Abstract
The sugar N-acetylglucosamine (GlcNAc) plays an important role in nutrient sensing and cellular regulation in a wide range of organisms from bacteria to humans. In the fungal pathogen Candida albicans, GlcNAc induces a morphological transition from budding to hyphal growth. Proteomic comparison of plasma membrane proteins from buds and from hyphae induced by GlcNAc identified a novel hyphal protein (Ngt1) with similarity to the major facilitator superfamily of transporters. An Ngt1-GFP fusion was detected in the plasma membrane after induction with GlcNAc, but not other related sugars. Ngt1-GFP was also induced by macrophage phagocytosis, suggesting a role for the GlcNAc response in signaling entry into phagolysosomes. NGT1 is needed for efficient GlcNAc uptake and for the ability to induce hyphae at low GlcNAc concentrations. High concentrations of GlcNAc could bypass the need for NGT1 to induce hyphae, indicating that elevated intracellular levels of GlcNAc induce hyphal formation. Expression of NGT1 in Saccharomyces cerevisiae promoted GlcNAc uptake, indicating that Ngt1 acts directly as a GlcNAc transporter. Transport mediated by Ngt1 was specific, as other sugars could not compete for the uptake of GlcNAc. Thus, Ngt1 represents the first eukaryotic GlcNAc transporter to be discovered. The presence of NGT1 homologues in the genome sequences of a wide range of eukaryotes from yeast to mammals suggests that they may also function in the cellular processes regulated by GlcNAc, including those that underlie important diseases such as cancer and diabetes.
INTRODUCTION
N-acetylglucosamine (GlcNAc) is an amino sugar that carries out important roles in a broad range of cells from bacteria to humans. One aspect of GlcNAc function is to mediate cellular signaling. In bacteria, GlcNAc induces components that are important for colonization of human hosts, including fimbrins that mediate adhesion to host cells (Sohanpal et al., 2004), multidrug exporter genes (Hirakawa et al., 2006) and Curli fibers that promote biofilm formation (Barnhart et al., 2006). In mammals, GlcNAc is a key sensor of nutrient status that is involved in insulin signaling, cell cycle control, and other essential processes. Nutritional effects that lead to increased GlcNAc levels result in its conversion to UDP-GlcNAc and subsequent attachment to proteins on serine or threonine residues in a dynamic manner that is analogous to modification of proteins by phosphorylation (Slawson and Hart, 2003; Zachara and Hart, 2006). In fact, the interplay between O-GlcNAc modification and phosphorylation of proteins regulates many critical transcription factors, such as c-myc and p53 (Chou and Hart, 2001; Yang et al., 2006). O-GlcNAc modification also regulates other processes including proteosome function (Zachara and Hart, 2004). In addition to these regulatory roles, GlcNAc contributes to the N-linked glycosylation, glycophosphatidylinositol (GPI) anchor addition to proteins and is polymerized into chitin, which forms part of the fungal cell wall and the exoskeletons of parasites, insects, and other organisms.
GlcNAc induces the fungal pathogen Candida albicans to switch from forming round budding cells to instead growing as long thin hyphal cells. This morphological transition is thought to be an important virulence factor (Sudbery et al., 2004; Whiteway and Oberholzer, 2004). C. albicans is one of the leading causes of nosocomial bloodstream infections and is capable of causing severe systemic infections (Edmond et al., 1999; Mavor et al., 2005; Spellberg et al., 2006). C. albicans can be detected at sites of infection in various morphologies including rounded buds, pseudohyphae (chains of elongated buds), and hyphae (chains of long thin cells with parallel walls) (Odds, 1988). Each form is thought to confer a distinct advantage during infection. For example, small buds are more likely to disseminate in the bloodstream and hyphal cells may be better suited for invasive growth into tissues. The distinct cell types also differ in their production of virulence factors, such as the adhesin proteins that mediate attachment to host cells and secreted hydrolytic enzymes that facilitate invasive growth (Whiteway and Oberholzer, 2004; Kumamoto and Vinces, 2005).
In vitro studies have identified a variety of different stimuli that can induce C. albicans to switch morphologies. One of the strongest known inducers of hyphal growth is serum. Screens for chemically defined inducers have found that nutrients, such as the amino sugar GlcNAc (Simonetti et al., 1974) and certain amino acids (Odds, 1988; Maidan et al., 2005), will also stimulate hyphal growth. Environmental conditions also contribute to hyphal induction, as it occurs optimally at 37°C and can be stimulated by alkaline pH and 5% CO2 (Davis, 2003; Sudbery et al., 2004; Klengel et al., 2005). The upstream components of the hyphal signal pathway have not been clearly identified, but many of the downstream signaling components are known (Berman and Sudbery, 2002). For example, production of cAMP by adenylyl cyclase plays a key role in inducing hyphal formation. In contrast to Saccharomyces cerevisiae, adenylyl cyclase is not needed for growth of C. albicans and instead plays an essential role in promoting hyphal formation (Rocha et al., 2001). cAMP stimulation of protein kinase A results in phosphorylation of downstream targets, including the transcription factor Efg1, which is needed for hyphal growth (Stoldt et al., 1997; Tebarth et al., 2003). In addition, MAP kinase signaling activates the Cph1 transcription factor that is also involved in hyphal signaling (Lo et al., 1997). Hyphal growth can also be promoted by proteolytic activation of the Rim101 transcription factor at alkaline pH (Davis et al., 2000). The hyphal pathway is also negatively regulated by the repressor Tup1 (Braun and Johnson, 1997).
To better define the upstream components involved in hyphal signaling, as well as the differences between buds and hyphae, we carried out a proteomic comparison of plasma membrane proteins from budding and hyphal cells. GlcNAc was selected as the inducer of hyphal growth to better understand its role in this process, because the teleological basis for the effects of GlcNAc has not been understood from previous studies. GlcNAc was first reported to induce hyphae over 30 years ago in a screen for chemically defined inducers of hyphal growth in vitro (Simonetti et al., 1974). Subsequent studies revealed C. albicans is capable of taking up GlcNAc and using it as an energy source, whereas the nonpathogenic S. cerevisiae lacks the proteins needed to transport and catabolize GlcNAc (Kumar et al., 2000; Singh et al., 2001). Mutational analysis indicates that the enzymes needed to catabolize GlcNAc in C. albicans also contribute to virulence (Singh et al., 2001; Yamada-Okabe et al., 2001). These include the enzymes that phosphorylate (Hxk1), deacetylate (Dac1), and deamidate (Nag1) GlcNAc, leading to its conversion to fructose-6-PO4 (Kumar et al., 2000; Singh et al., 2001; Yamada-Okabe et al., 2001). The GlcNAc transporter had not been identified previously, but in this study we report that one of the proteins identified by mass spectrometry (Ngt1) acts as a GlcNAc transporter, thus permitting direct analysis of its role in GlcNAc transport and signaling. Ngt1 is also significant in that it represents the first eukaryotic GlcNAc transporter to be identified and will therefore facilitate the analysis of the role of the homologous proteins in other organisms for their role in cellular regulation by GlcNAc.
MATERIALS AND METHODS
Strains and Media
The C. albicans and S. cerevisiae strains used in this study are described in Table 1. Strains were propagated on rich YPD medium or on synthetic medium essentially as described (Sherman, 1991). Uridine, 80 mg/l, was added to C. albicans cultures. To induce hyphal growth with different concentrations of GlcNAc, cells grown overnight in basal yeast medium (BYNB) containing galactose and buffered to pH 6.8 with 10 mM PIPES were washed and inoculated into 5 ml fresh medium at a density of 2 × 106 cells/ml. After 5 h of growth, the cells were washed with buffered BYNB medium and inoculated into buffered BYNB containing different concentrations of GlcNAc. Cells were grown at 37°C for 2 h, and then the cell morphologies were examined microscopically. The buffered medium and the growth regimen described above were used to maintain a low spontaneous level of hyphal formation that can been seen under other conditions due to farnesol depletion and pH changes when adjusting the cultures (Enjalbert and Whiteway, 2005).
Table 1.
Strains used in this study
| Strain | Parent | Genotype |
|---|---|---|
| C. albicans strains | ||
| Sc5314 | Wild type | |
| BWP17 | Sc5314 | ura3Δ::λimm434/ura3Δ::λimm434 his::hisG/his1::hisG arg4::hisG/arg4::hisG |
| DIC185 | BWP17 | ura3Δ::λimm434/URA3 his::hisG/HIS1 arg4::hisG/ARG4 |
| YJA1 | CAI4 | ura3Δ::imm434/ura3Δ::imm434 NGT1-GFP-URA3/NGT1 |
| YJA2 | BWP17 | ngt1Δ::ARG4/ngt1Δ::HIS1 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA3 | YJA2 | ngt1Δ::ARG4/ngt1Δ::HIS1 ura3Δ::λimm434/URA3 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA4 | YJA2 | ngt1Δ::ARG4/ngt1Δ::HIS1::NGT1-URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| S. cerevisiae strain | ||
| W303 | MATa/MATα his3-11,15/his3-11,15 can1-100/can1-100, leu2-3,112/leu2-3,112 trp1-1/trp1-1, ade2-1/ade2-1, ura3-1/ura3-1 |
The C. albicans NGT1-GFP strain was constructed by homologous recombination. PCR was used to add ∼70 base pairs of sequence homologous to the 3′ end of the NGT1 open reading frame (ORF) to a cassette that contains GFP and a URA3 selectable marker (Gerami-Nejad et al., 2001). Ura+ colonies resulting from the transformation of the PCR product into C. albicans were then grown on GlcNAc medium and screened for GFP-positive cells by fluorescence microscopy. The ngt1Δ homozygous deletion strain was constructed by successive transformation of strain BWP17 with PCR-generated constructs containing either the ARG4 or HIS1 selectable marker genes flanked by ∼70-base pair regions that were homologous to the sequences flanking the ORF of NGT1 (Wilson et al., 1999). Proper integration of the GFP and deletion cassettes was verified by PCR.
Mass Spectrometry Analysis of Plasma Membrane Proteins
Cells were grown in budding phase by diluting a fresh overnight culture to 8 × 105 cells/ml in YPD medium and then incubating at 30°C until the cultures reached 107 cells/ml. To harvest cells initiating hyphal phase growth (germ tubes), a fresh overnight culture grown in YPD was diluted to 8 × 105 cells/ml in YPD medium and then grown to 107 cells/ml at 30°C. Cells were washed, resuspended in minimal medium containing 2.5 mM GlcNAc, and incubated at 37°C for 75–90 min. Germ tube emergence was confirmed by microscopy. Budding cells and germ tubes were harvested by centrifugation and washed with water, and then pellets of 109 cells were frozen for later use. The cell pellets were lysed by agitation with glass beads in cold TNE buffer with protease inhibitors (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, 2.5 mM benzamidine, and 30 μM pepstatin). The plasma membrane fraction was then isolated by density gradient centrifugation, essentially as described previously (Schandel and Jenness, 1994; Dosil et al., 1998). In brief, cell extract was loaded at the bottom of a density gradient that ranged from 38 to 22% Renocal-76 (Bracco Diagnostic, Princeton, NJ). Tubes were spun in a Beckman SW40Ti rotor (Fullerton, CA) for 37 h at 35,000 rpm. The gradient was partitioned into 14 fractions by taking samples from the top, and then a 10-μl aliquot of each sample was run on a Western blot and probed with anti-Pma1 antibodies in order to detect the plasma membrane fraction. The plasma membrane fractions were subsequently extracted with ice-cold 1% Triton X-100 in order to isolate the detergent-resistant membrane fraction and thereby purify plasma membrane proteins away from contaminating cytoplasmic proteins, such as the abundant ribosomal proteins. Plasma membrane fractions extracted with cold 1% Triton X-100 were loaded at the bottom of a 40–5% sucrose gradient and spun in a SW41Ti rotor for 25 h at 35,000 rpm. The gradient was split into different fractions and run on Western blots probed with anti-Pma1 antibodies to identify the detergent resistant membrane fraction that floated to the top of the gradient, which was then removed for analysis.
Approximately 500 μg of detergent-resistant membrane fraction from budding or germ tube cells was then separated by SDS-PAGE and prepared for mass spectrometry. 10-kDa ranges of the gel lanes were cut out, diced up, washed with 50% (vol/vol) methanol and 5% (vol/vol) acetic acid, dehydrated with acetonitrile, rehydrated with 10 mM DTT to reduce the samples, and then treated with 100 mM iodoacetamide to alkylate the proteins. The gel slices were washed with 100 mM ammonium bicarbonate, dehydrated with acetonitrile, and then dried. The samples were then rehydrated in 100 mM ammonium bicarbonate buffer and then 30 μl of 20 ng/ml sequencing grade TPCK-treated trypsin (Promega, Madison, WI) was added, and samples were incubated overnight at 37°C. Tryptic peptides were recovered with extraction buffer (50% acetonitrile and 50% formic acid) and analyzed at the Stony Brook University Proteomics Center by liquid chromatography followed by tandem mass spectrometry with electrospray ionization (nano-LC/ESI/MSMS) on an API Qstar Pulsar i quadrupole-time of fly (Q-TOF) tandem mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA). Mass spectrometry results were then compared with the ORF19 release of the Candida genome database to identify the corresponding proteins (http://www.candidagenome.org/).
Plasmid Construction
A plasmid carrying NGT1 was constructed by PCR amplification of the genomic sequence from 974 base pairs upstream of the initiator ATG to 354 bases downstream of the terminator codon. This DNA fragment was then inserted between the SacI and SacII restriction sites of the URA3 plasmid pDDB57 (Wilson et al., 2000). The resulting plasmid was linearized in the promoter region by digestion with EcoRV, and integrated into the ngt1Δ strain YJA2 to create a complemented strain called YJA4. A plasmid designed to express NGT1 in S. cerevisiae under control of the GAL1 promoter was constructed by PCR amplifying the NGT1 ORF and inserting it at the SpeI site in the polylinker region of plasmid pRS426GAL1 (Mumberg et al., 1994).
GlcNAc Uptake Assays
To assay GlcNAc uptake in C. albicans strains, overnight cultures were harvested by centrifugation, washed in BYNB, and then inoculated into fresh BYNB containing either dextrose or GlcNAc. Cells were grown for 4 h at 30°C to induce the GlcNAc pathway, harvested by centrifugation, washed with BYNB, and then resuspended at 1.1 × 108 cells/ml. Uptake assays were carried out in 96-well plates and were initiated by adding 107 cells to 10 μl of radioactive [3H]GlcNAc/well (200 μM final concentration; 5 mCi/mmol). Reactions were terminated by collecting cells onto Whatman GF/C glass microfiber filters (Clifton, NJ) using a Brandel Cell Harvester (Gaithersburg, MD). Filters were dried, and then the [3H]GlcNAc taken up by the cells was quantified using a scintillation counter.
To assay GlcNAc uptake in S. cerevisiae strains, W303 cells containing either pRS426GAL1-NGT1 or the control empty vector were grown overnight in synthetic medium lacking uracil and containing either 2% dextrose or galactose. Cells were then washed, inoculated into fresh medium, and grown for 4 h at 30°C. [3H]GlcNAc uptake assays were then carried out as described above. The ability of different sugars to compete for the uptake of GlcNAc was assayed in S. cerevisiae strain W303 containing pRS426GAL1-NGT1 that was prepared as described above. Cells, 107, were added per well of 96-well plates that also contained [3H]GlcNAc (200 μM final) plus either 2 mM (10×) or 20 mM (100×) final concentration of one of the following cold competitor sugars: GlcNAc, dextrose, galactose, fructose, glucosamine or N-acetylmannosamine.
Macrophage Infection Assay
J774 macrophage cells were prepared for infection by plating 105 cells/well on coverslips in 24-well plates containing DMEM medium with 10% FBS. Cells were incubated overnight and then the medium was replaced with fresh DMEM lacking FBS. The C. albicans strains were prepared for infection assays by growing overnight in YPD, and then washing and resuspending the cells in phosphate-buffered saline (PBS) at 107 cells/ml. Infections were initiated by adding the appropriate C. albicans strain to a well of J774 cells at an MOI of 1. Cells were incubated for 40 min at 37°C, and then the J774 cells were washed and fresh medium was added. Assays were stopped at different time points by adding paraformaldehyde to 2.5% final concentration for 20 min. Samples were then washed, mounted on slides, and examined microscopically as described below.
Microscopy
Ngt1-GFP fluorescence was analyzed in cells that were grown overnight in synthetic complete medium containing glucose but lacking uracil. Cells were washed with BYNB and then incubated with minimal medium containing different sugars or serum for 2–4 h at 37°C. Cells were washed with BYNB and analyzed immediately by microscopy. To compare the morphogenesis of wild-type versus ngt1Δ cells, cells were grown overnight in synthetic medium containing dextrose, washed, adjusted to 2–3 × 105 cells/ml in BYNB, and incubated in the presence or absence of 2 mM GlcNAc at 37°C. Cells were analyzed by fluorescence microscopy to detect GFP and by differential interference (DIC) microscopy to detect cell morphology. Images were captured using an Olympus BH2 microscope (Melville, NY) equipped with a Zeiss AxioCam digital camera (Thornwood, NY). Fluorescence intensity of the images was quantified using Openlab 3.0.8 software from Improvision (Lexington, MA).
Analysis of NGT1 Homologues
BLAST searches (Altschul et al., 1990) were carried out to identify NGT1 homologues in the genome sequences of other organisms present at the Web site of the National Center for Biotechnology (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignments of the predicted Ngt1 proteins and an evolutionary tree of their relatedness were carried out using Clustal W (Thompson et al., 1994).
RESULTS
Identification of GlcNAc-induced Plasma Membrane Proteins by Mass Spectrometry
A proteomics approach aimed at identifying plasma membrane proteins that are unique to the bud or hyphal phases of C. albicans was carried out to identify new proteins related to the regulation of morphogenesis and virulence. Plasma membranes were purified by density gradient centrifugation from budding cells grown in standard medium (YPD) or from cells induced with GlcNAc to form hyphae. The detergent-resistant fraction of the plasma membrane was then isolated to help remove contaminating cytoplasmic proteins (see Materials and Methods). The purified membrane proteins were then separated by SDS gel electrophoresis, trypsin-digested, and analyzed by liquid chromatography followed by tandem mass spectrometry. Comparison of the mass spectrometry data with the ORFs predicted in the C. albicans genome sequence identified 137 different proteins. Approximately one third were detected only in buds, one third in hyphal cells, and one third were common to both buds and hyphae. A representative set of the proteins that were identified is shown in Table 2, and the full set of proteins is described in the Supplementary Data.
Table 2.
Representative subset of proteins identified by mass spectrometry
| Cell type | ORF19a | C. albicans gene | S. cerevisiae homologb | Description of functionc |
|---|---|---|---|---|
| Bud | 5759 | SNQ2 | SNQ2 | ABC transporter protein involved in multidrug resistance |
| 1415 | FRE10 | FRE1 | Cell-surface ferric reductase | |
| 4118 | CNT | None | Cation-coupled nucleoside transporter | |
| 3765 | RAX2 | RAX2 | Transmembrane protein involved in bipolar bud site selection | |
| 7001 | — | YCK1,2 | Casein kinase I | |
| Germ tube | 7094 | HGT12 | SNF3 | MFS superfamily glucose sensor/transporter |
| 4335 | TNA1 | TNA1 | Nicotinic acid transporter | |
| 2614 | RSR1 | RSR1 | RAS-related protein | |
| 176 | OPT4 | OPT2 | Oligopeptide transporter | |
| 7030 | SSR1 | CCW14 | GPI anchor protein with a role in cell wall structure | |
| Both | 5383 | PMA1 | PMA1,2 | Plasma membrane H(+)-ATPase |
| 4456 | GAP4 | GAP1 | Putative amino acid permease; hyphal induced | |
| 3829 | PHR1 | GAS1,2 | GPI-anchored glycoside; role in cell wall structure | |
| 3646 | CTR1 | CTR1 | Copper transporter | |
| 1760 | RAS1 | RAS1,2 | RAS signal transduction GTPase |
a Open reading frame designation from ORF19 release of the C. albicans genome database.
b Closest homolog found in the S. cerevisiae genome.
c Function as indicated by experimental work or by close similarity to known genes in other organisms.
Transporters comprised the major class of proteins, which accounted for 40% of the total. The majority of the proteins were thus membrane-spanning proteins, although ∼5% of the proteins were GPI anchored and another 5% were acylated. Another major group of proteins were those involved in iron uptake (∼5%), which is critical for C. albicans virulence. About 15% of the proteins did not have an obvious homolog in S. cerevisiae, and a similar fraction did not have an obvious prediction for their function. Most types of proteins were represented at roughly similar levels in the different groups. However, it was interesting that the proteins lacking a homolog in S. cerevisiae represented 19% of those found only in buds, 18% of those found in germ tubes, but only 6% of those found in both buds and germ tubes. Altogether there were 20 proteins that lack a homolog in the nonpathogenic S. cerevisiae and are thus candidates for carrying out virulence functions in C. albicans.
Ngt1-GFP Is Specifically Induced by GlcNAc
A protein we named Ngt1 that corresponds to ORF orf19.5392 in the C. albicans genome was selected for further study, because it was identified with very high confidence by mass spectrometry and initially appeared to be specific to hyphae. Subsequent analyses detected Ngt1 in budding cells, albeit with lower confidence scores, suggesting that Ngt1 was likely to be induced by GlcNAc. Ngt1 is also predicted to contain multiple transmembrane domains and was therefore unlikely to be a cytoplasmic contaminant. It was also interesting that homologues of Ngt1 were absent from S. cerevisiae and its close relatives, but present in a wide range of other fungi, including the pathogens Cryptococcus neoformans and Aspergillus fumigatis, suggesting that Ngt1 could be related to virulence.
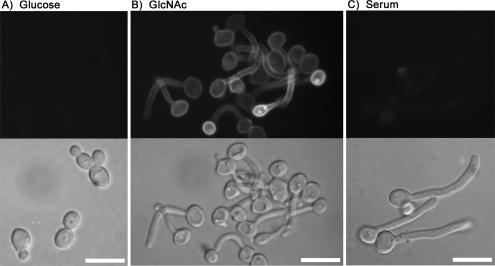
To verify that Ngt1 encodes a plasma membrane protein present in GlcNAc-induced hyphal cells, an NGT1-GFP fusion gene was constructed in the genome of C. albicans by introducing a GFP tag at the C terminus. Fluorescence microscopy readily detected Ngt1-GFP at the periphery of GlcNAc-induced hyphae, consistent with plasma membrane localization. In contrast, Ngt1-GFP was not detected in budding cells grown in glucose medium (Figure 1). Ngt1-GFP was also not detected in cells that were induced to form hyphae by treatment with serum, one of the strongest known inducers of hyphal morphogenesis. These results indicate that NGT1 is induced by GlcNAc and is not generally a hyphal-induced gene.
Figure 1.
Ngt1-GFP induction by GlcNAc. C. albicans strain YJA1 carrying an NGT1-GFP fusion gene was grown in the indicated medium for 2 h at 37°C, and then fluorescence and light microscope images were recorded. Cells were incubated in minimal medium containing glucose, GlcNAc, or glucose plus 10% bovine serum as indicated. Fluorescent microscope images of Ngt1-GFP are shown in the top panels and light microscope (DIC) images are shown below. Bar, 10 μm.
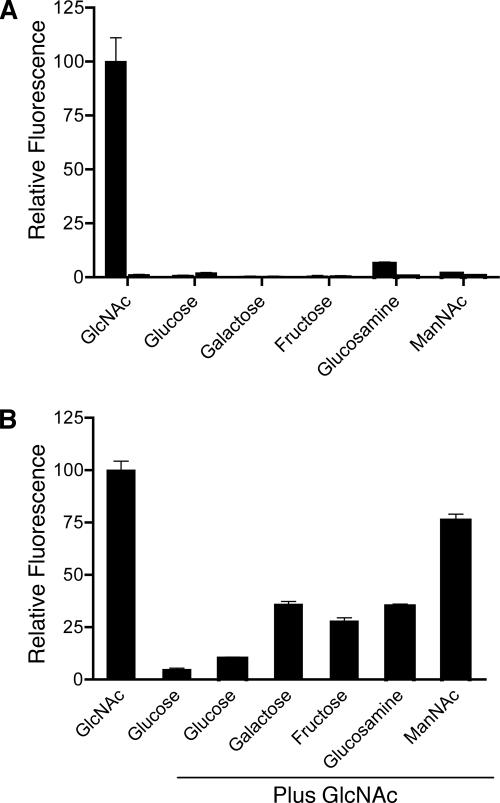
The specificity of NGT1 regulation was analyzed further by growing cells in medium containing different sugars (Figure 2A). Ngt1-GFP was only highly induced by GlcNAc and not by other related sugars such as glucose, galactose, fructose, glucosamine, or N-acetylmannosamine. Quantitation of the fluorescence signal indicated that Ngt1-GFP was induced >50-fold in GlcNAc medium. Growth of the cells in medium containing a second sugar in addition to GlcNAc demonstrated that glucose strongly repressed the expression of NGT1. The glucose repression was not complete as very weak Ngt1-GFP fluorescence could be detected above the background seen in cells grown only in glucose (Figure 2B). Galactose, fructose, glucosamine and N-acetylmannosamine still permitted high-level induction of Ngt1-GFP, although they did cause a partial reduction in Ngt1-GFP fluorescence. Thus, NGT1 is specifically induced by GlcNAc and is repressed by glucose.
Figure 2.
Ngt1-GFP is specifically induced by GlcNAc and is repressed by glucose. (A) Relative fluorescence of cells incubated in medium containing the indicated sugar for 4 h. For each sugar, the first bar represents the NGT1-GFP strain YJA1, and the second bar is the untagged strain SC5314. Fluorescence was very low under all conditions for the untagged strain. (B) Relative fluorescence of cells induced with the indicated sugar or combination of GlcNAc and another sugar. Note that glucose repressed induction of NGT1-GFP. Relative fluorescence was determined from analysis of digital microscope images (see Materials and Methods).
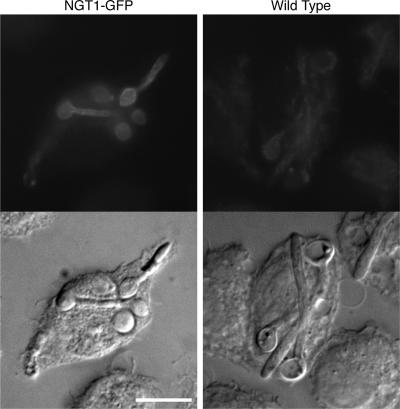
Ngt1-GFP Is Induced by Macrophage Phagocytosis
The role of GlcNAc signaling in C. albicans has been unclear, because GlcNAc was initially identified as an inducer of hyphal growth in a screen for a chemically defined alternative to serum (Simonetti et al., 1974). Therefore, we took advantage of NGT1-GFP as a reporter gene to examine a possible biological role for GlcNAc signaling. In particular, the ability of macrophage phagocytosis to induce Ngt1-GFP was examined because phagocytosis is known to induce hyphal growth (Lo et al., 1997; Lorenz et al., 2004). Phagolysosomes are also low in glucose and are likely to contain GlcNAc due to the degradation carbohydrate chains on glycoproteins or as a result of the cleavage of Candida cell wall chitin by acidic mammalian chitinase to release the GlcNAc subunits (see Discussion). As shown in Figure 3, Ngt1-GFP was induced after phagocytosis by the murine macrophage cell line J774. Although there was detectable nonspecific fluorescence in J774 cells that had phagocytosed untagged wild-type cells, the level of Ngt1-GFP was readily detected above this background in three independent experiments. The Ngt1-GFP fluorescence was also qualitatively different in that it appeared more evenly across the plasma membrane than did the nonspecific fluorescence. Consistent with this, microarray analysis of gene expression found that NGT1 and the genes needed for GlcNAc catabolism are induced after phagocytosis (Lorenz et al., 2004). These results indicate that the GlcNAc pathway is activated after phagocytosis and suggest that one aspect of the biological role for the response to GlcNAc is to signal cells to form hyphae after phagocytosis.
Figure 3.
Ngt1-GFP is induced after macrophage phagocytosis. The murine macrophage cell line J774 was infected with C. albicans NGT1-GFP strain YJA1 or wild-type strain SC5314 at an MOI of 1 for 40 min. The cells were incubated for 2 h and then fixed with paraformaldehyde. Fluorescent microscope images of Ngt1-GFP are shown on top and light microscope (DIC) images are shown below. Bar, 10 μm.
NGT1 Is Required for Efficient Growth on GlcNAc and Hyphal Induction
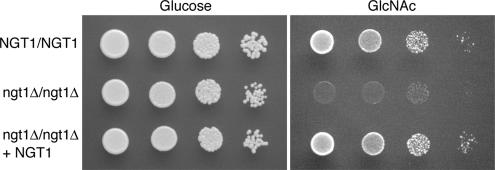
Hydropathy analysis of the Ngt1 protein indicates that it contains 12 transmembrane domains, which is characteristic of transporter proteins and suggests that Ngt1 may act as a GlcNAc transporter. To test this possibility, a homozygous deletion strain was constructed in which both copies of NGT1 were deleted (C. albicans is a diploid organism). The deletion mutations were constructed in the BWP17 strain (Wilson et al., 1999) by replacing one allele of NGT1 with ARG4 and the other allele with HIS1. As shown in Figure 4, the ngt1Δ mutant grew normally in glucose medium, but was defective for growth in the GlcNAc medium commonly used for hyphal induction (2.5 mM GlcNAc).
Figure 4.
NGT1 is required for growth on GlcNAc medium. Wild-type strain SC5314, ngt1Δ homozygous deletion strain YJA3, and strain YJA4 in which ngt1Δ was complemented by reintegration of NGT1, were adjusted to 5 × 106 cells/ml, and then 10-fold serial dilutions of cells were spotted onto solid medium plates containing either glucose or GlcNAc. The plates were incubated for 2 d at 30°C and then photographed.
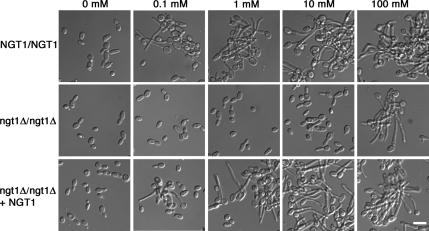
The ngt1Δ mutant was also defective in forming hyphae under the standard conditions of 2.5 mM GlcNAc. The defect was examined further by testing whether cells can form hyphae at higher concentrations of GlcNAc. The rationale for this is based on the previous observation that cells with a mutation in the galactose transporter can grow at high concentrations of galactose, but not at lower doses that are still permissive for growth of wild-type cells (Douglas and Condie, 1954). Alternatively, some transporter-type proteins have been found to transduce a separate signal pathway (e.g., glucose or ammonium; Holsbeeks et al., 2004; Wu et al., 2006). To distinguish between these possibilities, cells were examined after growth in different concentrations of GlcNAc. Interestingly, ngt1Δ mutants started forming hyphae in response to 100 mM GlcNAc, which was ≥1000-fold higher than the concentration required for wild-type cells to form hyphae (Figure 5). GlcNAc, 100 mM, also caused clumping of the ngt1Δ cells, which indicates that the adhesin proteins were induced in the hyphal cell walls (Sundstrom, 2002). Hyphal formation at 100 mM was still specific to GlcNAc and was not seen with dextrose or galactose (data not shown). GlcNAc, 100 mM, is not an unusually high level of sugar; yeast medium typically contains ∼111 mM dextrose (2% wt/vol). The mutant phenotypes were complemented by reintroduction of a wild-type copy of NGT1, and the ngt1Δ mutant could form hyphae efficiently in response to serum, which demonstrate that the effects of this mutation are specific to the GlcNAc response.
Figure 5.
High levels of GlcNAc can bypass the role of Ngt1 in stimulating hyphal growth. Wild-type strain DIC185, ngt1Δ homozygous deletion strain YJA3, and strain YJA4 in which ngt1Δ was complemented by reintegration of NGT1 were grown to log phase, resuspended in medium containing the indicated concentration of GlcNAc, and then incubated at 37°C for 2 h. Hyphal cells clumped, presumably due to induction of the cell surface adhesin proteins. Bar, 10 μm.
NGT1 Is Required for GlcNAc Transport in C. albicans
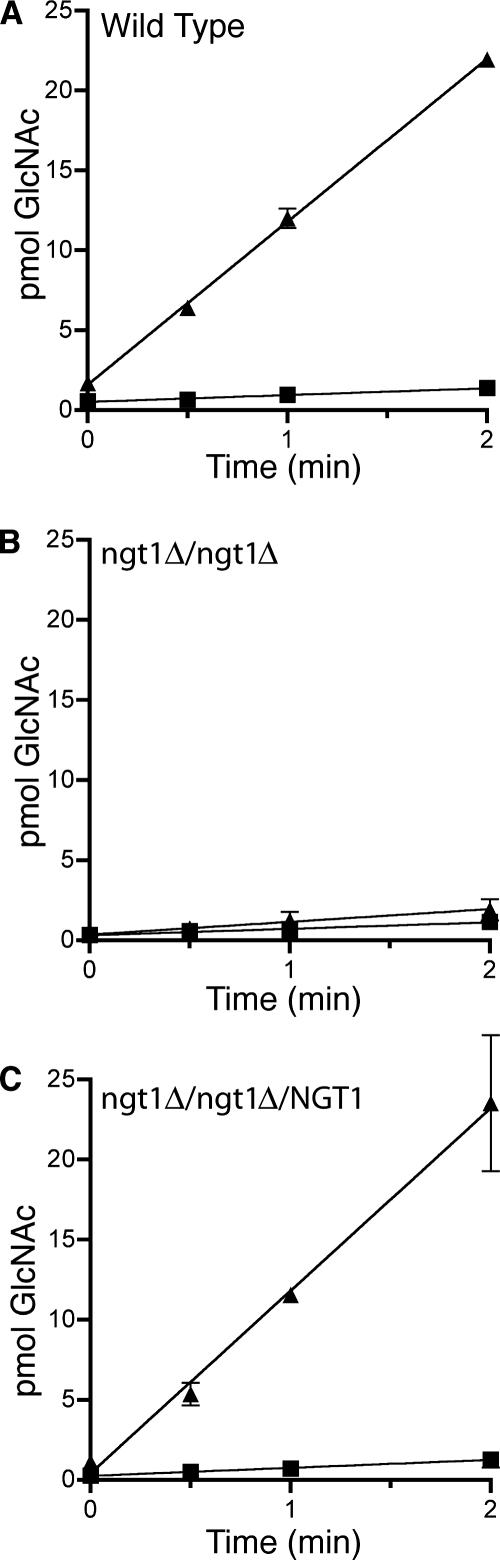
To gain further support for the idea that Ngt1 mediates GlcNAc transport, we examined the ability of the mutant cells to take up radioactive GlcNAc. Previous studies have demonstrated that glucose-grown cells do not display significant GlcNAc uptake and that cells must first be pregrown in GlcNAc for several hours to induce the transporter function (Singh and Datta, 1978, 1979). Therefore, we analyzed the ability of wild-type and of ngt1Δ cells to take up [3H]GlcNAc after being incubated for 4 h in medium containing either glucose or GlcNAc. Cells were then washed and resuspended in basal yeast medium without sugar (BYNB). Aliquots of cells were incubated for different times with [3H]GlcNAc and then rapidly harvested by filtration. Quantitation of the results by scintillation counting clearly demonstrated that GlcNAc-grown wild-type cells take up GlcNAc very efficiently, but that the ngt1Δ mutants only took up very low levels of GlcNAc (Figure 6). Complementation of the ngt1Δ mutation by reintroducing a wild-type copy of NGT1 restored the ability to take up GlcNAc with high efficiency.
Figure 6.
NGT1 is needed for efficient GlcNAc uptake by C. albicans. (A) Wild-type strain SC5314, (B) ngt1Δ homozygous deletion strain YJA3, and (C) strain YJA4 in which ngt1Δ complemented by reintegration of NGT1 were incubated for 4 h at 30°C in minimal medium containing GlcNAc (▲) to induce NGT1, and were also grown in glucose medium (■) as a control. Cells were harvested and washed, and then 107 cells were then incubated in 200 μM [3H]GlcNAc for the indicated time. Cells were collected on filters, and the [3H]GlcNAc taken up was determined by scintillation counting. Error bars, SE.
NGT1 Promotes GlcNAc Uptake When Expressed in S. cerevisiae
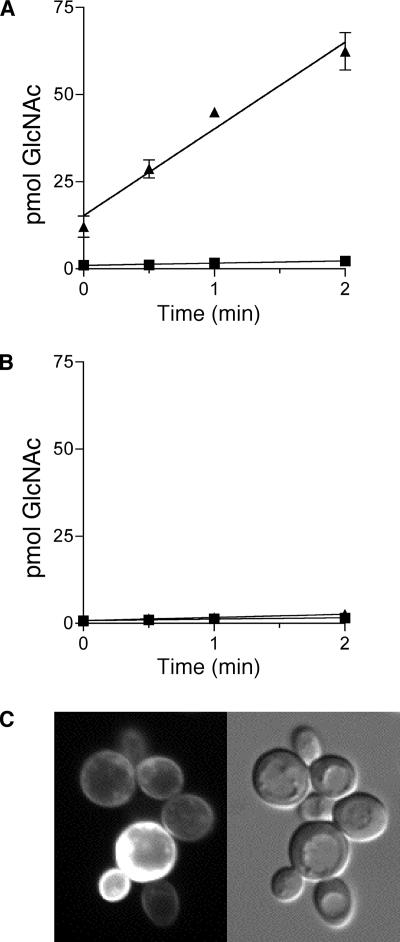
To more directly confirm that Ngt1 mediates GlcNAc uptake, we took advantage of the fact that S. cerevisiae lacks the ability to transport GlcNAc (Singh and Datta, 1979). The NGT1 gene was therefore placed under control of the galactose-regulated GAL1 promoter in plasmid p426GAL1 and transformed into S. cerevisiae strain W303. Cells were then cultured in glucose or galactose medium and then prepared for GlcNAc uptake assays. The results demonstrated that S. cerevisiae cells carrying the GAL1-NGT1 plasmid rapidly took up high levels of GlcNAc when grown in galactose medium (Figure 7A). Significant uptake was even observed in samples that were processed quickly for the time zero control. In contrast, cells that were grown in glucose to repress NGT1 did not display significant uptake of GlcNAc. In addition, cells carrying the empty vector control did not show significant GlcNAc uptake when pregrown in either medium (Figure 7B). As an additional control, an NGT1-GFP fusion gene was expressed in S. cerevisiae, and the GFP fluorescence was detected primarily at the plasma membrane (Figure 7C). Thus, expression of NGT1 in S. cerevisiae is sufficient to promote GlcNAc uptake, indicating that the Ngt1 protein directly promotes the uptake of GlcNAc.
Figure 7.
NGT1 expression in S. cerevisiae is sufficient to promote GlcNAc uptake. S. cerevisiae strain W303 carrying a plasmid designed to (A) express NGT1 under control of the galactose-inducible promoter GAL1 or (B) the empty vector plasmid (pRS426GAL1). Cells were grown in galactose medium (▲) overnight to induce NGT1, or in glucose medium (■) to repress NGT1. Cells (n = 107) were incubated with 200 μM [3H]GlcNAc for the indicated time and then the [3H]GlcNAc taken up was determined by scintillation counting. (C) Fluorescent microscope image and corresponding light microscope image of S. cerevisiae cells induced with galactose to express NGT1-GFP. Variable levels of Ngt1-GFP observed in different cells are expected due the cells containing different levels of the multicopy plasmid vector pRS426GAL1 used to express NGT1-GFP. Error bars, SE.
Ngt1 Is Specific for GlcNAc Transport
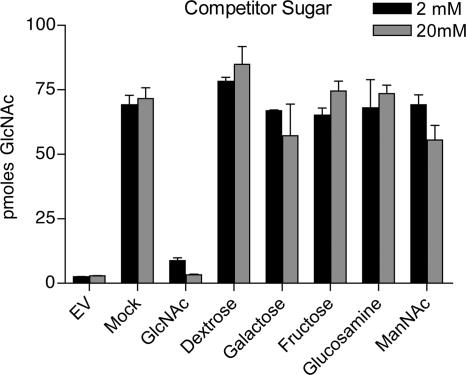
The specificity of transport by Ngt1 was examined by assaying the ability of various sugars to compete for the uptake of radioactive GlcNAc in S. cerevisiae cells that were induced to express NGT1. For this analysis, cells were incubated in a reaction mix that contained 200 μM [3H]GlcNAc and either a 10-fold (2 mM) or 100-fold (20 mM) excess of a nonradioactive competitor sugar. Addition of a 10-fold excess of nonradioactive GlcNAc was sufficient to strongly reduce the uptake of the [3H]GlcNAc, as expected (Figure 8). However, addition of even 100-fold excess of other competitor sugars did not have significant effects on [3H]GlcNAc uptake. The competitor sugars tested were closely related to GlcNAc and included glucose, galactose, fructose glucosamine, and N-acetylmannosamine. These results indicate that Ngt1 is a specific transporter for GlcNAc.
Figure 8.
Ngt1 is specific for GlcNAc uptake. The specificity of the transport activity promoted by Ngt1 was examined by testing the ability of other sugars to compete with the uptake of [3H]GlcNAc. Assays were carried out with S. cerevisiae strain W303 that expressed NGT1 under control of the galactose-inducible promoter GAL1 as described in the legend to Figure 7. Reactions contained 200 μM [3H]GlcNAc plus either 2 mM or 20 mM of the indicated nonradioactive sugar and were carried out for 2 min. The mock sample received only water. The results demonstrate that only nonradioactive GlcNAc, and not other related sugars, competed for the uptake of [3H]GlcNAc. Error bars, SE.
Ngt1 Homologues
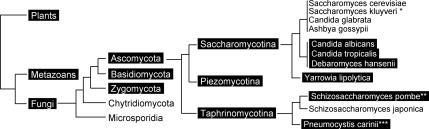
Because there have been no previous reports of a eukaryotic GlcNAc transporter, we performed BLAST searches and found Ngt1 homologues in diverse organisms including plants and metazoans. Interestingly, C. albicans Ngt1 showed much higher conservation with the homologues from the plants Arabidopsis thaliana and Oryza sativa (>41% identity) than it did with the metazoan homologues (≤ 25% identity). The metazoan homologues were mainly conserved between residues 100–235, which includes transmembrane domains 2–6 that play a major role in solute recognition in the lacY lactose transporter (Abramson et al., 2004). The closer similarity of fungal and plant Ngt1 homologues was very unexpected, because fungi are much more closely related to metazoans than they are to plants (Figure 9; James et al., 2006). This suggests that there may have been selective pressure to maintain the similarity of Ngt1 in plants and fungi. Alternatively, it is intriguing to speculate that there may have been horizontal gene transfer. The plant Ngt1 homologues clustered most closely with those from plant pathogenic fungi (e.g., Ustilago maydis) in a phylogenetic tree generated by Clustal W.
Figure 9.
Evolutionary conservation of NGT1. BLAST searches (Altschul et al., 1990) were used to identify NGT1 homologues in the genome sequences of other organisms. White letters in black boxes indicate the presence of NGT1 and black letters indicate the absence. *S. kluyveri is marked by an asterisk because it lacks NGT1 and DAC1, but contains NAG1. **S. pombe is marked because it contains an NGT1 homolog but lacks the other catabolic genes (e.g., DAC1 and NAG1). ***P. carinii contains an NGT1 homolog, but the genome sequence is not complete so it is not clear if homologues of NAG1 and DAC1 will be present. The evolutionary tree was constructed based on a recent phylogenetic analysis of fungi (James et al., 2006) and also on an analysis of Ascomycota (Dujon, 2006). The branches in the figure were sized to clearly display the relationship of the different lineages and are not sized to represent predicted evolutionary distance.
Analysis of the Fungal Kingdom indicates that the ability to utilize GlcNAc was lost independently in several lineages. Ngt1 homologues were detected in three of the five major divisions of the fungi (Figure 9). In the Ascomycota division, NGT1 appears to have been lost in two out of three of the classes. In the Saccharomycotina class that includes the budding yeasts (e.g., C. albicans and S. cerevisiae), the 6 organisms most closely related C. albicans contain an NGT1 homolog, and all 10 yeast that were more related to S. cerevisiae lack an NGT1 homolog. A similar pattern was detected for the GlcNAc catabolic enzymes DAC1 and NAG1. The loss of NGT1 coincides with an evolutionary split that occurred before the appearance of the triplicated mating cassettes and short centromeres but after the appearance of retroposons Ty3 and Ty5 (Dujon, 2006). In a second class of the Ascomycota (Taphrinomycotina), an NGT1 homolog was detected in the fission yeast Schizosaccharomyces pombe but not in the related Schizosaccharomyces japonica. Neither contains homologues of the GlcNAc catabolic genes, and S. pombe was not able to grow in GlcNAc medium. In contrast, an NGT1 homolog was readily detected in all 22 members of the Pezizomycotina class (e.g., A. fumigatis and Neurospora crassa), which includes many plant and animal pathogens. Although many pathogenic fungi contain NGT1, the correlation is not 100% because the human pathogen Candida glabrata and the plant pathogen Ashbya gossypii lack homologues for NGT1 and the GlcNAc catabolic genes.
DISCUSSION
GlcNAc plays important roles in cell signaling in a wide range of cells in addition to serving as a source of nutrition. In metazoans, GlcNAc is a sensor of nutrient status that is involved in insulin signaling, cell cycle control, and other essential processes. Modulation of GlcNAc levels promotes O-linked attachment of GlcNAc to serine or threonine residues on proteins as a regulatory modification that is analogous to phosphorylation (Slawson and Hart, 2003; Zachara and Hart, 2006). In C. albicans, GlcNAc induces the formation of hyphal cells, which is an underlying virulence trait as budding and hyphal cells are thought to carry out unique roles during infection. In addition to different morphologies, budding and hyphal cells differ in the expression of virulence factors (Nantel et al., 2002; Lorenz et al., 2004), such as adhesin proteins, secreted hydrolytic enzymes, and antioxidant enzymes (Hube, 2004; Whiteway and Oberholzer, 2004; Kumamoto and Vinces, 2005). Therefore, in this study we used mass spectrometry to identify plasma membrane proteins in budding cells and GlcNAc-stimulated hyphae, which have the potential to mediate interaction with the host environment and contribute to the infectious process. One protein was selected for detailed analysis and was subsequently named Ngt1 for its role as an N-acetylglucosamine transporter. Ngt1 was also found to mediate GlcNAc-induced hyphal formation in C. albicans. These results are also significant in that Ngt1 represents the first eukaryotic GlcNAc transporter to be identified. This will make it possible to examine the role of homologues present in a wide range of organisms from yeast to plants and animals for their function in GlcNAc transport and signaling.
Ngt1-GFP Induction by GlcNAc
Ngt1-GFP was specifically induced by GlcNAc, and not by the switch to hyphal growth, because Ngt1-GFP was not induced in serum-stimulated hyphae (Figure 1). In addition, Ngt1-GFP was highly induced by GlcNAc at 23 or 30°C, even though C. albicans cells do not switch to hyphal growth under these lower temperature growth conditions (F.J.A. and J.B.K, unpublished data). Only GlcNAc induced Ngt1-GFP, and not other related sugars such as glucosamine and N-acetylmannosamine. Glucose repressed the ability of GlcNAc to induce Ngt1-GFP. Altogether these studies indicate that NGT1 expression is specifically regulated by GlcNAc, similar to the enzymes needed for GlcNAc catabolism in C. albicans (Kumar et al., 2000; Singh et al., 2001; Yamada-Okabe et al., 2001). However, NGT1 is on Chromosome 6 and is thus not linked to the other catabolic genes (NAG1, DACX1, and HXK1), which are present in a cluster on Chromosome 3 (Kumar et al., 2000; Yamada-Okabe et al., 2001). Attempts to identify a common DNA sequence motif in the upstream regions of these genes using MEME (Bailey and Elkan, 1994) were inconclusive.
Ngt1 Functions as a GlcNAc Transporter
The predicted amino acid sequence of Ngt1 indicates that it is a transporter of the major facilitator superfamily (MFS; Pao et al., 1998; Abramson et al., 2004). One hallmark of MFS transporters is that they contain 12 transmembrane domains. Hydrophathy analysis of Ngt1 readily identified 11 transmembrane domains, and a twelfth transmembrane domain was identified as an amphipathic helix in which the polar side presumably faces the helix bundle to shield it from the nonpolar membrane environment. Ngt1 also contains a close match to the consensus sequence (GTXXNXXGXRXXL) that is commonly found in intracellular loop 1 of MFS transporters (Pao et al., 1998).
A homozygous ngt1Δ deletion mutant was defective in growing on GlcNAc medium (Figure 4) and in GlcNAc transport (Figure 6). Although this implicated Ngt1 as a GlcNAc transporter, it was not clear from these studies whether Ngt1 is directly involved in transport. In S. cerevisiae, the Snf3 and Rgt2 glucose sensors display sequence similarity to transporters, yet they are inactive as transporters and function instead as receptors that activate a signal pathway leading to induction of active glucose transporters (Moriya and Johnston, 2004; Kim and Johnston, 2006). Therefore, we took advantage of the fact that S. cerevisiae lacks the ability to transport GlcNAc (Singh and Datta, 1978, 1979) to use it as a heterologous expression system. S. cerevisiae cells expressing NGT1 displayed rapid GlcNAc uptake that was similar to the levels seen in C. albicans. The specificity of Ngt1 transport function was confirmed in a competition assay in which closely related sugars such as glucosamine and N-acetylmannosamine did not outcompete the uptake of [3H]GlcNAc. These results, combined with the similarity of Ngt1 to MFS transporters, demonstrate that Ngt1 is a GlcNAc-specific transporter.
Role of Ngt1 in Hyphal Growth Stimulated by GlcNAc
The initial observation that ngt1Δ cells were defective in forming hyphae under standard GlcNAc induction conditions (∼2 mM) demonstrated that Ngt1 is involved in promoting this morphological change. However, it was not clear from this whether Ngt1 acts only as a transporter and that cells respond to increased intracellular levels of GlcNAc or if Ngt1 activates a separate signal pathway to induce hyphal formation. Many transporter proteins have been shown recently to also act as signal transducers (Holsbeeks et al., 2004; Moriya and Johnston, 2004; Wu et al., 2006). For example, the C. albicans Mep2 ammonium permease activates the Cph1 MAP kinase and the cAMP kinase pathways that induce hyphal growth (Biswas and Morschhauser, 2005). However, it is not necessary for Ngt1 to transduce a separate signal. An ngt1Δ mutant could be induced to form hyphae by exposing cells to 1000-fold higher concentrations of GlcNAc than were required to stimulate hyphal formation in the wild type (Figure 5), indicating that cells are responding to increased intracellular levels of GlcNAc. GlcNAc uptake under these conditions presumably occurs via an low-affinity process that has been observed in C. albicans (Singh and Datta, 1978). Thus, it appears that the primary role of Ngt1 is to facilitate high-affinity transport of GlcNAc.
The ability of C. albicans cells to form hyphae in response to elevated GlcNAc, but not to exogenously added glucosamine or fructose, suggests that the intracellular signal is an upstream component such as GlcNAc itself. GlcNAc taken up by C. albicans is converted to GlcNAc-6-PO4 and then deacetylated to glucosamine-6-PO4 and deamidated to fructose-6-PO4. One model for how GlcNAc signaling may occur is based on galactose regulation in S. cerevisiae. Cells respond to elevated intracellular galactose, which acts together with ATP and Gal3 to relieve the inhibition of the Gal4 transcription factor by its negative regulator Gal80 (Sil et al., 1999). Another possibility is based on GlcNAc signaling in metazoan cells, in which elevated GlcNAc levels stimulate signal transduction via O-GlcNAc modification of proteins (Slawson and Hart, 2003; Zachara and Hart, 2006). However, this modification has not been reported in fungi, so it remains to be determined how the GlcNAc signal is propagated in C. albicans.
Role of Ngt1 during Macrophage Phagocytosis
The biological role for GlcNAc signaling in C. albicans has not been well defined; the ability of GlcNAc to induce hyphae was discovered in a search for a chemically defined inducers in vitro (Simonetti et al., 1974). Subsequent studies revealed that the GlcNAc catabolism genes (HXK1, DAC1, and NAG1) contribute to the virulence of C. albicans in a mouse tail-vein injection model of pathogenesis (Singh et al., 2001; Yamada-Okabe et al., 2001). The GlcNAc catabolic genes are repressed in serum, which contains glucose, but C. albicans also grows in low glucose environments such as the macrophage phagolysosome (Lorenz et al., 2004). Interestingly, Ngt1-GFP was induced after macrophage phagocytosis (Figure 3). NGT1 and the genes needed for GlcNAc catabolism were also found to be induced after macrophage phagocytosis by microarray analysis of gene expression (Lorenz et al., 2004). The correlation between GlcNAc induction and phagocytosis is also strengthened by the observation that 6 of the 10 GlcNAc-induced proteins identified with highest confidence by mass spectrometry correspond to genes that were induced by macrophage phagocytosis. However, the ngt1Δ mutant was still capable of forming hyphae after phagocytosis (F.J.A. and J.B.K., unpublished data), indicating either that GlcNAc levels are very high or that other hyphal inducers are present in the phagolysosome. Perhaps multiple signals stimulate hyphal growth after phagocytosis to ensure an effective response to the hostile phagolysosomal environment. The importance of evading host defenses may also explain why such a variety of different stimuli can induce hyphal growth in vitro. Future studies on GlcNAc signaling in C. albicans will help define how various stimuli are integrated to regulate hyphal growth and will also serve as a model for defining the role of GlcNAc regulation in other organisms.
Ngt1 Homologues
The identification of Ngt1 as a GlcNAc transporter made it possible to search for homologues in other organisms (Figure 9). Ngt1 homologues were present in at least 3 of the 5 divisions of the fungal kingdom. It appears that NGT1 and the GlcNAc catabolic enzymes (HXK1, DAC1, and NAG1) were independently lost in several branches of the fungal lineage, including the branch between S. cerevisiae and C. albicans in the Ascomycota. The galactose utilization genes have also been lost several independent times in the Ascomycota, although the losses of the galactose and GlcNAc genes do not coincide (Dujon, 2006). Many human and plant fungal pathogens contain NGT1 and the GlcNAc catabolic genes, but they are not required for all pathogens since C. glabrata and A. gossypii lack homologues for these genes.
NGT1 homologues were also detected in two very divergent branches of multicellular organisms, plants and metazoans. Surprisingly, C. albicans Ngt1 showed much higher conservation with the plants homologues, given that fungi are much more closely related to metazoans (James et al., 2006). Current models assume that GlcNAc levels in metazoans are regulated indirectly by other nutritional effects. Thus, the discovery of Ngt1 homologues in metazoans broadens the possible functions of GlcNAc signaling to include intercellular communication. In this regard it is interesting that as part of a full-genome RNAi study in Caenorhabditis elegans interference of an Ngt1 homolog caused defects in early embryogenesis (Sonnichsen et al., 2005). Other metazoan homologues have not been studied, but they are apparently expressed since they are represented in cDNA libraries. It will therefore be important to determine the role of Ngt1 homologues, given the essential role that GlcNAc signaling plays in biomedically important processes such as insulin signaling, cell cycle control, and proteosome regulation (Slawson and Hart, 2003; Zachara and Hart, 2004, 2006).
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Filler for providing plasmids, strains and advice; James Bliska, Hana Fukuto, and Kate Klein for assistance with growing J774 cells; and Lois Douglas for advice and comments on the manuscript. We also thank the reviewers for their helpful suggestions. This work was supported by Grant RO1 AI47837 from the National Institutes of Health that was awarded to J.B.K.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0931) on December 27, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abramson J., Iwata S., Kaback H. R. Lactose permease as a paradigm for membrane transport proteins. Mol. Membr. Biol. 2004;21:227–236. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey T., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Barnhart M. M., Lynem J., Chapman M. R. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J. Bacteriol. 2006;188:5212–5219. doi: 10.1128/JB.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J., Sudbery P. E. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 2002;3:918–930. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- Biswas K., Morschhauser J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 2005;56:649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- Braun B. R., Johnson A. D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Chou T. Y., Hart G. W. O-linked N-acetylglucosamine and cancer: messages from the glycosylation of c-Myc. Adv. Exp. Med. Biol. 2001;491:413–418. doi: 10.1007/978-1-4615-1267-7_26. [DOI] [PubMed] [Google Scholar]
- Davis D. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 2003;44:1–7. doi: 10.1007/s00294-003-0415-2. [DOI] [PubMed] [Google Scholar]
- Davis D., Edwards J. E., Jr., Mitchell A. P., Ibrahim A. S. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosil M., Giot L., Davis C., Konopka J. B. Dominant-negative mutations in the G protein-coupled α-factor receptor map to the extracellular ends of the transmembrane segments. Mol. Cell. Biol. 1998;18:5981–5991. doi: 10.1128/mcb.18.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Condie F. The genetic control of galactose utilization in Saccharomyces. J. Bacteriol. 1954;68:662–670. doi: 10.1128/jb.68.6.662-670.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Edmond M. B., Wallace S. E., McClish D. K., Pfaller M. A., Jones R. N., Wenzel R. P. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Enjalbert B., Whiteway M. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell. 2005;4:1203–1210. doi: 10.1128/EC.4.7.1203-1210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad M., Berman J., Gale C. A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- Hirakawa H., Inazumi Y., Senda Y., Kobayashi A., Hirata T., Nishino K., Yamaguchi A. N-acetyl-d-glucosamine induces the expression of multidrug exporter genes, mdtEF, via catabolite activation in Escherichia coli. J. Bacteriol. 2006;188:5851–5858. doi: 10.1128/JB.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde S., Thevelein J. M. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 2004;29:556–564. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr. Opin. Microbiol. 2004;7:336–341. doi: 10.1016/j.mib.2004.06.003. [DOI] [PubMed] [Google Scholar]
- James T. Y., et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:26144–26149. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- Klengel T., et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto C. A., Vinces M. D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Kumar M. J., Jamaluddin M. S., Natarajan K., Kaur D., Datta A. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. USA. 2000;97:14218–14223. doi: 10.1073/pnas.250452997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Bender J. A., Fink G. R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidan M. M., De Rop L., Serneels J., Exler S., Rupp S., Tournu H., Thevelein J. M., Van Dijck P. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-PKA pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell. 2005;16:1971–1986. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavor A. L., Thewes S., Hube B. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr. Drug Targets. 2005;6:863–874. doi: 10.2174/138945005774912735. [DOI] [PubMed] [Google Scholar]
- Moriya H., Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A., et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C. Philadelphia: Bailliere Tindall; 1988. Candida and Candidosis. [Google Scholar]
- Pao S. S., Paulsen I. T., Saier M. H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha C. R., Schroppel K., Harcus D., Marcil A., Dignard D., Taylor B. N., Thomas D. Y., Whiteway M., Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandel K. A., Jenness D. D. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol. Cell. Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sil A. K., Alam S., Xin P., Ma L., Morgan M., Lebo C. M., Woods M. P., Hopper J. E. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol. Cell. Biol. 1999;19:7828–7840. doi: 10.1128/mcb.19.11.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti N., Strippoli V., Cassone A. Yeast-mycelial conversion induced by N-acetyl-d-glucosamine in Candida albicans. Nature. 1974;250:344–346. doi: 10.1038/250344a0. [DOI] [PubMed] [Google Scholar]
- Singh B., Datta A. Regulation of N-acetylglucosamine uptake in yeast. Biochim. Biophys. Acta. 1979;557:248–258. doi: 10.1016/0005-2736(79)90107-x. [DOI] [PubMed] [Google Scholar]
- Singh B. R., Datta A. Glucose repression of the inducible catabolic pathway for N-acetylglucosamine in yeast. Biochem. Biophys. Res. Commun. 1978;84:58–64. doi: 10.1016/0006-291x(78)90262-0. [DOI] [PubMed] [Google Scholar]
- Singh P., Ghosh S., Datta A. Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect. Immun. 2001;69:7898–7903. doi: 10.1128/IAI.69.12.7898-7903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Hart G. Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr. Opin. Struct. Biol. 2003;13:631–636. doi: 10.1016/j.sbi.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Sohanpal B. K., El-Labany S., Lahooti M., Plumbridge J. A., Blomfield I. C. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA. 2004;101:16322–16327. doi: 10.1073/pnas.0405821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B., et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Spellberg B. J., Filler S. G., Edwards J. E., Jr. Current treatment strategies for disseminated Candidiasis. Clin. Infect. Dis. 2006;42:244–251. doi: 10.1086/499057. [DOI] [PubMed] [Google Scholar]
- Stoldt V. R., Sonneborn A., Leuker C. E., Ernst J. F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P., Gow N., Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sundstrom P. Adhesion in Candida spp. Cell. Microbiol. 2002;4:461–469. doi: 10.1046/j.1462-5822.2002.00206.x. [DOI] [PubMed] [Google Scholar]
- Tebarth B., Doedt T., Krishnamurthy S., Weide M., Monterola F., Dominguez A., Ernst J. F. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 2003;329:949–962. doi: 10.1016/s0022-2836(03)00505-9. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Oberholzer U. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 2004;7:350–357. doi: 10.1016/j.mib.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Mitchell A. P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Ottow K., Poulsen P., Gaber R. F., Albers E., Kielland-Brandt M. C. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 2006;173:327–331. doi: 10.1083/jcb.200602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada-Okabe T., Sakamori Y., Mio T., Yamada-Okabe H. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur. J. Biochem. 2001;268:2498–2505. doi: 10.1046/j.1432-1327.2001.02135.x. [DOI] [PubMed] [Google Scholar]
- Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell. Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- Zachara N., Hart G. Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zachara N. E., Hart G. W. O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell. Biol. 2004;14:218–221. doi: 10.1016/j.tcb.2004.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.