Abstract
Targeted disruption of the mouse Hus1 cell cycle checkpoint gene results in embryonic lethality and proliferative arrest in cultured cells. To investigate the essential functions of Hus1, we developed a system for the regulated inactivation of mouse Hus1 in primary fibroblasts. Inactivation of a loxP site-flanked conditional Hus1 allele by using a cre-expressing adenovirus resulted in reduced cell doubling, cell cycle alterations, and increased apoptosis. These phenotypes were associated with a significantly increased frequency of gross chromosomal abnormalities and an S-phase–specific accumulation of phosphorylated histone H2AX, an indicator of double-stranded DNA breaks. To determine whether these chromosomal abnormalities occurred randomly or at specific genomic regions, we assessed the stability of common fragile sites, chromosomal loci that are prone to breakage in cells undergoing replication stress. Hus1 was found to be essential for fragile site stability, because spontaneous chromosomal abnormalities occurred preferentially at common fragile sites upon conditional Hus1 inactivation. Although p53 levels increased after Hus1 loss, deletion of p53 failed to rescue the cell-doubling defect or increased apoptosis in conditional Hus1 knockout cells. In summary, we propose that Hus1 loss leads to chromosomal instability during DNA replication, triggering increased apoptosis and impaired proliferation through p53-independent mechanisms.
INTRODUCTION
Cell cycle checkpoints monitor the fidelity of chromosome replication and segregation. In response to genome damage, checkpoint signaling induces cell cycle arrest and promotes DNA repair, or alternatively, it triggers apoptosis to eliminate damaged cells. Mammalian DNA damage responses are coordinated by two primary checkpoint pathways that center on the phosphatidyl inositol kinase-like protein kinases ataxia telangiectasia mutated (Atm) and Atm- and Rad3-related (Atr) (Bakkenist and Kastan, 2004). An Atm-dependent pathway responds to double-stranded DNA breaks (DSBs) such as those caused by ionizing radiation, whereas an Atr-dependent pathway is activated by a variety of DNA lesions, including bulky DNA lesions and replication stress as well as DSBs.
Optimal Atr signaling requires its binding partner, Atrip, as well as additional accessory factors TopBP1, Brca1, Claspin, and the Rad9–Rad1–Hus1 (9-1-1) complex (Shechter et al., 2004b). The 9-1-1 complex shares predicted structural similarity with the sliding clamp proliferating cell nuclear antigen and is loaded onto chromatin at damage sites by a clamp loader containing the checkpoint protein Rad17 (Parrilla-Castellar and Karnitz, 2003). 9-1-1 promotes the phosphorylation of Atr substrates such as Chk1, Rad17, and Rad9 itself (Weiss et al., 2002; Zou et al., 2002; Roos-Mattjus et al., 2003; Bao et al., 2004) and is required for an intra-S cell cycle checkpoint that represses DNA synthesis after DNA damage (Roos-Mattjus et al., 2003; Weiss et al., 2003; Bao et al., 2004; Wang et al., 2004b). Additional evidence indicates that the 9-1-1 complex also has a direct role in DNA repair. The 9-1-1 complex physically associates with multiple translesion DNA polymerases (Kai and Wang, 2003; Sabbioneda et al., 2005) as well as base excision repair factors, including the MYH DNA glycosylase, DNA polymerase β, flap endonuclease I, and DNA ligase I (Toueille et al., 2004; Wang et al., 2004a. 2006a; Chang and Lu, 2005; Friedrich-Heineken et al., 2005; Smirnova et al., 2005; Shi et al., 2006). The 9-1-1 complex additionally is required for homologous recombinational repair (Pandita et al., 2006; Wang et al., 2006b). Consistent with its important roles in cell cycle control and DNA repair, impaired 9-1-1 function is associated with cellular hypersensitivity to replication inhibitors and DNA damaging agents (Weiss et al., 2000, 2003; Kinzel et al., 2002; Roos-Mattjus et al., 2003; Hopkins et al., 2004; Wang et al., 2004b, 2006b).
Targeted disruption of components of the Atr-dependent checkpoint pathway in mice causes embryonic lethality. Deletion of Atr or Chk1 results in peri-implantation lethality (Brown and Baltimore, 2000; de Klein et al., 2000; Liu et al., 2000; Takai et al., 2000), whereas inactivation of Hus1, Rad9, or Rad17 causes midgestational embryonic lethality (Weiss et al., 2000; Budzowska et al., 2004; Hopkins et al., 2004). The essential nature of these genes highlights the critical, yet poorly understood, function of this pathway during an unperturbed cell cycle. In the course of a normal cell cycle, the 9-1-1 complex and other checkpoint components can be detected in association with chromatin (Guo et al., 2000; Hekmat-Nejad et al., 2000; Roos-Mattjus et al., 2002; You et al., 2002; Zou et al., 2002; Jiang et al., 2003; Lee et al., 2003; Dart et al., 2004). Even in the absence of extrinsic stress, checkpoint signaling inhibits the cell cycle phosphatases Cdc25A and Cdc25B, regulates origin firing, and suppresses premature entry into mitosis (Miao et al., 2003; Shechter et al., 2004a; Sorensen et al., 2004; Niida et al., 2005; Syljuasen et al., 2005; Schmitt et al., 2006).
The Atr-dependent checkpoint pathway is also thought to play a critical role in stabilizing stalled replication forks and promoting fork restart (Lopes et al., 2001; Tercero and Diffley, 2001; Sogo et al., 2002; Trenz et al., 2006). Possibly due to failure of these important processes, certain yeast checkpoint mutants show defects in the elongation step of DNA replication and accumulate chromosomal breaks at particular, nonrandom genomic regions (Cha and Kleckner, 2002; Raveendranathan et al., 2006). These sites may be analogous to vertebrate common fragile sites (CFSs), chromosomal regions where gaps and breaks frequently arise in metaphase chromosomes prepared from cells under conditions of replication stress. Recent studies indicate that several components of the DNA damage checkpoint machinery, including Atr (Casper et al., 2002), Chk1 (Durkin et al., 2006), Brca1 (Arlt et al., 2004), and TopBP1 (Kim et al., 2005), among others, are essential for maintaining CFS stability. No primary sequence conservation has been identified at CFSs, but generally these sites are relatively AT rich, highly flexible, and late replicating (Glover et al., 2005). These properties suggest that CFSs might be prone to form secondary structures that inhibit the progression of replication forks, creating a requirement for cell cycle delay and replication fork stabilization or repair by the checkpoint machinery (Cimprich, 2003). Understanding the molecular basis for fragile site stability has important implications, because these regions are frequently deleted or rearranged in cancer cells (Arlt et al., 2006).
Previous attempts at molecular analysis of the essential functions of Hus1 by using a conventional gene targeting approach were complicated by severe phenotypes, including midgestational lethality in embryos and proliferative arrest in mouse embryonic fibroblasts (MEFs) (Weiss et al., 2000). Successful culturing of Hus1-deficient cells from a constitutive knockout mouse model additionally required deletion of the checkpoint genes p21 or p53 (Weiss et al., 2000; our unpublished data). Furthermore, because embryos lacking both Hus1 and either p21 or p53 remained undersized and developmentally delayed, sufficient numbers of cells for experimental analysis could be obtained only with immortalized cultures. In this report, we describe a system for the regulated deletion of Hus1 in primary cultured cells, for use in dissecting the immediate consequences of Hus1 inactivation. By infecting primary MEFs containing a loxP site-flanked conditional Hus1 allele with a cre-expressing recombinant adenovirus (Ad-cre), we generated and analyzed large populations of Hus1-deficient and control cells in vitro. Our results indicate that Hus1 inactivation results in impaired cell proliferation and apoptosis associated with CFS expression and S-phase–specific DSB accumulation.
MATERIALS AND METHODS
Mouse Strains and Cell Culture
Previously described Hus1flox and Hus1Δ1 mice were maintained on an 129S6 inbred genetic background (Weiss et al., 2000; Levitt et al., 2005). p53+/− mice harboring the Trp53tm1Tyj allele were maintained on a C57BL/6J background (Jacks et al., 1994). Mice were housed in accordance with institutional animal care and use guidelines. MEFs were prepared from 13.5 dpc embryos from timed matings between Hus1flox/flox and Hus1+/Δ1 mice or from Hus1flox/flox p53+/− and Hus1+/Δ1 p53+/− mice. Briefly, embryos were dissected from the deciduum, mechanically disrupted, and cultured in DMEM supplemented with 10% fetal bovine serum, 1.0 mM l-glutamine, 0.1 mM minimal essential medium nonessential amino acids, 100 μg/ml streptomycin sulfate, and 100 U/ml penicillin. The initial plating was defined as passage zero (p0). MEFs at p1 or p2 were used for all experiments.
Viral Infections
Ad-cre, an adenovirus that expresses Cre from the cytomegalovirus promoter (University of Iowa Gene Transfer Vector Core, Iowa City, IA) (Stec et al., 1999), was prepared in 293 cells. Briefly, cells were harvested at 48–72 h postinfection, and viral lysate was subjected to CsCl gradient ultracentrifugation at 63,000 rpm at 14°C for 7 h. Virus was further purified with a PD-10 desalting column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and virus titer was estimated by spectrophotometry according to the formula: 1 OD280 = 1012 virus particles/ml. For infections, 1 × 106 MEFs were plated into a 10-cm culture dish and grown for 1 d. Cells were infected with 1.95 × 1011 Ad-cre particles in 2.5 ml of culture medium at 37°C for 6 h, after which time the virus was removed and fresh medium was added. Unless otherwise specified, cells were passaged at 1 d postinfection (dpi) and then maintained on a 3T3 culture schedule in which 1 × 106 cells were passaged onto a 10-cm culture dish every 3 d (Todaro and Green, 1963).
Southern and Northern Blotting
Genomic DNA for Southern blotting was isolated from MEFs by proteinase K digestion and precipitation with ethanol. DNA was digested with NheI, run through a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled 190-base pair EagI fragment from plasmid pCR2.1-5′UTR-Δ2,3 (Levitt et al., 2005). For Northern blotting, total RNA was prepared from MEFs by using RNA STAT-60 reagent (Tel-Test, Friendswood, TX), and poly(A)+ mRNA was isolated with biotinylated oligo(dT) (Promega. Madison, WI). Purified mRNA was resolved on a 1% agarose/formaldehyde gel, transferred to a nylon membrane, and hybridized with a 32P-labeled cDNA probe containing the entire mouse Hus1 open reading frame as described previously (Weiss et al., 1999). After stripping, the membrane was hybridized to a 32P-labeled mouse Gapdh cDNA probe.
Cell Proliferation Assays and Cell Cycle Analysis
For cell proliferation assays, triplicate cultures were maintained on a 3T3 culture schedule. Population doublings (PDLs) were calculated using the formula ΔPDL = log(nf/n0)/log2, where n0 is the initial number of cells and nf is the final number of cells (Blasco et al., 1997). For cell cycle analysis, 1 × 106 cells were plated per 10-cm culture dish 24 h before analysis. The next day, the cells were incubated with 10 μM bromodeoxyuridine (BrdU) for 45 min, harvested by trypsinization, washed once in phosphate-buffered saline (PBS), and fixed in 70% ethanol at −20°C. The cells were then incubated in 2 N HCl, 0.5% Triton X-100, washed twice with 0.1 M Na2B4O7·10H2O, pH 8.5, incubated with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU (BD Biosciences, Franklin Lakes, NJ) for 30 min at room temperature (RT), washed, treated with RNAse A, and stained with propidium iodide (PI). Flow cytometry was performed on a FACScan flow cytometer (BD Biosciences).
Apoptosis Assays
Cells grown in 10-cm culture dishes were collected by trypsinization along with floating cells in the culture medium, washed twice with PBS at 4°C, and resuspended in 1× binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) at a concentration of 1 × 106 cells/ml. Cells (1 × 105) were then incubated with Annexin V-FITC (BD Biosciences) and PI for 15 min at RT. Flow cytometry was performed on a FACScan flow cytometer (BD Biosciences) within 1 h.
Indirect Immunofluorescence Assays (IFAs)
Cells grown on coverslips were fixed in 2% paraformaldehyde in TBS for 35 min at 4°C (for γ-H2AX IFA) or in methanol at −20°C for 30 min followed by ice-cold acetone for two seconds (for p53 IFA). Cells were then incubated in 3% bovine serum albumin (BSA), 0.01% skim milk, 0.2% Triton X-100 in Tris-buffered saline (TBS) for 20 min at RT. For γ-H2AX IFA, cells were incubated with primary anti-γ-H2AX antibody (JBW301; Upstate Biotechnology, Lake Placid, NY) at 1:500 for 45 min, followed by secondary goat anti-mouse Ig (H+L)-FITC (Southern Biotechnology Associates, Birmingham, AL) at 1:60 for 35 min. For p53 IFA, cells were incubated with primary anti-p53 antibody (FL393; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:60 dilution at RT for 1 h, followed by secondary goat anti-rabbit Ig (H+L)-FITC (Southern Biotechnology Associates) at 1:60 at RT for 35 min. Cells were counterstained with 33 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) for 1 min.
Fluorescence-activated Cell Sorting (FACS) Analysis of γ-H2AX
Cells (1 × 106) were fixed in ice-cold 70% ethanol, incubated with 1% BSA, 0.25% Triton X-100 in TBS for 15 min on ice, and stained with primary anti-γ-H2AX antibody (JBW301, Upstate Biotechnology) at 1:500 overnight at 4°C. The next day, the cells were stained with secondary goat anti-mouse Ig (H+L)-FITC (Southern Biotechnology Associates) at 1:400 for 30 min at RT and counterstained with 5 μg/ml PI containing RNAse A for 30 min at RT. Flow cytometry was performed on an LSR II flow cytometer (BD Biosciences).
Metaphase Chromosome Preparation and Fluorescence In Situ Hybridization (FISH)
Metaphase spreads were prepared as described previously (Weiss et al., 2000). Briefly, cells were incubated in culture medium containing 0.15 μg/ml colcemid for 1 h and then trypsinized, incubated in hypotonic buffer (0.05 M KCl, 0.0034 M trisodium citrate) for 12 min at 37°C, and fixed for at least 20 min on ice in 75% methanol, 25% acetic acid. Cells were then spotted onto microscope slides and stained with 2% Giemsa in Gurr buffer, pH 7.0. Metaphase chromosomes were scored under a 100× oil objective lens according to standard guidelines (Savage, 1976; Mitelman, 1995). For FISH, unstained metaphase chromosomes on slides were denatured in 70% formamide/2× standard saline citrate (SSC) at 70°C for 2 min. Bacterial artificial chromosomes (BACs) containing mouse genomic sequence mapped to fragile site regions were used as probes in FISH analysis. Probe BAC-CITB-57C24 (Open Biosystems, Huntsville, AL) was used to detect mouse Fra8E1 (Krummel et al., 2002), and BAC-CITB-316M9 and BAC-CITB-513J1 (Open Biosystems) were used to detect mouse Fra6C1 (Rozier et al., 2004). Probes were labeled with SpectrumGreen-dUTP (Vysis, Downers Grove, IL) by nick translation, ethanol precipitated in the presence of mouse Cot-1 DNA (Invitrogen, Carlsbad, CA), resuspended in deionized formamide, and incubated in hybridization buffer (20% dextran sulfate/2× SSC, pH 7.0) at 37°C for 10 min. Probes were then denatured at 75°C for 10 min, incubated at 42°C for 30 min, and hybridized with metaphase chromosomes at 37°C for 24 h. After hybridization, slides were washed three times each in 50% formamide/2× SSC at 42°C, 2× SSC at 37°C, and 0.1× SSC at 60°C, followed by a single wash in 4× SSC/0.1% Tween 20 at RT. Slides were then counterstained with 33 ng/ml DAPI for 1 min. FISH signal was examined using a DMRE fluorescence microscope (Leica Microsystems, Deerfield, IL).
RESULTS
Rapid and Complete Deletion of Hus1 in Primary Cultured Cells by Cre-mediated Recombination
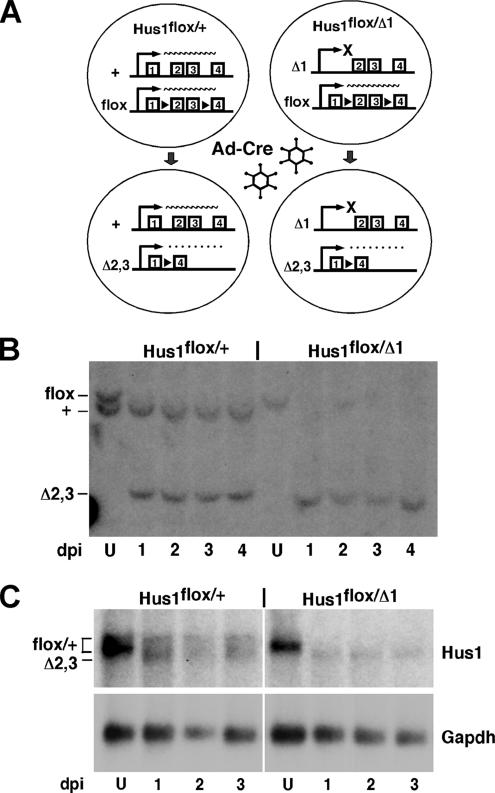
To examine the effects of acute Hus1 loss in primary cultured cells, we established a genetic system for the regulated inactivation of Hus1 in MEFs. This system is based on a conditional allele, Hus1flox, in which exons 2 and 3 are flanked by loxP sites, recognition sequences for the cre recombinase (Figure 1A). Previous studies indicated that cre-mediated recombination at Hus1flox deletes exons 2 and 3, producing a null allele, Hus1Δ2,3, which has the capacity to encode only the first 19 of 281 Hus1 amino acids (Levitt et al., 2005). For conditional Hus1 inactivation, we generated MEFs containing Hus1flox and either Hus1Δ1, a constitutive null allele in which exon 1 and the start codon have been deleted (Weiss et al., 2000), or wild-type Hus1 as a control, and then performed infections with Ad-cre. In both Hus1flox/+ and Hus1flox/Δ1 MEFs, cre-mediated recombination was predicted to convert the conditional allele into the null allele Hus1Δ2,3. Ad-cre–infected Hus1flox/+ cells would continue to express Hus1 from the remaining wild-type allele, whereas Ad-cre–infected Hus1flox/Δ1 cells would produce no functional Hus1 transcripts (Figure 1A).
Figure 1.
Conditional inactivation of Hus1 in primary MEFs by using Ad-cre. (A) Schematic of the system for conditional inactivation of Hus1. The various Hus1 alleles used are indicated, with the first several exons shown. Hus1flox is a conditional Hus1 allele in which exons 2 and 3 are flanked by loxP sites (black triangles). After Ad-cre infection, exons 2 and 3 of the Hus1flox allele are deleted, producing the null allele Hus1Δ2,3. A nonfunctional Hus1 transcript lacking exons 2 and 3 is produced from Hus1Δ2,3 and is represented by a dotted line. Hus1Δ1 is a constitutive null allele that lacks exon 1 and additional upstream sequences. Ad-cre–infected Hus1flox/+ MEFs continue to express wild-type Hus1 from Hus1+, whereas Ad-cre–infected Hus1flox/Δ1 MEFs fail to produce any functional Hus1 transcripts. (B) Southern blot analysis of Hus1 deletion after Ad-cre infection. Genomic DNA was prepared from Hus1flox/+ and Hus1flox/Δ1 MEFs at 1, 2, 3, or 4 d postinfection with Ad-cre or at 2 d after mock infection (U, uninfected) and then subjected to Southern blot analysis. The cells used in this experiment were not passaged after Ad-cre or mock infection The positions of Hus1flox, Hus1+, and Hus1Δ2,3 bands are indicated. The Hus1Δ1 allele is not detected in this assay. (C) Northern blot analysis of mRNA prepared from cells prepared Ad-cre or mock-infected Hus1flox/+ and Hus1flox/Δ1 MEFs. The positions of the transcripts produced from Hus1flox, Hus1+, and Hus1Δ2,3 are indicated.
Initially, the minimal dose of Ad-cre required for complete deletion of the Hus1flox allele was determined to minimize the cellular toxicity caused by cre (Loonstra et al., 2001; Silver and Livingston, 2001). Hus1flox/+ cells were infected with various doses of Ad-cre, and genomic DNA was isolated at 2 dpi and subjected to Southern blot analysis. Once the minimal effective Ad-cre dose was determined (data not shown), the kinetics of Hus1 inactivation were examined. By 1 d after infection with the minimal Ad-cre dose, the Hus1flox allele was fully converted to Hus1Δ2,3 in both Hus1flox/+ and Hus1flox/Δ1 MEFs (Figure 1B). Northern blot analysis also was performed to evaluate Hus1 expression at the corresponding times after Ad-cre infection. As shown in Figure 1C, mock-infected Hus1flox/+ and Hus1flox/Δ1 cells both expressed wild-type Hus1 transcripts, with the expression level being higher in Hus1flox/+ cells than Hus1flox/Δ1 cells as expected. As rapidly as 1 dpi, wild-type Hus1 transcripts were no longer detectable in Ad-cre–infected Hus1flox/Δ1 cells and were replaced with a lower-molecular-weight mRNA corresponding to the nonfunctional Hus1Δ2,3 transcript. Ad-cre–infected Hus1flox/+ cells expressed both wild-type Hus1 and Hus1Δ2,3 transcripts as anticipated. In short, these results establish that Ad-cre infection of conditional knockout MEFs allows for the rapid and efficient inactivation of Hus1.
Hus1 Loss Results in Decreased Cell Proliferation and Increased Apoptosis
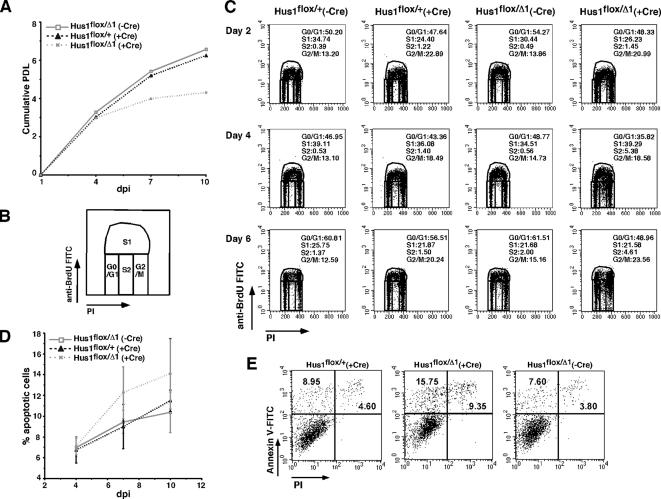
Fibroblasts derived from Hus1-deficient embryos fail to proliferate in culture (Weiss et al., 2000). However, these experiments are complicated by the fact that the Hus1-null cells must be obtained from morphologically abnormal embryos at a much earlier developmental stage than is typical for MEF culture. The conditional Hus1 knockout system offered the opportunity to rapidly inactivate Hus1 in a large population of normal MEFs and to test the immediate impact of Hus1 deficiency in a controlled setting. To examine the effect of Hus1 loss on cell proliferation, we first quantified PDLs for mock-infected and Ad-cre–infected Hus1flox/+ and Hus1flox/Δ1 MEFs cultured on a conventional 3T3 passage schedule. As shown in Figure 2A, MEFs of all genotypes initially doubled similarly. However, after 4 dpi Ad-cre–infected Hus1flox/Δ1 MEFs accumulated significantly fewer PDLs than control Ad-cre–infected Hus1flox/+ MEFs or mock-infected Hus1flox/Δ1 MEFs. These results identify an important role for Hus1 in cell doubling under normal growth conditions.
Figure 2.
Conditional inactivation of Hus1 results in impaired cell proliferation and increased apoptosis. (A) Analysis of cell proliferation. Hus1flox/+ and Hus1flox/Δ1 MEFs at passage one were infectectd with Ad-cre or mock infected and then cultivated following a 3T3 culture schedule as described in Materials and Methods. Plot shows the number of accumulated PDLs. (B) Schematic representation of a FACS dot plot of cell cycle distribution. Staining intensity for PI (x-axis) is plotted versus that for anti-BrdU-FITC (y-axis). The S-phase population is divided into S1 (BrdU positive) and S2 (BrdU negative). (C) Effects of Hus1 loss on cell cycle distribution. MEFs of the indicated genotypes were labeled with BrdU at the indicated times postinfection or mock infection, stained with anti-BrdU and PI, and analyzed by flow cytometry. Cells were passaged 1 d before BrdU labeling. The percentage of cells in each phase of the cell cycle is indicated. (D and E) Increased apoptosis after conditional inactivation of Hus1. MEFs of the indicated genotypes were stained with Annexin V-FITC and PI at the indicated times postinfection or mock infection and analyzed by flow cytometry. (D) Plot shows the percentage of apoptotic cells (Annexin V positive, PI negative). Values are the mean of three independent experiments, with error bars representing the SD. (E) Representative FACS dot plots showing apoptosis in cells of the indicated genotypes at 7 d postinfection or mock infection. Staining intensity for PI (x-axis) is plotted versus that for Annexin V-FITC (y-axis). The percentage of cells categorized as apoptotic (top left quadrant; Annexin V positive, PI negative) or necrotic (top right quadrant; Annexin V positive, PI positive) is indicated.
The reduced cell doubling in Hus1 conditional knockout cells could be due to cell cycle arrest, apoptosis, or both. To differentiate between these possibilities, we first monitored the progression of Hus1flox/+ and Hus1flox/Δ1 MEFs through the cell cycle after Ad-cre infection. Asynchronous cultures were harvested after labeling with BrdU at 2, 4, or 6 dpi. Analysis of cell cycle distribution by bivariate FACS revealed no significant differences between cells of the various Hus1 genotypes at 2 dpi (Figure 2, B and C). However, at both 4 and 6 dpi a small population of cells that possessed an S-phase DNA content but were BrdU negative (designated S2) was consistently observed specifically in Ad-cre–infected Hus1flox/Δ1 cultures (5.38% at 4 dpi and 4.61% at 6 dpi). These abnormal S-phase cells were rare in control Ad-cre–infected Hus1flox/+ cultures (1.40% at 4 dpi and 1.50% at 6 dpi) as well as uninfected cultures. The distribution of cells in other stages of the cell cycle was largely unaffected by Hus1 loss. A slight accumulation of Ad-cre–infected Hus1flox/Δ1 MEFs in G2/M was noted, but similar results were observed for Ad-cre–infected Hus1flox/+ MEFs.
The contribution of apoptosis to the reduced doubling of conditional Hus1 knockout cells was also investigated. Apoptosis was measured by flow cytometric analysis of cells stained with Annexin V, a phospholipid binding protein that can be used to identify cells in which phosphatidylserine has translocated from the inner to the outer leaflet of the plasma membrane, a marker of apoptosis (Vermes et al., 1995). The cells were also stained with PI as a measure of membrane integrity, to distinguish intact cells in the initial stages of apoptosis from cells undergoing necrosis or other forms of cell death. The percentage of cells in the early stages of apoptosis (Annexin V+ PI−) in conditional Hus1 knockout and control cultures is shown in Figure 2D. Beyond 4 dpi, Ad-cre–infected Hus1flox/Δ1 cultures consistently contained a greater percentage of apoptotic cells than Ad-cre–infected Hus1flox/+ and mock-infected Hus1flox/Δ1 cultures, although these differences were not statistically significant. For example, at 7 dpi 12.3 ± 2.5% of Ad-cre–infected Hus1flox/Δ1 cells were apoptotic, versus 9.0 ± 2.1% of Ad-cre–infected Hus1flox/+ cells or 9.5 ± 1.1% of mock-infected Hus1flox/Δ1 cells. Representative plots showing FACS analysis of apoptosis in conditional Hus1 knockout cultures are presented in Figure 2E. Together, the results suggest that Hus1 inactivation causes reduced cell doubling through subtle cell cycle alterations and increased apoptosis.
Spontaneous Chromosome Abnormalities after Conditional Inactivation of Hus1
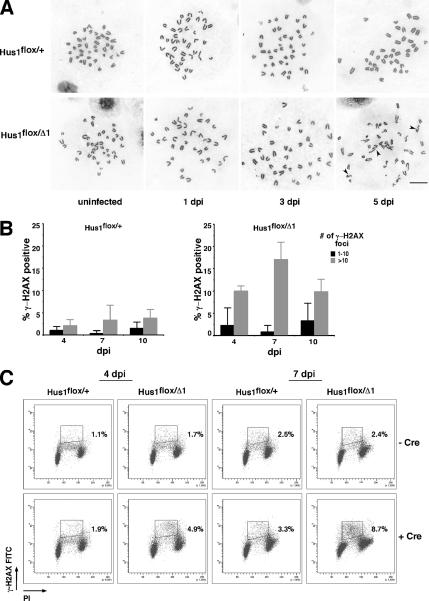
Hus1 acts in a pathway that is essential for the maintenance of genomic integrity. To quantify the extent of genome damage in conditional Hus1 knockout cells, we determined the frequency of gross chromosomal abnormalities in metaphase spreads prepared from Ad-cre–infected Hus1flox/+ and Hus1flox/Δ1 MEFs. Ad-cre–infected Hus1flox/Δ1 cells displayed a dramatic increase in chromosome abnormalities that was first detectable at 5 dpi (Table 1). By this time point, 55% of conditional Hus1 knockout cells had at least one abnormality, and 45% had greater than two abnormalities, including several cells with such extensive genome damage that it could not be quantified. By contrast, only 11.8% of control Ad-cre–infected Hus1flox/+ MEFs contained chromosomal abnormalities at 5 dpi, and none had greater than two abnormalities. Chromosome breaks and gaps affecting a single chromatid were the most common abnormalities in conditional Hus1 knockout cells (Figure 3A and Table 1). Chromatid interchanges were also observed at a low frequency.
Table 1.
Conditional inactivation of Hus1 results in increased chromosomal abnormalitiesa
| Genotype | dpib | Total cells | Breaks/gaps |
Exchanges |
Extensive damagec | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Chromatid | Total | Avg. | Total | Avg. | ||||
| Hus1flox/+ | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 5 | 17 | 0 | 0 | 0 | 0 | 2 | 0.12 | 0 | |
| 7 | 17 | 0 | 1 | 1 | 0.06 | 0 | 0 | 0 | |
| Hus1flox/Δ1 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 20 | 0 | 0 | 0 | 0 | 1 | 0.05 | 0 | |
| 5 | 20 | 0 | 47 | 47 | 2.35 | 4 | 0.2 | 4 | |
| 7 | 18 | 0 | 10 | 10 | 0.56 | 6 | 0.33 | 0 | |
a Metaphase chromosomes were prepared from cells of the indicated genotypes, and the occurrence of chromosomal abnormalities was quantified.
b Days post-Ad-cre infection.
c Extensive damage refers to metaphases that contained too many chromosome abnormalities to be counted.
Figure 3.
Conditional Hus1 inactivation results in chromosomal abnormalities and H2AX phosphorylation in primary MEFs. (A) Increased gross chromosomal abnormalities in Hus1 conditional knockout cells. Metaphase spreads were prepared from Hus1flox/+ and Hus1flox/Δ1 MEFs at 1, 3, and 5 d after Ad-cre infection or at 2 d after mock infection (U, uninfected). Cells analyzed at 3 or 5 dpi were passaged 2 d before metaphase spread preparation. Representative images are shown, with arrowheads indicating chromosomal abnormalities. (B and C) Accumulation of γ-H2AX in Hus1 conditional knockout cells. (B) The percentage of γ-H2AX-positive cells was determined by indirect immunofluorescence assay. Values are the mean number of cells with the indicated number of γ-H2AX foci from three independent 40× microscope fields, with error bars representing the SD. (C) The cell cycle distribution of γ-H2AX–positive cells was determined by FACS analysis. MEFs of the indicated genotypes were stained with anti-γ-H2AX antibody and PI at 4 or 7 d after Ad-cre infection or mock infection and analyzed by flow cytometry. The percentage of γ-H2AX–positive cells in S phase is indicated.
To further characterize the spontaneous genome damage that occurs after regulated Hus1 inactivation, we performed immunofluorescence assays to detect γ-H2AX, the phosphorylated form of histone H2AX that accumulates at DSBs. Although H2AX phosphorylation requires checkpoint signaling, previous studies indicated that Hus1 is dispensable for γ-H2AX accumulation after replication stress (Ward and Chen, 2001). As shown in Figure 3B, there was a slight increase in staining for γ-H2AX in Hus1flox/Δ1 cells at 4 d after Ad-cre infection, and by 7 dpi 16.9 ± 3.9% of Ad-cre–infected Hus1flox/Δ1 cells contained >10 γ-H2AX–positive foci, compared with only 3.1 ± 3.4% of Ad-cre–infected Hus1flox/+ MEFs. Mock-infected Hus1flox/+ and Hus1flox/Δ1 cells displayed a low background level of γ-H2AX staining similar to that observed for Ad-Cre infected Hus1flox/+ cells (data not shown). Thus, Hus1 loss specifically results in increased H2AX phosphorylation and the appearance of chromosomal abnormalities. We next investigated whether these DNA lesions arose in a particular stage of the cell cycle. This was accomplished by bivariate FACS analysis of mock-infected and Ad-cre–infected Hus1flox/+ and Hus1flox/Δ1 cells after staining with anti-γ-H2AX antibody and PI. The results confirmed increased H2AX phosphorylation in Ad-cre–infected Hus1flox/Δ1 cells and further indicated that the γ-H2AX accumulation after Hus1 loss was largely restricted to cells with S-phase DNA content (Figure 3C). Importantly, this finding was not due to an inability of this method to detect γ-H2AX in other stages of the cell cycle, because positive staining was observed in G1, S, and G2/M populations after treatment of cells with exogenous genotoxins (data not shown; Marti et al., 2006). In sum, these results indicate that conditional Hus1 inactivation causes increased genomic instability and DSB accumulation in S phase of the cell cycle.
Increased Common Fragile Site Expression after Hus1 Inactivation
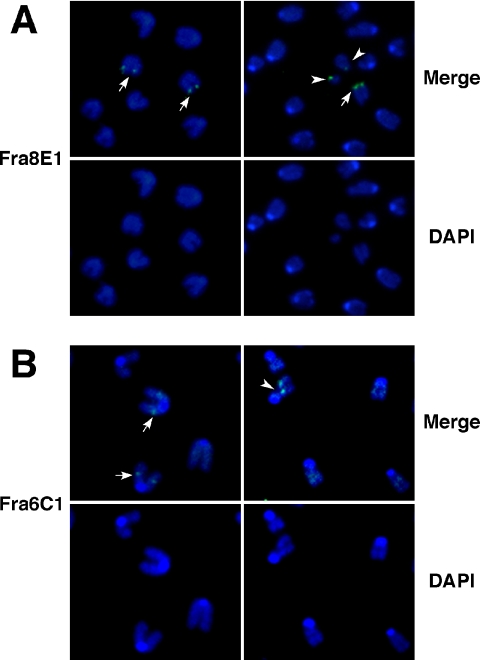
The spontaneous DNA lesions in conditional Hus1 knockout cells could arise randomly throughout the genome or at specific genomic regions. Because Hus1 is required for S-phase checkpoint function and also is essential for cellular responses to extrinsic replication stresses (Weiss et al., 2000, 2003), we tested whether the spontaneous chromosome breaks in Hus1-deficient cells occurred preferentially at CFSs, genomic regions prone to breakage under conditions of replication stress (Glover et al., 2005). For this purpose, FISH assays were performed to quantify how often chromosomal breaks localized to CFSs in conditional Hus1 knockout cells. CFSs were detected using probes prepared from bacterial artificial chromosomes containing mouse genomic sequence from fragile sites Fra8E1 and Fra6C1. Consistent with the results reported in Figure 3A and Table 1, Ad-cre–infected Hus1flox/Δ1 cells displayed an average of 1.60–2.30 breaks per cell, whereas Ad-cre–infected Hus1flox/+ cells displayed only 0.18–0.21 chromosomal breaks per cell on average (Table 2). Notably, a significant increase in the frequency of spontaneous CFS breakage was observed in conditional Hus1 knockout cells. At CFS Fra8E1, nine breaks were identified among the 233 loci analyzed in Ad-cre–infected Hus1flox/Δ1 cells, compared with no breaks at 264 loci analyzed in control Ad-cre–infected Hus1flox/+ cells. Nearly 10% (9 of 91) of the spontaneous breaks and gaps in Hus1-deficient cells localized to this fragile site. Similarly, five breaks were identified among 151 CFS Fra6C1 loci analyzed in Ad-cre infected Hus1flox/Δ1 cells, accounting for 5.7% of observed breaks (5 of 87), but none were detected in Ad-cre–infected Hus1flox/+ cells. Representative FISH images are shown in Figure 4. These results indicate that the spontaneous breaks that arise upon Hus1 loss occur preferentially at CFSs, establishing a new role for Hus1 in the maintenance of CFS stability.
Table 2.
Conditional inactivation of Hus1 leads to increased common fragile site expressiona
| CFS | Genotype | No. of cells | No. of breaks | Avg. breaks/cell | No. of breaks at CFS | % CFS loci with a break | % of breaks at CFS |
|---|---|---|---|---|---|---|---|
| Fra8E1 | Hus1flox/+ | 57 | 12 | 0.21 | 0 | (0 (0/264b) | 0 (0/12) |
| Hus1flox/Δ1 | 57 | 91 | 1.60 | 9 | 3.4 (9/233) | 9.9 (9/91) | |
| Fra6C1 | Hus1flox/+ | 38 | 7 | 0.18 | 0 | 0 (0/154) | 0 (0/7) |
| Hus1flox/Δ1 | 38 | 87 | 2.3 | 5 | 3.3 (5/151) | 5.7 (5/87) |
a FISH was used to detect CFS loci Fra8E1 or Fra6C1 as described in Materials and Methods.
b Individual cells occasionally contained fewer or more than the expected four FISH signals because of chromosome loss or polyploidy, respectively; therefore, the total number of CFS loci analyzed was not exactly 4 times the number of cells analyzed.
Figure 4.
Increased expression of common fragile sites after Hus1 inactivation. Representative images of CFS expression in metaphase spreads prepared from Ad-cre–infected Hus1flox/Δ1 MEFs. Cells were passaged at 2 dpi, harvested for metaphase spread preparation at 4dpi, and then stained with probes specific to CFS Fra8E1 (A) or Fra6C1 (B) (green) and counterstained with DAPI (blue). Shown are merged images of CFS and DAPI staining (top) or the corresponding images with DAPI staining alone (bottom). Arrows indicate CFS at intact chromosomal sites, and arrowheads indicate colocalization of CFS and chromosomal breaks or gaps.
p53 Accumulates after Conditional Hus1 Inactivation, but the Impaired Proliferation and Increased Apoptosis Phenotypes Persist in Its Absence
The observation of increased spontaneous chromosomal abnormalities following regulated Hus1 deletion raised the possibility that the accompanying reduced proliferation and increased apoptosis could be due to a Hus1-independent DNA damage response. We previously reported that p53 becomes activated in Hus1-null embryos, triggering increased expression of p53 target genes (Weiss et al., 2002), and furthermore that p21 or p53 inactivation allows for the serial culture of Hus1-deficient MEFs (Weiss et al., 2000; our unpublished data). We therefore hypothesized that conditional Hus1 inactivation triggers p53 activation and subsequent checkpoint responses. Because p53 activation is associated with p53 protein stabilization and accumulation (Harris and Levine, 2005), we first measured p53 levels at the single cell level by immunofluorescence assay. p53 is normally at very low levels in wild-type cells; accordingly, <3% of Ad-cre–infected Hus1flox/+ cells were found to be p53 positive at 4, 7, or 10 dpi. By contrast, 12.7 ± 5.8 and 14.3 ± 1.4% of Hus1flox/Δ1 cells were p53 positive at 7 and 10 d post-Ad-cre infection, respectively (Figure 5A). We conclude that conditional Hus1 inactivation results in p53 accumulation.
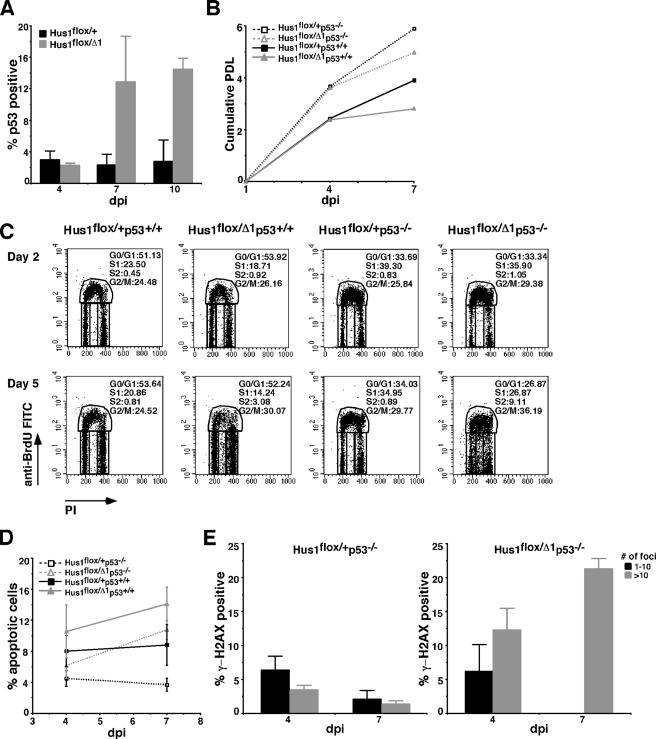
Figure 5.
p53 accumulates after conditional Hus1 inactivation, but its deletion fails to fully rescue the impaired cell doubling or increased apoptosis in Hus1 conditional knockout cells. (A) p53 levels in Hus1flox/+ and Hus1flox/Δ1 MEFs after Ad-cre infection. The percentage of p53-positive cells was determined at the indicated times postinfection by immunofluorescence assay. Values are the mean number of p53-positive cells from three independent 40× microscope fields, with error bars representing the SD. (B) Analysis of cell doubling following Hus1 inactivation in a p53-deficient background. MEFs of the indicated genotypes at passage two were infected with Ad-cre or mock infected and then cultured according to a 3T3 passage schedule. Cumulative PDLs are plotted. (C) Enhanced cell cycle defects following Hus1 inactivation in a p53-deficient background. Ad-cre infected MEFs of the indicated genotypes were labeled with BrdU and analyzed by flow cytometry as described in the legend to Figure 2. The percentage of cells in each phase of the cell cycle is indicated. (D) Apoptosis after conditional Hus1 inactivation in a p53-null background. MEFs of the indicated genotypes were stained with Annexin V-FITC and PI and analyzed by flow cytometry. Plots show the percentage of cells undergoing apoptosis (Annexin V positive, PI negative). Values are the mean of three independent experiments, with error bars representing the SD. (E) Accumulation of γ-H2AX in Hus1flox/+p53−/− and Hus1flox/Δ1p53−/− MEFs after Ad-cre infection. The percentage of γ-H2AX–positive cells was determined by immunofluorescence assay as described in the legend to Figure 3.
We next directly tested the role of p53 in the cell proliferation defects and increased apoptosis that occur after conditional Hus1 knockout. For this purpose, we cultured Hus1 conditional knockout cells in a p53-deficient background. Hus1flox/Δ1p53+/+, Hus1flox/Δ1p53−/−, Hus1flox/+p53+/+, and Hus1flox/+p53−/− MEFs were infected with Ad-cre and cultured following a 3T3 passage schedule. Similar to the results of Figure 2B, Ad-cre–infected Hus1flox/Δ1p53+/+ cells initially doubled as well as Ad-cre–infected Hus1flox/+p53+/+ MEFs, but they showed reduced doubling after 4 dpi (Figure 5B). The effect of Hus1 inactivation was similar in a p53-deficient background, although in general the p53-null cells doubled more rapidly than their p53 wild-type counterparts. Specifically, Ad-cre–infected Hus1flox/Δ1p53−/− cells doubled as well as Ad-cre–infected Hus1flox/+p53−/− MEFs until 4 dpi but then showed a reduction in doubling that paralleled that observed in the p53+/+ cells. Thus, p53 deletion did not rescue the cell-doubling defect in conditional Hus1 knockout cells. These experiments could not be extended beyond 7 dpi, because the cultures became overgrown with Hus1flox/Δ1p53−/− cells that failed to undergo Hus1 deletion. By Southern blot, these cells in which the conditional allele remained unrecombined were very rare immediately after Ad-cre infection, but they accounted for the majority of the culture by 7 dpi (data not shown).
To determine whether p53 mediated other responses to genomic instability after Hus1 loss, we next examined the effects of p53 deficiency on cell cycle distribution and apoptosis in Hus1 conditional knockout cells. Consistent with the results in Figure 2B, no differences were apparent in the cell cycle profiles of Hus1flox/Δ1p53+/+ and Hus1flox/+p53+/+ cells at 2 dpi (Figure 5C). However, as observed previously, a small but significant population of BrdU-negative S-phase (S2) cells (3.08%) was present in Ad-cre–infected Hus1flox/Δ1p53+/+ cultures by 5 dpi. Rather than suppressing the accumulation of S2 cells, p53 deficiency actually significantly increased their frequency, as 9.11% of Ad-cre–infected Hus1flox/Δ1p53−/− cells were present in the S2 fraction at 5 dpi. p53 deletion also failed to prevent the relative increase in apoptosis associated with conditional Hus1 inactivation, although it did cause an overall reduction in apoptosis, regardless of Hus1 status (Figure 5D). Consistent with the results of Figure 2D for p53-expressing MEFs, 14.1 ± 2.2% of Ad-cre–infected Hus1flox/Δ1p53+/+ cells were apoptotic at 7 dpi, whereas 8.8 ± 2.6% of Ad-cre–infected Hus1flox/+p53+/+ cells were apoptotic at the same time point. Similarly, 10.8 ± 1.2% of Ad-cre–infected Hus1flox/Δ1p53−/− cells were apoptotic at 7 dpi, compared with only 3.7 ± 0.8% of Ad-cre–infected Hus1flox/+p53−/− cells. In short, p53 loss failed to suppress the increased apoptosis in conditional Hus1 knockout cells and exacerbated the cell cycle defects associated with Hus1 inactivation.
Finally, we tested whether p53 deficiency would impact the extent of DSB accumulation in conditional Hus1 knockout cells. There was a low frequency of γ-H2AX–positive cells in Ad-cre–infected Hus1flox/+p53−/− cultures. 1.2 ± 0.5% of these cells contained more than 10 γ-H2AX foci at 7 dpi (Figure 5E), comparable with the results for Ad-cre–infected Hus1flox/+p53+/+ cultures (Figure 3B). Mock-infected cultures of all genotypes showed a similar low level of γ-H2AX staining (data not shown). Interestingly, combining p53 deficiency with conditional Hus1 inactivation did not diminish but instead slightly increased γ-H2AX staining relative to the effect of Hus1 loss alone. We determined that 21.1 ± 1.5% of Ad-cre–infected Hus1flox/Δ1p53−/− MEFs contained >10 γ-H2AX foci at 7 dpi (Figure 5E) compared with 16.9 ± 3.9% of Ad-cre–infected Hus1flox/Δ1p53+/+ MEFs (Figure 3B). Together, the results indicate that p53 loss does not reduce the accumulation of genome damage in conditional Hus1 knockout cells or suppress the resulting cell proliferation defects and apoptosis.
DISCUSSION
DNA damage checkpoints are best known for their roles in responding to genome damage that arises when a cell is exposed to an extrinsic genotoxic stress. However, it has emerged that many of the same checkpoint mechanisms also have critical functions during an unperturbed cell cycle. Mouse Hus1, like other components of the Atr-dependent checkpoint pathway, is essential for embryonic development and is required for the proliferation of primary fibroblasts. To investigate the essential functions of Hus1, we established a system in which a loxP site-flanked conditional Hus1 allele could be inactivated by cre-mediated recombination. Infection of Hus1flox/Δ1 MEFs with Ad-cre resulted in the disappearance of wild-type Hus1 transcripts within 1 d. Because of an inability to detect the endogenous Hus1 polypeptide in MEFs with available antibody reagents, we were unable to track the loss of Hus1 protein after Ad-cre infection. Conditional Hus1 knockout cells remained apparently normal for ∼4 d after Ad-cre infection. Similar results were reported for the cre-mediated deletion of Atr in MEFs, which allowed for one to two rounds of normal cell division before defects emerged (Brown and Baltimore, 2003). It is not clear whether this lag before the emergence of phenotypes reflects the time needed for complete depletion of the target protein or whether it is an indication that other events, such as the accumulation of some type of genome damage, must occur before the requirement for this checkpoint pathway becomes apparent.
Previous studies indicated that it is not possible to establish long-term cultures of cells in which Atr (Cortez et al., 2001; Brown and Baltimore, 2003), Chk1 (Liu et al., 2000), or RAD17 (Wang et al., 2003) have been inactivated by cre-mediated recombination. Similarly, Hus1 deletion in MEFs resulted in a significant impairment of cell doubling. At late time points postinfection, Ad-cre–infected Hus1flox/Δ1 cultures were overtaken by cells that failed to undergo recombination at the conditional Hus1 allele (data not shown). Thus, Hus1-deficient cells were at a selective disadvantage and were outcompeted by cells with intact Hus1 function. Hus1 loss was associated with fairly modest changes in cell cycle distribution and a slight increase in apoptosis. Similar results were reported for conditional Atr knockout cells (Brown and Baltimore, 2003). These findings might indicate a broad role for checkpoints throughout the cell cycle, and they also may reflect technical limitations of the conditional knockout system, in which the population of cells under study is asynchronous with respect to the time postinfection when the target protein falls below a critical threshold. Individual cells also likely would be at different cell cycle stages when the target protein becomes fully depleted. Although RAD17-deleted HCT116 cells undergo endoreduplication (Wang et al., 2003), we failed to observe this phenotype in conditional Hus1 knockout cells. This may suggest that this Rad17 function is independent of 9-1-1 loading or that primary MEFs possess redundant regulatory mechanisms that prevent endoreduplication.
The most notable effect of Hus1 inactivation on the cell cycle was the accumulation of a small population of cells with S-phase DNA content that failed to incorporate BrdU (S2 cells). Because these cells comprised only a small fraction of the total culture, this cell cycle abnormality probably does not fully account for the impaired doubling of Hus1-deficient cells. The percentage of S2 cells did not increase between 4 and 6 dpi, possibly because some of these abnormal cells were cleared by apoptosis, which was increased at 7 dpi compared with 4 dpi. Based on their staining properties, these cells seemed to be impaired for DNA replication. The mammalian 9-1-1 complex has been linked to important S-phase functions previously. For example, this checkpoint trimer is required for an intra-S DNA damage checkpoint that represses DNA synthesis after genome damage (Bao et al., 2001; Roos-Mattjus et al., 2003; Weiss et al., 2003; Wang et al., 2004b). In addition, Rad1 is required for the resumption of DNA synthesis following hydroxyurea treatment, implying a role for the 9-1-1 complex in the stabilization or repair of stalled replication forks (Bao et al., 2004). Further evidence for an important S-phase function for Hus1 comes from the finding in this study that conditional Hus1 knockout cells accumulated DSBs specifically within this stage of the cell cycle. One interesting possibility is that DSB accumulation and impaired BrdU incorporation are two features of the same subpopulation of conditional Hus1 knockout cells. Because more γ-H2AX–positive cells were detected than S2 cells, individual S2 cells might contain multiple DSBs.
Inhibition of Chk1 (Syljuasen et al., 2005) or knockdown of the Chk1 regulator Claspin by RNA interference (Liu et al., 2006) similarly results in S-phase–specific H2AX phosphorylation. These findings are consistent with current models of checkpoint signaling in which Claspin and the 9-1-1 complex function to promote Chk1 phosphorylation and activation by Atr. The origin of the spontaneous DSB in S-phase cells defective for Hus1, Claspin, or Chk1 remains unknown. Syljuasen and colleagues have proposed two models by which impaired Chk1 activation could lead to DSB formation during S phase (Syljuasen et al., 2005). Given that Chk1 negatively regulates Cdc25A, one possibility is that Chk1 loss causes Cdk2 activation and increased initiation of DNA replication, leading to the accumulation of stretches of single-stranded DNA that are prone to breakage. Alternatively, Chk1 inhibition could lead to DSB formation via the aberrant processing or collapse of stalled replication forks. That at least some of the DSB in conditional Hus1 knockout cells occurred preferentially at CFSs rather than randomly favors the latter possibility in the case of Hus1 deficiency. CFSs are genomic regions that are prone to breakage under conditions of replication stress. Although the precise molecular basis for CFS breakage has not been determined, one model is that these regions form secondary structures that impair replication fork progression and trigger checkpoint activation for fork stabilization and/or repair (Cimprich, 2003; Glover et al., 2005). Our findings add Hus1 to a growing list of checkpoint factors that are required for CFS integrity, because ∼15% of all gaps and breaks in conditional Hus1 knockout cells localized to the two fragile sites analyzed. It should be noted that enforcement of the G2/M DNA damage checkpoint, rather than the preservation of replication forks, also has been proposed as the critical checkpoint function for CFS maintenance (Arlt et al., 2004). However, the 9-1-1 complex seems to be dispensable for the G2/M DNA damage checkpoint (Weiss et al., 2003; Bao et al., 2004), whereas other checkpoint factors such as Atm are required for G2/M checkpoint function but not CFS stability (Casper et al., 2002). That spontaneous DSB formation occurs in S phase in Hus1-deficient cells also argues against the possibility that the breaks arise when cells enter mitosis with incompletely replicated DNA, although it remains unclear how DSB arising in S phase in conditional Hus1 knockout cells would escape detection at the G2/M transition. Clearly, additional experiments are necessary to definitively establish the molecular basis for S-phase–specific DSB formation and CFS expression in conditional Hus1 knockout cells.
In mouse embryos, Hus1 deficiency triggers p53-dependent induction of p53 target genes such as p21 and Perp (Weiss et al., 2002). Furthermore, p21 or p53 deletion allows for the serial culture of fibroblasts derived from Hus1-null embryos, which fail to proliferate otherwise (Weiss et al., 2000; our unpublished data). These results suggest that Hus1 inactivation leads to genome damage that activates p53. Accordingly, we observed p53 accumulation in conditional Hus1 knockout cells. To examine the contribution of p53 to the cell proliferation defects and apoptosis in conditional Hus1 knockout MEFs, we compared the effects of Hus1 inactivation in p53+/+ and p53−/− backgrounds. Interestingly, p53 deletion failed to rescue the relative reduction in cell doubling in Ad-cre–infected Hus1flox/Δ1 cells compared with Hus1flox/+ cells. Apoptosis also remained elevated in Ad-cre–infected Hus1flox/Δ1 cultures relative to controls, even in the absence of p53. In light of these new data, we speculate that in our previous studies p21 and p53 deletion enabled the culturing of Hus1-null MEFs indirectly, by facilitating the accumulation of additional mutations that permitted long-term cell proliferation in the absence of Hus1. These results are consistent with the previous finding that deletion of p53 or p21 failed to affect the embryonic lethality associated with Hus1 deficiency (Weiss et al., 2000). Similarly, Chk1-deficient embryos die of p53-independent apoptosis (Liu et al., 2000). p53 deficiency actually worsened some of the phenotypes in conditional Hus1 knockout cells. The frequency of BrdU-negative S-phase cells was significantly increased in Ad-cre–infected Hus1flox/Δ1p53−/− MEFs compared with their p53+/+ counterparts. Spontaneous DSBs, as measured by H2AX phosphorylation, were also slightly elevated. p53 previously has been shown to suppress DSB formation at stalled replication forks (Kumari et al., 2004; Squires et al., 2004), possibly through the direct regulation of homologous recombination (Sengupta et al., 2005). Thus, the increased occurrence of S-phase defects and DSB formation in cells lacking Hus1 and p53 is consistent with a model in which both gene products function in the preservation of replication fork stability. Future studies aimed at dissecting the precise molecular roles of these factors at the replication fork promise to provide important new insights into the maintenance of genomic stability.
ACKNOWLEDGMENTS
We thank Dr. Beth Sullivan for advice on FISH assays; The Biomedical Sciences Flow Cytometry Core Laboratory for assistance with FACS analyses; and Eric Alani, Cyrus Vaziri, Peter Levitt, Gabriel Balmus, and Xia Xu for helpful discussions and suggestions. This work was supported by National Institutes of Health grants CA-108773 and ES-012917.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0957) on January 10, 2007.
REFERENCES
- Arlt M. F., Durkin S. G., Ragland R. L., Glover T. W. Common fragile sites as targets for chromosome rearrangements. DNA Repair. 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Arlt M. F., Xu B., Durkin S. G., Casper A. M., Kastan M. B., Glover T. W. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Bao S., Lu T., Wang X., Zheng H., Wang L. E., Wei Q., Hittelman W. N., Li L. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23:5586–5593. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- Bao S., Tibbetts R. S., Brumbaugh K. M., Fang Y., Richardson D. A., Ali A., Chen S. M., Abraham R. T., Wang X. F. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]
- Blasco M. A., Lee H. W., Hande M. P., Samper E., Lansdorp P. M., DePinho R. A., Greider C. W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Brown E. J., Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M., et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper A. M., Nghiem P., Arlt M. F., Glover T. W. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Cha R. S., Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Chang D. Y., Lu A. L. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- Cimprich K. A. Fragile sites: breaking up over a slowdown. Curr. Biol. 2003;13:R231–R233. doi: 10.1016/s0960-9822(03)00158-1. [DOI] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S. J. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Dart D. A., Adams K. E., Akerman I., Lakin N. D. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J. Biol. Chem. 2004;279:16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- de Klein A., Muijtjens M., van Os R., Verhoeven Y., Smit B., Carr A. M., Lehmann A. R., Hoeijmakers J. H. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- Durkin S. G., Arlt M. F., Howlett N. G., Glover T. W. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25:4381–4388. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- Friedrich-Heineken E., Toueille M., Tannler B., Burki C., Ferrari E., Hottiger M. O., Hubscher U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Glover T. W., Arlt M. F., Casper A. M., Durkin S. G. Mechanisms of common fragile site instability. Hum. Mol. Genet. 2005;14:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- Guo Z., Kumagai A., Wang S. X., Dunphy W. G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. L., Levine A. J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Hekmat-Nejad M., You Z., Yee M. C., Newport J. W., Cimprich K. A. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Hopkins K. M., Auerbach W., Wang X. Y., Hande M. P., Hang H., Wolgemuth D. J., Joyner A. L., Lieberman H. B. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B. O., Schmitt E. M., Halachmi S., Bronson R. T., Weinberg R. A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jiang K., Pereira E., Maxfield M., Russell B., Goudelock D. M., Sanchez Y. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J. Biol. Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- Kai M., Wang T. S. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. E., McAvoy S. A., Smith D. I., Chen J. Human TopBP1 ensures genome integrity during normal S phase. Mol. Cell. Biol. 2005;25:10907–10915. doi: 10.1128/MCB.25.24.10907-10915.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzel B., Hall J., Natt F., Weiler J., Cohen D. Downregulation of Hus1 by antisense oligonucleotides enhances the sensitivity of human lung carcinoma cells to cisplatin. Cancer. 2002;94:1808–1814. doi: 10.1002/cncr.10383. [DOI] [PubMed] [Google Scholar]
- Krummel K. A., Denison S. R., Calhoun E., Phillips L. A., Smith D. I. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer. 2002;34:154–167. doi: 10.1002/gcc.10047. [DOI] [PubMed] [Google Scholar]
- Kumari A., Schultz N., Helleday T. p53 protects from replication-associated DNA double-strand breaks in mammalian cells. Oncogene. 2004;23:2324–2329. doi: 10.1038/sj.onc.1207379. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Levitt P. S., Liu H., Manning C., Weiss R. S. Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics. 2005;86:212–224. doi: 10.1016/j.ygeno.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Liu Q., et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell. Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra A., Vooijs M., Beverloo H. B., Allak B. A., van Drunen E., Kanaar R., Berns A., Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C. S., Foiani M. The DNA replication checkpoint stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- Marti T. M., Hefner E., Feeney L., Natale V., Cleaver J. E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Seiler J. A., Burhans W. C. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J. Biol. Chem. 2003;278:4295–4304. doi: 10.1074/jbc.M204264200. [DOI] [PubMed] [Google Scholar]
- Mitelman F. Basel, Switzerland: S. Karger; 1995. ISCN (1995): An International System for Human Cytogenetic Nomenclature. [Google Scholar]
- Niida H., Tsuge S., Katsuno Y., Konishi A., Takeda N., Nakanishi M. Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J. Biol. Chem. 2005;280:39246–39252. doi: 10.1074/jbc.M505009200. [DOI] [PubMed] [Google Scholar]
- Pandita R. K., et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar E. R., Karnitz L. M. Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J. Biol. Chem. 2003;278:45507–45511. doi: 10.1074/jbc.C300418200. [DOI] [PubMed] [Google Scholar]
- Raveendranathan M., Chattopadhyay S., Bolon Y. T., Haworth J., Clarke D. J., Bielinsky A. K. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J. 2006;25:3627–3639. doi: 10.1038/sj.emboj.7601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos-Mattjus P., Hopkins K. M., Oestreich A. J., Vroman B. T., Johnson K. L., Naylor S., Lieberman H. B., Karnitz L. M. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J. Biol. Chem. 2003;278:24428–24437. doi: 10.1074/jbc.M301544200. [DOI] [PubMed] [Google Scholar]
- Roos-Mattjus P., Vroman B. T., Burtelow M. A., Rauen M., Eapen A. K., Karnitz L. M. Genotoxin-induced Rad9-Hus1-Rad1 (9-1-1) chromatin association is an early checkpoint signaling event. J. Biol. Chem. 2002;277:43809–43812. doi: 10.1074/jbc.M207272200. [DOI] [PubMed] [Google Scholar]
- Rozier L., El-Achkar E., Apiou F., Debatisse M. Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C 1: possible association with an inherited disease and cancer. Oncogene. 2004;23:6872–6880. doi: 10.1038/sj.onc.1207809. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S., Minesinger B. K., Giannattasio M., Plevani P., Muzi-Falconi M., Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with polζ and is partially required for spontaneous polζ-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- Savage J. R. Classification and relationships of induced chromosomal structural changes. J. Med. Genet. 1976;13:103–122. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E., Boutros R., Froment C., Monsarrat B., Ducommun B., Dozier C. CHK1 phosphorylates CDC25B during the cell cycle in the absence of DNA damage. J. Cell Sci. 2006;119:4269–4275. doi: 10.1242/jcs.03200. [DOI] [PubMed] [Google Scholar]
- Sengupta S., et al. Tumor suppressor p53 represses transcription of RECQ4 helicase. Oncogene. 2005;24:1738–1748. doi: 10.1038/sj.onc.1208380. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004a;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair. 2004b;3:901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Shi G., Chang D. Y., Cheng C. C., Guan X., Venclovas C., Lu A. L. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D. P., Livingston D. M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Toueille M., Markkanen E., Hubscher U. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Lopes M., Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Sorensen C. S., Syljuasen R. G., Lukas J., Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- Squires S., Coates J. A., Goldberg M., Toji L. H., Jackson S. P., Clarke D. J., Johnson R. T. p53 prevents the accumulation of double-strand DNA breaks at stalled-replication forks induced by UV in human cells. Cell Cycle. 2004;3:1543–1557. doi: 10.4161/cc.3.12.1272. [DOI] [PubMed] [Google Scholar]
- Stec D. E., Davisson R. L., Haskell R. E., Davidson B. L., Sigmund C. D. Efficient liver-specific deletion of a floxed human angiotensinogen transgene by adenoviral delivery of Cre recombinase in vivo. J. Biol. Chem. 1999;274:21285–21290. doi: 10.1074/jbc.274.30.21285. [DOI] [PubMed] [Google Scholar]
- Syljuasen R. G., Sorensen C. S., Hansen L. T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Tominaga K., Motoyama N., Minamishima Y. A., Nagahama H., Tsukiyama T., Ikeda K., Nakayama K., Nakanishi M., Nakayama K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Todaro G. J., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toueille M., El-Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M. O., Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz K., Smith E., Smith S., Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I., Haanen C., Steffens-Nakken H., Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Wang W., Brandt P., Rossi M. L., Lindsey-Boltz L., Podust V., Fanning E., Sancar A., Bambara R. A. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA. 2004a;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lindsey-Boltz L. A., Sancar A., Bambara R. A. Mechanism of stimulation of human DNA ligase I by the Rad9-Rad1-Hus1 checkpoint complex. J. Biol. Chem. 2006a;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- Wang X., Guan J., Hu B., Weiss R. S., Iliakis G., Wang Y. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res. 2004b;32:767–775. doi: 10.1093/nar/gkh243. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang X., Hu B., Weiss R. S., Wang Y. The effect of Hus1 on ionizing radiation sensitivity is associated with homologous recombination repair but is independent of nonhomologous end-joining. Oncogene. 2006b;25:1980–1983. doi: 10.1038/sj.onc.1209212. [DOI] [PubMed] [Google Scholar]
- Wang X., Zou L., Zheng H., Wei Q., Elledge S. J., Li L. Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev. 2003;17:965–970. doi: 10.1101/gad.1065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I. M., Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Weiss R. S., Enoch T., Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- Weiss R. S., Kostrub C. F., Enoch T., Leder P. Mouse Hus1, a homolog of the Schizosaccharomyces pombe hus1+ cell cycle checkpoint gene. Genomics. 1999;59:32–39. doi: 10.1006/geno.1999.5865. [DOI] [PubMed] [Google Scholar]
- Weiss R. S., Leder P., Vaziri C. Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Mol. Cell. Biol. 2003;23:791–803. doi: 10.1128/MCB.23.3.791-803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. S., Matsuoka S., Elledge S. J., Leder P. Hus1 acts upstream of Chk1 in a mammalian DNA damage response pathway. Curr. Biol. 2002;12:73–77. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- You Z., Kong L., Newport J. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 2002;277:27088–27093. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- Zou L., Cortez D., Elledge S. J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]