaction of alpha-crystallin with its substrates and its implications for the molecular chaperone function. J. Biol. Chem. 2004;279:42648–42657. doi: 10.1074/jbc.M404444200. [DOI] [PubMed] [Google Scholar]
- Boelens W. C., Croes Y., de Jong W. W. Interaction between alphaB-crystallin and the human 20S proteasomal subunit C8/alpha7. Biochim. Biophys. Acta. 2001;1544:311–319. doi: 10.1016/s0167-4838(00)00243-0. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Lawrence J. G., Caplan A. J. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Casagrande R., Stern P., Diehn M., Shamu C., Osario M., Zuniga M., Brown P. O., Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- Chang Z., Primm T. P., Jakana J., Lee I. H., Serysheva I., Chiu W., Gilbert H. F., Quiocho F. A. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J. Biol. Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Choo-Kang L. R., Zeitlin P. L. Induction of HSP70 promotes DeltaF508 CFTR trafficking. Am. J. Physiol. 2001;281:L58–L68. doi: 10.1152/ajplung.2001.281.1.L58. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Conconi M., Djavadi-Ohaniance L., Uerkvitz W., Hendil K. B., Friguet B. Conformational changes in the 20S proteasome upon macromolecular ligand binding analyzed with monoclonal antibodies. Arch. Biochem. Biophys. 1999;362:325–328. doi: 10.1006/abbi.1998.1037. [DOI] [PubMed] [Google Scholar]
- Conconi M., Petropoulos I., Emod I., Turlin E., Biville F., Friguet B. Protection from oxidative inactivation of the 20S proteasome by heat-shock protein 90. Biochem. J. 1998;333:407–415. doi: 10.1042/bj3330407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- De A. K., Kodys K. M., Yeh B. S., Miller-Graziano C. Exaggerated human monocyte IL-10 concomitant to minimal TNF-alpha induction by heat-shock protein 27 (Hsp27) suggests Hsp27 is primarily an antiinflammatory stimulus. J. Immunol. 2000;165:3951–3958. doi: 10.4049/jimmunol.165.7.3951. [DOI] [PubMed] [Google Scholar]
- den Engelsman J., Bennink E. J., Doerwald L., Onnekink C., Wunderink L., Andley U. P., Kato K., de Jong W. W., Boelens W. C. Mimicking phosphorylation of the small heat-shock protein alphaB-crystallin recruits the F-box protein FBX4 to nuclear SC35 speckles. Eur. J. Biochem. 2004;271:4195–4203. doi: 10.1111/j.1432-1033.2004.04359.x. [DOI] [PubMed] [Google Scholar]
- den Engelsman J., Keijsers V., de Jong W. W., Boelens W. C. The small heat-shock protein alpha B-crystallin promotes FBX4-dependent ubiquitination. J. Biol. Chem. 2003;278:4699–4704. doi: 10.1074/jbc.M211403200. [DOI] [PubMed] [Google Scholar]
- Du K., Sharma M., Lukacs G. L. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- Ehrnsperger M., Graber S., Gaestel M., Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Farinha C. M., Amaral M. D. Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol. Cell Biol. 2005;25:5242–5252. doi: 10.1128/MCB.25.12.5242-5252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha C. M., Nogueira P., Mendes F., Penque D., Amaral M. D. The human DnaJ homologue (Hdj)-1/heat-shock protein (Hsp) 40 co-chaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem. J. 2002;366:797–806. doi: 10.1042/BJ20011717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell S. W., Travers K. J., Weissman J. S., Brodsky J. L. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- Fu L., Sztul E. Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J. Cell Biol. 2003;160:157–163. doi: 10.1083/jcb.200210086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnann A., Riordan J. R., Wolf D. H. Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htm1p/EDEM and the Cdc48 protein complex in yeast. Mol. Biol. Cell. 2004;15:4125–4135. doi: 10.1091/mbc.E04-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Braun N., Stromer T., Richter B., Model N., Weinkauf S., Buchner J. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23:638–649. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Haslbeck M., Walke S., Stromer T., Ehrnsperger M., White H. E., Chen S., Saibil H. R., Buchner J. Hsp 26, a temperature-regulated chaperone. EMBO J. 1999;18:6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Cooper A. A. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M. M., Finger A., Schweiger M., Wolf D. H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble lumenal protein. J. Biol. Chem. 2004;297:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Ito H., Kamei K., Iwamoto I., Inaguma Y., Garcia-Mata R., Sztul E., Kato K. Inhibition of proteasomes induces accumulation, phosphorylation, and recruitment of HSP27 and alphaB-crystallin to aggresomes. J. Biochem. 2002;131:593–603. doi: 10.1093/oxfordjournals.jbchem.a003139. [DOI] [PubMed] [Google Scholar]
- Jakob U., Gaestel M., Engel K., Buchner J. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- Johnston J. A., Ward C. L., Kopito R. R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Beckerich J. M., Brodsky J. L. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe G., Franck E., Verschuure P., Boelens W. C., Leunissen J. A., de Jong W. W. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1–10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser G. L., Gentzsch M., Kloser A. K., Balzi E., Wolf D. H., Goffeau A., Riordan J. R. Expression and degradation of the cystic fibrosis transmembrane conductance regulator in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2001;390:195–205. doi: 10.1006/abbi.2001.2385. [DOI] [PubMed] [Google Scholar]
- Kleizen B., van Vlijmen T., de Jonge H. R., Braakman I. Folding of CFTR is predominantly cotranslational. Mol. Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteiche H. A., McHaourab H. S. Mechanism of chaperone function in small heat-shock proteins. Phosphorylation-induced activation of two-mode binding in alphaB-crystallin. J. Biol. Chem. 2003;278:10361–10367. doi: 10.1074/jbc.M211851200. [DOI] [PubMed] [Google Scholar]
- Kuchler K., Sterne R. E., Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. J., Roseman A. M., Saibil H. R., Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis H. A., et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D., Tam A., Schmidt W. K., Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD) Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- McHaourab H. S., Dodson E. K., Koteiche H. A. Mechanism of chaperone function in small heat shock proteins. Two-mode binding of the excited states of T4 lysozyme mutants by alphaA-crystallin. J. Biol. Chem. 2002;277:40557–40566. doi: 10.1074/jbc.M206250200. [DOI] [PubMed] [Google Scholar]
- Meacham G. C., Lu Z., King S., Sorscher E., Tousson A., Cyr D. M. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Ng D. T., Spear E. D., Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Harada K., Takeya M., Yamahira K., Wada I., Shuto T., Suico M. A., Hashimoto Y., Kai H. Delta F508 CFTR pool in the endoplasmic reticulum is increased by calnexin overexpression. Mol. Biol. Cell. 2004;15:563–574. doi: 10.1091/mbc.E03-06-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcellier A., Schmitt E., Gurbuxani S., Seigneurin-Berny D., Pance A., Chantome A., Plenchette S., Khochbin S., Solary E., Garrido C. HSP27 is a ubiquitin-binding protein involved in I-kappaBalpha proteasomal degradation. Mol. Cell Biol. 2003;23:5790–5802. doi: 10.1128/MCB.23.16.5790-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcellier A., et al. HSP27 favors ubiquitination and proteasomal degradation of p27Kip1 and helps S-phase re-entry in stressed cells. FASEB J. 2006;20:E281–E293. doi: 10.1096/fj.05-4184fje. [DOI] [PubMed] [Google Scholar]
- Petko L., Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- Pilewski J. M., Frizzell R. A. Role of CFTR in airway disease. Physiol. Rev. 1999;79:S215–255. doi: 10.1152/physrev.1999.79.1.S215. [DOI] [PubMed] [Google Scholar]
- Pind S., Riordan J. R., Williams D. B. Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]
- Plemper R. K., Bohmler S., Bordallo J., Sommer T., Wolf D. H. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Qu B. H., Thomas P. J. Alteration of the cystic fibrosis transmembrane conductance regulator folding pathway. J. Biol. Chem. 1996;271:7261–7264. doi: 10.1074/jbc.271.13.7261. [DOI] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxo-Rosa M., da Costa G., Luider T. M., Scholte B. J., Coelho A. V., Amaral M. D., Penque D. Proteomic analysis of nasal cells from cystic fibrosis patients and non-cystic fibrosis control individuals: search for novel biomarkers of cystic fibrosis lung disease. Proteomics. 2006;6:2314–2325. doi: 10.1002/pmic.200500273. [DOI] [PubMed] [Google Scholar]
- Rubenstein R. C., Zeitlin P. L. Sodium 4-phenylbutyrate downregulates Hsc 70, implications for intracellular trafficking of DeltaF508-CFTR. Am. J. Physiol. 2000;278:C259–C267. doi: 10.1152/ajpcell.2000.278.2.C259. [DOI] [PubMed] [Google Scholar]
- Sathish H. A., Koteiche H. A., McHaourab H. S. Binding of destabilized betaB2-crystallin mutants to alpha-crystallin: the role of a folding intermediate. J. Biol. Chem. 2004;279:16425–16432. doi: 10.1074/jbc.M313402200. [DOI] [PubMed] [Google Scholar]
- Sayeed A., Ng D. T. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- Shashidharamurthy R., Koteiche H. A., Dong J., McHaourab H. S. Mechanism of chaperone function in small heat shock proteins: dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. J. Biol. Chem. 2005;280:5281–5289. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland E., Qu B. H., Millen L., Thomas P. J. The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1997;272:25421–25424. doi: 10.1074/jbc.272.41.25421. [DOI] [PubMed] [Google Scholar]
- Studer S., Narberhaus F. Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J. Biol. Chem. 2000;275:37212–37218. doi: 10.1074/jbc.M004701200. [DOI] [PubMed] [Google Scholar]
- Sun Y., MacRae T. H. The small heat shock proteins and their role in human disease. FEBS J. 2005;272:2613–2627. doi: 10.1111/j.1742-4658.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- Sun F., Zhang R., Gong X., Geng X., Drain P. F., Frizzell R. A. Derlin-1 promotes the efficient degradation of CFTR and CFTR folding mutants. J. Biol. Chem. 2006;281:36856–36863. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- Thibodeau P. H., Brautigam C. A., Machius M., Thomas P. J. Side chain and backbone contributions of Phe508 to CFTR folding. Nat. Struct. Mol. Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- van Montfort R., Slingsby C., Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv. Protein Chem. 2001;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B. J., Margolis J. W. Age-dependent association of isolated bovine lens multicatalytic proteinase complex (proteasome) with heat-shock protein 90, an endogenous inhibitor. Arch. Biochem. Biophys. 1995;323:455–462. doi: 10.1006/abbi.1995.0067. [DOI] [PubMed] [Google Scholar]
- Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang F., Kartner N., Lukacs G. L. Limited proteolysis as a probe for arrested conformational maturation of delta F508 CFTR. Nat. Struct. Biol. 1998;5:180–183. doi: 10.1038/nsb0398-180. [DOI] [PubMed] [Google Scholar]
- Zhang H., Peters K. W., Sun F., Marino C. R., Lang J., Burgoyne R. D., Frizzell R. A. Cysteine string protein interacts with and modulates the maturation of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2002a;277:28948–28958. doi: 10.1074/jbc.M111706200. [DOI] [PubMed] [Google Scholar]
- Zhang H., Schmidt B. Z., Sun F., Condliffe S. B., Butterworth M. B., Youker R. T., Brodsky J. L., Aridor M., Frizzell R. A. Cysteine string protein monitors late steps in CFTR biogenesis. J. Biol. Chem. 2006;281:11312–11321. doi: 10.1074/jbc.M512013200. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Michaelis S., Brodsky J. L. CFTR expression and ER-associated degradation in yeast. Methods Mol. Med. 2002b;70:257–265. doi: 10.1385/1-59259-187-6:257. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Nijbroek G., Sullivan M. L., McCracken A. A., Watkins S. C., Michaelis S., Brodsky J. L. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.