Abstract
The cell fate determination factor DACH1 plays a key role in cellular differentiation in metazoans. DACH1 is engaged in multiple context-dependent complexes that activate or repress transcription. DACH1 can be recruited to DNA via the Six1/Eya bipartite transcription (DNA binding/coactivator) complex. c-Jun is a critical component of the activator protein (AP)-1 transcription factor complex and can promote contact-independent growth. Herein, DACH1 inhibited c-Jun–induced DNA synthesis and cellular proliferation. Excision of c-Jun with Cre recombinase, in c-junf1/f1 3T3 cells, abrogated DACH1-mediated inhibition of DNA synthesis. c-Jun expression rescued DACH1-mediated inhibition of cellular proliferation. DACH1 inhibited induction of c-Jun by physiological stimuli and repressed c-jun target genes (cyclin A, β-PAK, and stathmin). DACH1 bound c-Jun and inhibited AP-1 transcriptional activity. c-jun and c-fos were transcriptionally repressed by DACH1, requiring the conserved N-terminal (dac and ski/sno [DS]) domain. c-fos transcriptional repression by DACH1 requires the SRF site of the c-fos promoter. DACH1 inhibited c-Jun transactivation through the δ domain of c-Jun. DACH1 coprecipitated the histone deacetylase proteins (HDAC1, HDAC2, and NCoR), providing a mechanism by which DACH1 represses c-Jun activity through the conserved δ domain. An oncogenic v-Jun deleted of the δ domain was resistant to DACH1 repression. Collectively, these studies demonstrate a novel mechanism by which DACH1 blocks c-Jun-mediated contact-independent growth through repressing the c-Jun δ domain.
INTRODUCTION
Homo- or heterodimeric transcription factors with basic region-leucine zipper structure, including Jun, Fos, ATF, and Maf subfamilies, regulate transcription of activator protein (AP)-1–responsive genes in a DNA sequence-specific manner. c-Jun encodes a critical component of the AP-1 transcription factor complex. Proliferative signals induce expression of both c-Fos and c-Jun (Karin et al., 1997; Schreiber et al., 1999). Mice genetically engineered to abolish either c-Jun or c-Jun NH2-terminal kinase (JNK) activity results in embryonic lethality (Jochum et al., 2001). Of the four mitogen-activated protein kinase (MAPK) subfamilies, c-Jun activity is regulated primarily by Jun kinase (Morton et al., 2003). The abundance of c-Jun is regulated by transcriptional induction and subsequent degradation through ubiquitination (Angel and Karin, 1991; Treier et al., 1994; Fuchs et al., 1996; Musti et al., 1997).
The c-Jun protein structure consists of multiple functional domains, including an amino-terminal transactivation domain, a regulatory (δ domain), a carboxy-terminal basic DNA binding domain, and a leucine zipper protein dimerization domain. The δ domain is critical for transcriptional activation by c-Jun. One of the first reported functions of the δ domain was its engagement of cell type-specific inhibitors of c-Jun (Bohmann and Tjian, 1989; Baichwal and Tjian, 1990; Baichwal et al., 1991). JNK docks to c-Jun on residues within the δ domain (Kallunki et al., 1995, 1996). Phosphorylation of c-Jun is thought to facilitate interaction of the c-Jun/AP-1 complex with coactivators. Transcriptional coactivators encoding histone acetyl transferase (HAT) activity, such as the CREB-binding protein (CBP) coactivator, bind to c-Jun facilitating the interaction between the AP-1 complex and the basal transcriptional machinery (Mayr and Montminy, 2001). JNK-mediated phosphorylation also accelerates c-Jun degradation by allowing recognition of the E3 ligase from the Fbw 7-containing Skp/Culin/F-box protein complex. In addition, JNK enhances activity of the E3 ligase, promoting degradation of c-Jun and JunB (Gao et al., 2004; Nateri et al., 2004).
c-Jun contributes to contact-independent growth and is essential for the development of chemically induced tumors in mice (Eferl et al., 2003). Both cellular Jun (c-Jun) and viral Jun (v-Jun) induce oncogenic transformation (Vogt, 2001). The retrovirally transduced allele of c-Jun, v-Jun, induces fibrosarcoma in chickens (for review, see Vogt, 2001). Oncogenic v-jun encodes a protein with complete deletion of the δ domain and fails to bind JNK. The role of the δ domain itself versus JNK phosphorylation sites within the δ domain, in cell cycle control, cellular proliferation, and oncogenesis is complex. Although c-Jun promotes G1/S phase progression independently of its phosphorylation status (Wisdom et al., 1999), c-Jun phosphorylation of serines 63 and 73 were required for Ha-Ras–induced cellular transformation in some (Smeal et al., 1991), but not all studies (Kennedy et al., 2003). Finally, although point mutation analysis demonstrated the oncogenic effects of the δ domain deletion can be uncoupled from JNK signaling (Sprowles and Wisdom, 2003), mice and cells harboring a mutant allele of c-jun show reduced tumorigenesis by activated Ras signaling (Bannister et al., 1991).
A variety of mechanisms attenuate c-Jun–mediated activity (Schutte et al., 1989). Factors regulating c-jun stability inhibit c-Jun function through reducing its abundance. These factors include macrophage migration inhibitory factor (MIF) (Kleemann et al., 2000), E3 ligase Fbw 7-containing Skp/Culin/F-box protein complex (SCFFbw) (Gao et al., 2004), and the E3 ligase Itch (Nateri et al., 2004). Jab1 association with MIF inhibits Jab1-mediated AP-1 activity. Thus, the cytokine MIF serves to transduce cytokine signaling to nuclear c-Jun function. Components of the AP-1 complex itself can inhibit c-Jun activity. The AP-1 transcription factor family is composed of Jun, Fos, and ATF subunits, which interact through their leucine zipper motif and bind DNA through a basic region. Jun proteins form homo- and heterodimers. Several other basic proteins heterodimerize with c-Jun, including Maf (v-Maf, c-Maf, MafB, MafG, and MafK) proteins and the neural retina leucine zipper gene product 1 (Nr1), which may thereby regulate the activity of c-Jun (Angel and Karin, 1991). Finally, c-Jun–binding repressor proteins have been described. The c-Jun–interacting proteins JDP1 and JDP2 regulate UV-mediated apoptosis (Piu et al., 2001). Recent genome-wide interrogation identified an inhibitor of AP-1–responsive target genes as the DACH1 gene, which is known to be involved in compound eye development (Wu et al., 2003).
Development of the compound eye in Drosophila is governed by a regulatory network of genes. The eyeless, sine oculis, eyes absent, and dachshund genes are required for normal eye development, and ectopic expression of eyeless, eyes absent, and dachshund induce ectopic eye formation. This regulatory pathway, conserved among metazoans and the vertebrate homologues of eyeless (Pax6), sine oculis (Six1-6), eyes absent (Eya1-4), and dachshund (DACH1-2), contributes to organism development (Kawakami et al., 2000). The six genes encode a conserved Six and Homeo domain sequence-specific DNA binding family of transcription factors. Eya functions as a coactivator for Six proteins, and functions in a phosphatase-dependent manner (Lee et al., 2000a). The dachshund genes encode cointegrator proteins recruited to Six binding sites at the promoters of target genes that promote cellular differentiation and cell cycle exit. A conserved domain (dac and ski/sno domain 1), which has significant identity with Ski/Sno ([DS] domain), is conserved from Drosophila to the human (Chen et al., 1997; Davis et al., 1999; Kawakami et al., 2000). The DACH1 protein binds the CBP coactivator or interacts directly with corepressors to regulate transcriptional activity (Ikeda et al., 2002; Li et al., 2003). The recruitment of DACH1 to Six binding sites is regulated through mechanisms that have as yet to be defined.
In view of the finding that c-Jun plays a key role in oncogene-induced transformation and that DACH1 functions as an inhibitor of AP-1–responsive gene expression, we investigated the potential role for DACH1 in c-Jun–mediated cellular growth. DACH1 inhibited c-Jun-mediated DNA synthesis and contact-independent growth. AP-1 activity was repressed by DACH1 requiring a conserved ([DS]) domain. Repression of c-Jun transcription and transactivation by DACH1 required the DACH1 DS domain. DACH1 bound a corepressor complex, including HDAC1/HDAC3 and repressed c-Jun transactivation through the c-Jun δ domain. c-Jun transactivation and contact-independent growth is controlled by the cell fate determination factor DACH1.
MATERIALS AND METHODS
Plasmid Constructions
The expression plasmids that include an N-terminal FLAG peptide for DACH1, DACH1 DS-domain alone (DS), or DACH1 DS-domain deleted (ΔDS) were described previously (Wu et al., 2003). The FLAG-tagged DACH1 c-DNA was subcloned into the MSCV-IRES-GFP and pLRT vector. The full-length DACH1 cDNA was subcloned in frame with the VP16 activation domain in the pVP16 vector to form a DACH1–VP16 fusion protein. DACH1 short hairpin RNA (shRNA) in the pSM2 expression vector was purchased from Open Biosystems (Huntsville, AL) and subcloned into the LMP retroviral vector. The Ski cDNAs was subcloned into the 3x Flag-CMV7.1 vector (Sigma-Aldrich, St. Louis, MO). The expression vector encoding adenovirus-induced Cre expression or control viruses were described previously (Wang et al., 1995). The cyclin A promoter (Katabami et al., 2005) and stathmin promoter (Kinoshita et al., 2003) reporter genes were described previously. The wt c-fos-LUC, serum response element (SRE), or ternary complex factor (TCF) mutants (PM12 and PM18) were described previously (Wang et al., 1998). AP-1 Luc (3TPlux), c-Jun Luc and JunB Luc were described previously (Watanabe et al., 1996). The expression vectors for c-Jun linked to Gal4 or E2 and truncated c-Jun were described previously (Baichwal and Tjian, 1990; Baichwal et al., 1991). The Ha-Ras expression vector was described previously (Albanese et al., 1995).
Cell Culture, DNA Transfection, and Luciferase Assays
Cell culture, DNA transfection, and luciferase assays were performed as described previously (Fu et al., 2000, 2002, 2003). The NIH3T3, HEK293T, and MCF-7 cell line were cultured in DMEM supplemented with 10% fetal calf serum, 1% penicillin, and 1% streptomycin. c-junfl/fl 3T3 cells were derived from mouse embryo fibroblast from c-junfl/fl transgenic mouse (Zenz et al., 2003) by using the standard 3T3 protocol. Rat1a-J4 cells express c-Jun in a doxycycline-controlled manner (Katabami et al., 2005). Cells were plated at a density of 1 × 105 cells in a 24-well plate on the day before transfection with Superfect according to the manufacturer's protocol (QIAGEN, Valencia, CA). At least two different plasmid preparations of each construct were used. In cotransfection experiments, a dose response was determined in each experiment with 50 and 200 ng of expression vector and the promoter reporter plasmids (1 μg). Luciferase activity was normalized for transfection by using β-galactosidase reporters as an internal control. Luciferase assays were performed at room temperature by using an Autolumat LB 953 (Berthold Technologies, Bad Wildbad, Germany). The -fold effect was determined for 50–200 ng of expression vector with comparison made to the effect of the empty expression vector cassette and statistical analyses was performed using the Mann–Whitney U-test.
Small-interfering RNA (siRNA) Transfection, Western Blot, and Immunoprecipitation Assays
The Dach1 siRNA target sequence is 5′-AAAGTGGCTTCCTTTACGGTG. Control siRNA was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Dach1-specific siRNAs or control siRNAs (100 or 200 nM) were transfected following the Oligofectamine protocol (Invitrogen, Carlsbad, CA). Transfection efficiency was monitored by no-silencing fluorescein siRNA from QIAGEN. 3T3 cells were infected with DACH1 shRNA or control retrovirus and selected by puromycin. Western blot analysis using antibodies to c-Jun (Santa Cruz Biotechnology), cyclin A (Santa Cruz Biotechnology), β-PAK (Santa Cruz Biotechnology), E2 tag (Abcam, Cambridge, MA), FLAG tag (Sigma-Aldrich), and the loading control guanine dissociation inhibitor (GDI) was conducted as described previously (Lee et al., 2000b; Wu et al., 2003). HEK293T cells were used for the detection of protein–protein interaction in vivo and immunoprecipitation was conducted as described previously using an anti-hemagglutinin (HA) antibody and immunoblotting with antibodies to HDAC1 (SC-7872), HDAC3 (SC-17795), NCoR (SC-8994), p300 (SC-585), and c-Jun (SC-44 and SC-1694).
Cell Cycle and DNA Synthesis Analysis
Cell cycle parameters were determined using laser scanning cytometry. Cells were processed by standard methods by using propidium iodide staining of cell DNA. Each sample was analyzed by flow cytometry with a FACScan flow cytometer (BD Biosciences, Mansfield, MA) by using a 488-nm laser. Histograms were analyzed for cell cycle compartments using ModFit version 2.0 (Verity Software House, Topsham, ME). A minimum of 20,000 events was collected to maximize statistical validity of the compartmental analysis. DNA synthesis was analyzed by [3H]thymidine (TdR) incorporation. Cells (105) were plated into 24-well plate and cultured for 36 h. Then, 1 μCi of [3H]TdR was added to each well, and culture was continued for 2 h. Cells were washed and fixed before measuring incorporated [3H]TdR by liquid scintillation.
Cell Proliferation Assays
For the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay, NIH3T3 cells infected with MSCV-IRES-GFP, MSCV-DACH1-IRES-GFP, or MSCV-DACH1ΔDS-IRES-GFP were seeded into 96-well plates in normal growth medium, and cell growth was measured every day by MTT bromide assays. c-Jun–expressing Rat1a-J4 cells were plated in growth medium in either the presence or absence of 2 μg/ml doxycycline. To measure growth curve, cells were seeded into 12-well plates and serially counted for 5–6 d.
Colony Formation Assay
Rat1a-J4 cells (4.0 × 103) were plated in triplicate in 3 ml of 0.3% agarose (sea plaque) in complete growth medium in the presence or absence of 2 μg/ml doxycycline overlaid on a 0.5% agarose base, also in complete growth medium. Two weeks after incubation, colonies >50 μm in diameter were counted using an Omnicon 3600 image analysis system. The colonies were visualized after staining with 0.04% crystal violet in methanol for 1–2 h.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed as described previously (Fu et al., 2004). Polymerase chain reaction (PCR) primers were as follows: for murine cyclin A: forward, 5′-CCTCAGGCTCCCGCCCTGTAAGATTCC and reverse, 5′-TCAAGTAGCCCGCGACTATTGAATAT; and for murine cyclin D1 AP-1 site: forward, 5′-CCGGTGGTCTGGTTCCTGGA and reverse, 5′-CCCCGAAAATTCCAGCAACA. The cells were cross-linked with formaldehyde buffer for 10 min at 37°C, and the procedure was followed as described in Fu et al. (2004). Double ChIP assay was performed first using polyclonal c-Jun antibody, and then elution was diluted with 10X volume of immunoprecipitation (IP) buffer and then immune-precipitated with anti-FLAG M2 antibody following standard protocol.
RESULTS
c-Jun Is Required for DACH1-mediated Inhibition of Cellular Proliferation
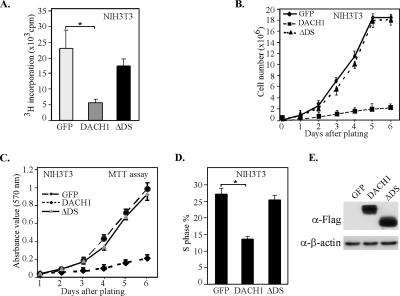
To determine the role of Dachshund1 in fibroblast cellular proliferation and growth, NIH3T3 cells were transduced with a retroviral expression plasmid encoding DACH1 linked to green fluorescent protein (GFP) through an internal ribosomal entry site. Homogeneously transduced populations of NIH3T3 cells were compared with cells transduced with an empty expression vector (MSCV-IRES-GFP). DACH1 expression inhibited tritiated thymidine uptake by 75% (Figure 1A). To determine the domain of DACH1 involved in cellular proliferation, a mutation of the conserved DS domain was assessed. NIH3T3 cells transduced with the mutant DACH1 protein, which was expressed in equal amounts by Western blotting, failed to inhibit tritiated thymidine uptake (Figure 1A). Cellular proliferation assays were conducted by serial counting of cell number (Figure 1B) and the MTT stain (Figure 1C). Cells transduced with DACH1 failed to proliferate, whereas cells transduced with either a mutant DACH1 deleted of the DS domain or the GFP vector alone proliferated with similar efficiency. Fluorescence-activated cell sorting (FACS) analysis of DACH1-transduced NIH3T3 cells demonstrated an inhibition of DNA synthesis that was dependent upon the DS domain (Figure 1D). Expression of the DACH1 mutant, DACH1ΔDS was similar to the DACH1 wild type as assessed by Western blot analysis of the FLAG epitope at the N terminus of each protein (Figure 1E).
Figure 1.
DACH1 inhibits cellular proliferation and DNA synthesis of NIH3T3 cells. (A) NIH3T3 cells were transfected with expression vectors encoding either DACH1, or a mutant of DACH1 deleting the DS domain (ΔDS) or a control vector (GFP). Forty-eight hours after transfection, tritiated thymidine incorporation was determined (*p < 0.01). The cell proliferation rate was assessed for NIH3T3 cells transduced with retroviral expression vectors encoding the GFP, DACH1, or ΔDS. Proliferation analyses were conducted by either counting cellular number (B) or MTT assay (C). (D) FACS analysis of NIH3T3 cells transduced with retroviral expression vectors. The proportion of cells in S phase is shown. Data are mean ± SEM of n = 6 separate experiments (*p < 0.01). (E) Western blot analysis to the FLAG-tag at the N terminus of DACH1 or DACH1ΔDS. β-Actin is used as a loading control showing similar level of expression of the wt and mutant DACH1 proteins.
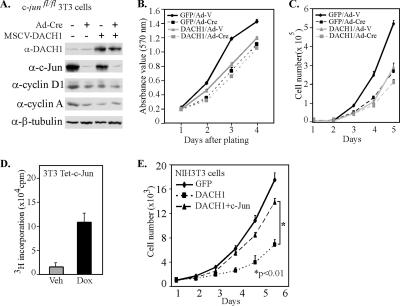
Previous studies had shown AP-1 reporter activity was inhibited by DACH1 (Wu et al., 2003). Endogenous AP-1 activity contributes to 3T3 cell proliferation. To examine the importance of c-Jun as an endogenous target of DACH1-mediated inhibition of DNA synthesis, we used c-jun–deficient 3T3 cells. c-jun−/− mice die in gestation from cardiovascular and hepatic defects (Eferl et al., 1999). Significant functional differences have been observed in mouse embryonic fibroblasts derived from embryonic stem cell knockout, compared with cells derived by acute target gene excision by using floxed alleles and Cre recombinase (Sage et al., 2003). We therefore derived c-junfl/fl 3T3 cells from c-junfl/fl mice (Zenz et al., 2003) by using a standard protocol (Todaro and Green, 1963). The c-junfl/fl 3T3 cells were transduced with Cre recombinase (Sage et al., 2003), and the deletion of c-Jun was confirmed by Western blot analysis (Figure 2A). These cells were transduced with either a DACH1 retroviral expression vector or equal amount of control viral vector, and Western blot analysis and cellular proliferation assays were conducted. c-jun deletion was associated with a reduction in cyclin D1 and cyclin A expression consistent with prior findings that c-jun induces cyclin D1 and cyclin A (Albanese et al., 1995). DACH1 expression in c-junfl/fl cells reduced cyclin D1 abundance 30%, and cyclin A expression 50% (Figure 2A). DACH1 expression in parental c-junfl/fl 3T3 cells reduced proliferation rates >35%. In cells deleted of c-jun (c-junfl/fl + Cre), expression of DACH1 failed to inhibit cellular proliferation (Figure 2B). A detailed time course was conducted to determine the role of c-jun in DACH1-mediated inhibition of cellular proliferation through daily counting of cells (Figure 2C). Cellular proliferation of wild-type cells was inhibited by DACH1 expression; however, the proliferation rate of cells deleted of c-jun was not affected by DACH1 expression (Figure 2C). To determine whether expression of c-Jun could overcome DACH1-mediated repression of cellular proliferation, a cell line encoding a doxycycline-inducible c-jun cDNA was used (3T3 Tet-c-Jun). The induction of c-Jun expression by the addition of doxycycline stimulated cellular proliferation (Figure 2D). Transduction of the NIH3T3 cells with DACH1 reduced cellular proliferation >60% at 6 d. Induction of c-Jun abundance, through the addition of doxycycline, restored cellular proliferation by >85% (Figure 2E). Together, these studies demonstrate DACH1 inhibition of cellular proliferation requires c-jun and can be overcome by c-Jun expression.
Figure 2.
DACH1 inhibition of cellular proliferation requires c-jun. (A) Western blot analysis of c-junfl/fl 3T3 cells transduced with adenovirus expressing Cre recombinase. Cells were transduced with the MSCV–DACH1 vector as indicated. Western blot analysis shows Ad-Cre excision of c-jun abrogated c-jun expression. Antibodies are to cyclin D1, cyclin A, and β-tubulin. Cellular proliferation assessed by MTT assay (B) and by cell counting of DACH1-transduced cells (C) shown in A. (D) Proliferation assay of 3T3 Tet-c-Jun cells, in which 2 μg/ml doxycycline for 48 h was used to induce c-Jun protein level. (E) NIH3T3 cells transduced with pLRT-GFP, pLRT-DACH1, or pLRT-DACH1 and pLRT-c-Jun expressing vector. Cellular number was determined daily for 6 d with 2 μg/ml doxycycline. c-Jun expression reversed DACH1-mediated inhibition of cellular proliferation (p < 0.01).
DACH1 Inhibits Colony Formation Requiring the Conserved DS Domain
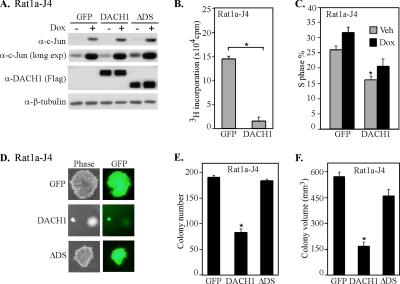
The expression of c-Jun is sufficient to induce contact-independent growth of Rat1a cells (Schutte et al., 1989). The stable cell line expressing doxycycline-inducible c-Jun was infected with an MSCV virus encoding either GFP, DACH1 wt or DACH1 mutant. Western blot analysis demonstrated the induction of c-Jun by doxycycline and transfected DACH1 protein was detected by an antibody directed to the N-terminal FLAG epitope (Figure 3A). DACH1 also inhibited basal proliferation of Rat1a-J4 cells (Figure 3B). Previous studies have demonstrated the importance of c-Jun in promoting cell cycle transition (Ryseck et al., 1988; Kovary and Bravo, 1991). The c-Jun–induced thymidine uptake, reflecting DNA synthesis, was attenuated by DACH1 expression (Figure 3C). Colony formation was induced by c-Jun expression (Figure 3D) as described previously (Katabami et al., 2005). Transduction of the inducible c-Jun stable cell line with a retroviral expression vector encoding DACH1 reduced the number of colonies by 40% (Figure 3E) and reduced the volume of individual colonies by >75% (Figure 3F). Expression of equal amount of a mutant DACH1 protein deleted of the DS domain (ΔDS) failed to inhibit c-Jun–induced contact-independent growth (Figure 3F). Together, these results suggest that DACH1 inhibits c-Jun induced cellular proliferation and blocks c-Jun–mediated contact-independent growth. These results further suggest c-Jun is a key molecular target of DACH1 inhibition of contact-independent growth.
Figure 3.
DACH1 inhibits c-Jun–mediated colony formation and cellular proliferation. (A) Rat1a cells stably expressing a doxycycline-inducible c-Jun cDNA were transduced with MSCV virus encoding either GFP, or DACH1 or ΔDS domain. Western blot analysis of doxycycline-treated cells (Dox+) show the induction of c-Jun protein and presence of DACH1 by using the Flag epitope to the N terminus of DACH1. β-Tubulin is a loading control. (B) Rat1a-HA-c-Jun cells were analyzed for cellular proliferation by tritiated thymidine incorporation after 48 h. (C) The DNA synthetic phase determined by FACS analysis in presence or absence of 2 μg/ml doxycycline. (D) Phase-contrast fluorescence microscopy of Rat1a-J4 cells infected with an MSCV vector encoding GFP, DACH1, or ΔDS in presence of 2 μg/ml doxycycline. Quantification of colony number (E) or colony volume (F) of Rat1a-J4 cells infected with MSCV vector encoding GFP, DACH1, or ΔDS analyzed after 14 d. Data represent the mean ± SEM of n = 6 separate experiments (*p < 0.01).
DACH1 Expression Inhibits Physiological Inducers of c-jun Expression and c-jun Target Gene Expression
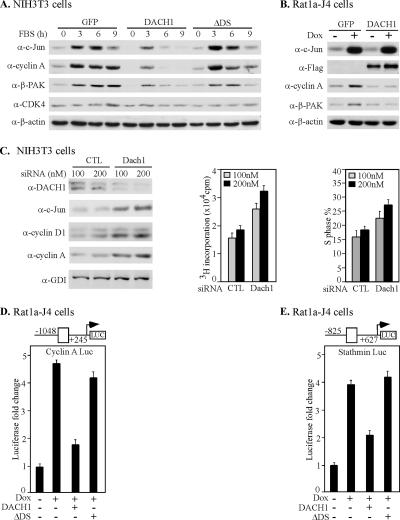
The c-jun gene is induced by diverse physiological stimuli, including serum and growth factors. To determine whether DACH1 expression was capable of inhibiting the induction of c-Jun by physiological stimuli, NIH3T3 cells were treated with serum. Cells were transduced with retroviral expression vectors encoding either DACH1 or the DACH1 mutant (ΔDS). Consistent with prior studies, serum induced c-Jun abundance approximately >10-fold. The c-Jun target genes cyclin A and β-PAK were also induced by serum (Figure 4A, lane l versus lane 2). NIH3T3 cells expressing DACH1 reduced serum-mediated expression of c-Jun by >50%. DACH1 expression reduced serum-induced expression of the c-Jun target genes cyclin A and β-PAK. Deletion of the DACH1 DS domain abrogated the DACH1-mediated inhibition of c-Jun, cyclin A, and β-PAK expression at 3 h. Expression of CDK4 and β-tubulin were unaffected by DACH1 expression, indicating the effect of DACH1, to inhibit c-Jun expression, is gene specific.
Figure 4.
DACH1 inhibits serum-induced expression of c-Jun and c-Jun target genes. (A) Western blot analysis of NIH3T3 transduced with MSCV-GFP, MSCV-DACH1 or MSCV-DACH1ΔDS. Cells were serum starved and then treated with serum for the time points indicated. Western blot, with the antibodies indicated, demonstrated DACH1 inhibits c-jun induction by serum. (B) The Rat1a-J4 cell line encoding tetracycline-inducible c-jun was treated with doxycycline for 48 h to induce c-Jun. Cells were transduced with retrovirus expressing GFP or DACH1. The M2 antibody detects the N-terminal FLAG-tag of DACH1. DACH1 inhibits c-Jun mediated induction of cyclin A and β-PAK. β-Actin is a loading control. (C) NIH3T3 cells were treated with DACH1 siRNA for 72 h. Western blot analysis was conducted with the antibodies indicated. Reduction in DACH1 abundance induces c-jun, and expression of the c-jun target genes cyclin D1 and cyclin A. [3H]Thymidine incorporation and percentage of cells in S phase is shown. The cyclin A (D) and stathmin promoter (E), linked to a luciferase reporter, were assessed in Rat1a-J4 cells. Cells were transduced with either expression vector for DACH1 or DACH1ΔDS. Data are shown as luciferase activity as mean ± SEM for n > 5 separate transfections.
To examine further the mechanism by which DACH1 repressed the c-jun target gene cyclin A, we deployed the tetracycline-inducible c-jun stable Rat1a-J4 cell line (Figure 4B). Doxycycline addition induced c-Jun abundance, associated with the induction of the c-jun target genes cyclin A and β-PAK. Transduction of Rat1a-J4 cells with DACH1 abrogated c-Jun-mediated induction of cyclin A and β-PAK, without affecting β-actin expression. Thus, DACH1 inhibits c-Jun–mediated induction of cyclin A. To determine whether endogenous DACH1 regulates c-Jun expression and c-Jun target gene expression, DACH1 siRNA was used. DACH1 siRNA reduced DACH1 abundance by ∼75% associated with 2.5-fold induction of c-Jun abundance (Figure 4C). The c-jun target gene cyclin D1 (Albanese et al., 1995) was induced threefold and cyclin A was induced two- to fivefold (Figure 4C). Consistent with a key role for cyclin D1 in 3T3 cell DNA synthesis (Albanese et al., 2003) the DACH1 siRNA-mediated induction of cyclin D1 was associated with an induction of DNA synthesis determined by [3H]thymidine uptake and S-phase distribution by FACS (Figure 4C).
To determine whether DACH1 directly regulated c-jun target genes, the cyclin A and stathmin promoters were assessed. Induction of c-Jun expression in the Rat1a-J4 cells by addition of doxycycline induced the activity of the cyclin A promoter activity fourfold (Figure 4D). Expression of DACH1 inhibited c-Jun–mediated induction of cyclin A promoter activity. Deletion of the DACH1 DS domain abolished the DACH1 repression function (Figure 4D). Similarly the stathmin promoter was induced by c-Jun, and c-Jun induction of the stathmin promoter was inhibited by DACH1, requiring the DACH1 DS domain (Figure 4E).
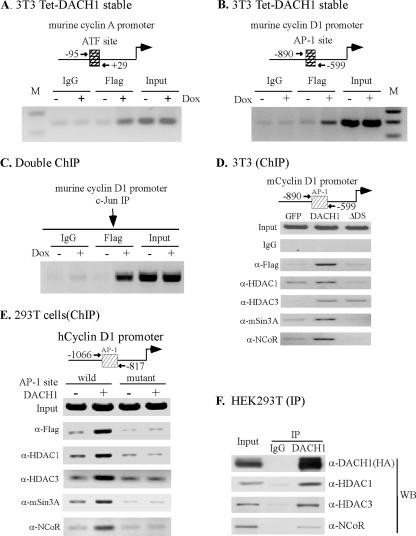
DACH1 Occupies the AP-1 Site of Endogenous c-Jun Target Genes in ChIP Assays
Because DACH1 repressed c-jun-mediated induction of several AP-1 target genes (Wu et al., 2003), we sought to determine whether DACH1 occupied AP-1 sites of endogenous genes in the context of their local chromatin structure. For these studies, NIH3T3 cells were stably transduced with a FLAG-tagged DACH1 expression vector. The cyclin A promoter, a known c-Jun target gene, was examined. Using oligonucleotides directed to the endogenous murine cyclin A promoter AP-1/ATF-1 site, ChIP assays, conducted with the anti-FLAG antibody, demonstrated the presence of DACH1 in the context of the local chromatin structure of the endogenous cyclin A promoter (Figure 5A). Control IgG did not result in similar amplification. Analysis of the endogenous murine cyclin D1 AP-1 site by using oligonucleotides directed to the murine promoter demonstrated the presences of DACH1 at the cyclin D1 promoter AP-1 site (Figure 5B). The complex formation of c-Jun and Dach1 protein at the AP-1 site of the cyclin D1 promoter was further confirmed by sequential immune-precipitation with c-Jun and FLAG antibodies (Figure 5C). Further analysis of the endogenous proteins associated with DACH1 at the murine cyclin D1 promoter was conducted. Oligonucleotides directed to the murine cyclin D1 AP-1 site evidenced the presence of the DACH1-associated corepressor proteins HDAC1, HDAC3, mSin3A, and NCoR recruited to the murine cyclin D1 promoter (Figure 5D). To determine the role of the cyclin D1 promoter AP-1 site in recruitment of DACH1, ChIP assays were conducted comparing the wild-type and mutant cyclin D1 AP-1 site promoters (Figure 5E). Expression of FLAG-tagged DACH1 enhanced recruitment of DACH1 (FLAG), together with HDAC1, HDAC3, mSin3A, and NCoR. Mutation of the cyclin D1 AP-1 site reduced recruitment of each of these proteins (Figure 5E). Immune-precipitation Western blot indicated these proteins form a cellular complex (Figure 5F).
Figure 5.
Chromatin immunoprecipitation assays demonstrate DACH1 within the AP-1 site of endogenous genes. ChIP assay conducted using oligonucleotides to the endogenous murine (A) cyclin A and (B) cyclin D1 promoter. Schematic representation of cyclin A and cyclin D1 promoter is shown. Amplification was conducted of products precipitated with indicated antibodies or control IgG. The cell line used is an NIH3T3 cell line expressing FLAG-tagged DACH1 under control of the Tet enhancer (Tet-DACH1). Doxycycline addition induced DACH1 protein abundance. A similar amount of DACH1 was used as a loading control. (C) The chromatin was first precipitated with antibody to c-Jun, and then the elution was diluted with IP buffer and immunoprecipitated with anti-FLAG. PCR amplification was conducted of the AP-1 site of the cyclin D1 promoter. (D) 3T3 cells were transduced with MSCV-GFP, MSCV-DACH1, and MSCV-DACH1ΔDS. ChIP assays were conducted of the endogenous murine cyclin D1 promoter AP-1 site with the indicated antibodies. The FLAG antibody is directed to the N-terminal tag of DACH1 or DACH1ΔDS. Equal amounts of DACH1 and DACH1ΔDS are detected by Western blot of the cells (data not shown). (E) ChIP assays of the transfected human cyclin D1 promoter, either wild type or mutant of the AP-1 binding site, was conducted using antibodies directed to endogenous HDAC1, HDAC3, mSin3A, and NCoR. (F) Immunoprecipitation Western blot analysis of DACH1 from HEK293T cells. The HA tagged DACH1 was used for immune-precipitation, with subsequent Western blot to the endogenous proteins indicated.
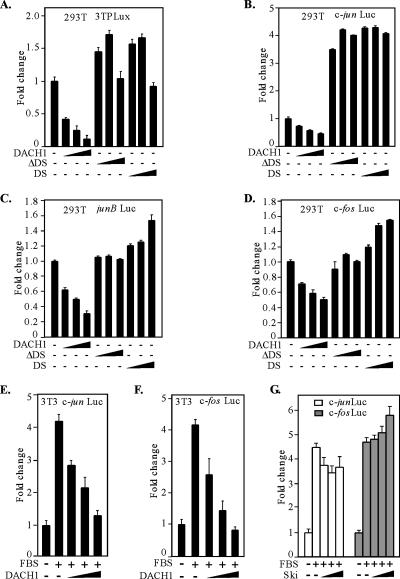
DACH1 Inhibition of AP-1 Family Members
To determine the domains of DACH1 involved in inhibiting c-Jun function, an AP-1–responsive reporter gene (3TP-LUX) was assessed in HEK293T cells. The activity of the AP-1 reporter gene was inhibited by DACH1 expression (Figure 6A). The deletion of the DACH1 DS domain abrogated repression of AP-1 reporter activity, and the DS domain alone was insufficient for repression of AP-1 activity (Figure 6A). We assessed the effect of DACH1 on the transcriptional activity of the key AP-1 genes, c-jun, junB, and c-fos. The transcriptional activity of the c-Jun promoter was repressed by DACH1 expression, and this transcriptional repression was abrogated by the deletion of the conserved DS domain (Figure 6B). Similarly, the junB and c-fos promoters were repressed by DACH1 expression, and the DS domain was required for repression (Figure 6, C and D).
Figure 6.
DACH1 inhibits serum-induced activation of AP-1 activity. (A) HEK293T cells were transfected with luciferase reporter plasmids encoding either a multimeric AP-1 site (A), the c-jun promoter (B), the junB promoter (C), or the c-fos promoter (D) and DACH1 mammalian expression vectors. A β-galactosidase report gene driven by a β-actin promoter was used to normalize the transfection deficiency. Data are shown as -fold change in luciferase activity. Data are mean ± SEM of n = 6 separate transfections. (E–G) 3T3 cells were transfected with a reporter plasmid encoding either the c-jun promoter (E) or c-fos promoter (F) and a DACH1 mammalian expression vector. Cells were serum starved and stimulated with 15% FBS for 6 h before the luciferase activity was determined. 3T3 cells were transfected with the luciferase reporter plasmid for the c-jun or c-fos promoter and a mammalian expression vector encoding Ski (G). Cells were treated as shown in E and F. The expression vector encoding Ski does not significantly reduce reporter activity. Data are mean ± SEM of n = 6 separate transfections.
AP-1 activity is serum inducible, and serum induction of the c-jun promoter was inhibited by DACH1 expression (Figure 6E). Similarly, the c-fos promoter induction by serum was inhibited by DACH1 expression (Figure 6F). DACH1 shares homology with Ski (Kozmik et al., 1999). To determine whether Ski functions as a transcriptional repressor of c-fos or c-jun promoter activity, serum induction experiments were conducted. Ski expression failed to repress serum-induced c-jun and c-fos promoter activity (Figure 6G). Thus, c-fos was repressed by DACH1 and not by Ski. Together, these results demonstrate distinct transcriptional repression profiles of DACH1 and the related Ski protein.
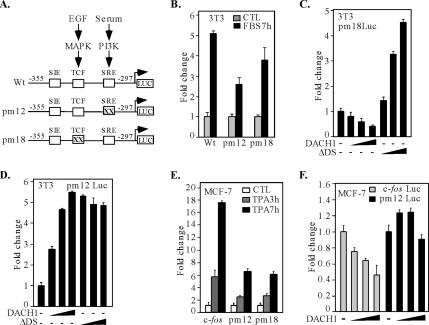
We examined further the DNA sequences of the c-fos promoter required for transcriptional repression by DACH1. A point mutant of the SCF binding site and the TCF binding site of the c-fos promoter were compared (Figure 7A). Consistent with previous findings (Hill et al., 1995), mutation of either the c-fos SRE or TCF binding site reduced serum-induced activation of the c-fos promoter (Figure 7B). The c-fos promoter encoding a point mutation of either the SRE or the TCF site was compared for repression by DACH1. DACH1 repressed activity of the c-fos promoter, encoding a mutation of the TCF site (c-fos PM18) (Figure 7C). In contrast, point mutation of the SRE binding site (c-fos PM12) abrogated repression by DACH1, inducing transactivation rather than repression by DACH1 (Figure 7D). These results demonstrate the SRE site of the c-fos promoter is a transcriptional target of DACH1 repression in 3T3 cells. To determine whether these effects were similar in other cell types, MCF-7 cells were assessed. The c-fos promoter was activated sixfold by 12-O-tetradecanoylphorbol-13-acetate (TPA) (Figure 7E). Point mutation of either the TCF or SRE site of the c-fos promoter reduced activation by TPA. Consistent with findings in NIH3T3 cells, DACH1 repressed the c-fos promoter and point mutation of the SRE site abrogated repression (Figure 7F). Collectively, these experiments demonstrate DACH1 inhibits the physiological induction of the promoters encoding AP-1 response genes, including c-jun, junB, and c-fos. The mechanism by which DACH1 inhibits gene expression is distinct from the related protein Ski.
Figure 7.
DACH1 inhibition of c-fos promoter activity requires the SRE, but not TCF site. (A) Schematic representation of c-fos promoter luciferase reporter vectors, indicating the TCF and SRE sites. (B) 3T3 cells were transiently transfected with a c-fos wild-type or mutant promoter reporter constructs and stimulated by serum for 7 h as shown. 3T3 cells were transiently transfected with c-fos promoter point mutant PM18 (C) or PM12 (D) luciferase reporter genes, and expression vectors encoding DACH1 wild-type or mutant. MCF-7 cells were transiently transfected with either wild-type or mutant c-fos promoter luciferase reporters and treated with TPA for 3 or 7 h (E) or cotransfected with a mammalian expression vector for DACH1 (F). All changes showing mean ± SEM of n = 6 separate transfections.
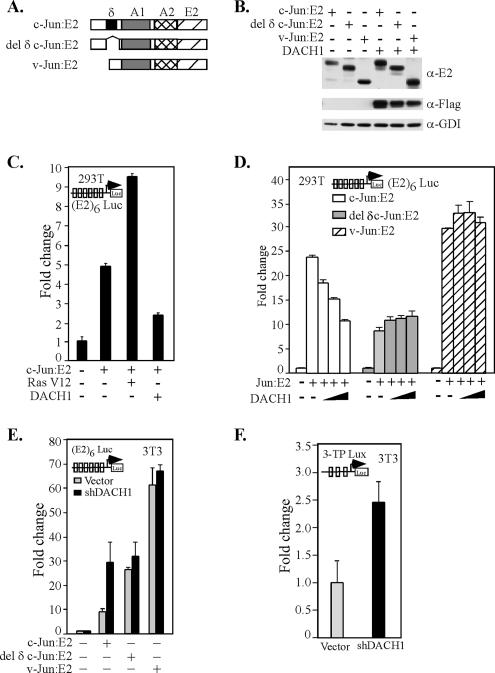
DACH1 Inhibition of c-Jun Transactivation Requires the δ Domain
To examine further the mechanisms by which DACH1 inhibited c-Jun transactivation, mammalian two-hybrid studies were undertaken. Comparison was made between either wild-type c-Jun or c-Jun proteins encoding either a deletion of the δ domain (del δ c-Jun) or the v-Jun protein. The v-Jun protein differs from the wild-type c-Jun primarily by the deletion of the δ domain. Previous studies with these expression vectors have demonstrated the presence of a c-Jun inhibitor functioning through the δ domain (Baichwal and Tjian, 1990). The heterologous transactivation domains of c-Jun linked to the E2 DNA binding domain were assessed using a multimeric E2 DNA binding domain, linked to a luciferase reporter gene (Figure 8A). The wild-type and mutant chimeric c-Jun proteins were expressed equally in DACH1-transfected cells (Figure 8B). Activity of the c-Jun protein was enhanced by oncogenic Ras (Ha-RasV12) consistent with previous findings that Ras alleviates repression of a c-Jun inhibitor (Baichwal et al., 1991). DACH1 expression inhibited c-Jun transactivation by Ha-RasV12 (Figure 8C). To determine the domains of c-Jun repressed by DACH1, comparison were made between c-Jun and the mutant c-Jun proteins. c-Jun E2 activity was repressed by DACH1 in a dose-dependent manner. In contrast, the del δ c-Jun and v-Jun activities were not repressed by DACH1 (Figure 8D). To determine the role of endogenous DACH1 as an inhibitor of c-jun transactivation function, DACH1 shRNA was used in 3T3 cells. The transactivation function of c-jun was enhanced by expression of DACH1 shRNA (Figure 8E). Activity of both the c-Jun protein deleted of the δ domain and v-Jun was constitutively more active than the corresponding c-Jun protein, and they were unaffected by depletion of DACH1 abundance (Figure 8E). Similarly, shRNA expression to DACH1 enhanced AP-1 activity using on AP-1–responsive reporter gene (Figure 8F). These findings suggest the δ domain of c-Jun is required for endogenous DACH1 repression of c-Jun transactivation activity.
Figure 8.
DACH1 inhibits c-Jun transactivation, requiring the δ domain. (A) Schematic representation of c-Jun expression vectors. (B) The HEK 293T cells transiently transfected with expression vectors encoding either wild-type or mutant c-Jun as an E2 fusion proteins or a DACH1 mammalian expression vector. Western blot analysis is shown for antibodies to either c-Jun (E2) or DACH1 (Flag). GDI is used as a protein loading control. (C and D) HEK293T cells were transiently transfected with a luciferase reporter encoding a multimeric E2 binding site linked to a luciferase reporter [(E2)6 LUC] and the mammalian expression vectors encoding either c-Jun-E2, Ha-Ras V12, or a DACH1 mammalian expression vector as indicated. Luciferase activity is shown as -fold change normalized to a β-galactosidase reporter gene used to normalize for transfection efficiency. Data are expressed as mean ± SEM for n = 7 separate transfections. (E) 3T3 cells were transfected with the vectors described in D, and the effect of shRNA to endogenous Dach1 was assessed. (F) Activity of an AP-1–responsive reporter gene was determined upon reduction of endogenous Dach1 through shRNA.
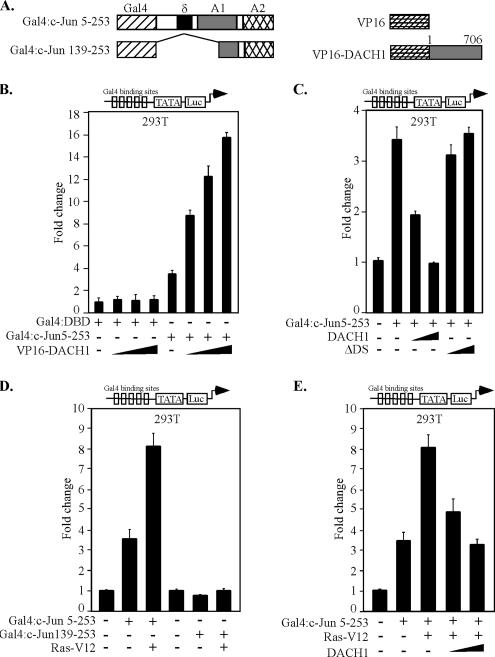
To examine further the mechanisms by which DACH1 repressed oncogenic Ras-induced activity of c-Jun, a series of Gal4-c-Jun expression vectors were used (Baichwal and Tjian, 1990) (Figure 9A). The interaction between DACH1 and c-Jun was assessed using a mammalian two-hybrid system, and activity of c-Jun was determined using a multimeric Gal4 DNA binding site linked to a luciferase reporter gene (Figure 9B). VP16-DACH1 demonstrated no functional interaction with the Gal4-DBD. Cointroduction of VP16-DACH1 enhanced transactivation with Gal4-c-Jun, indicating functional interaction between DACH1 and c-Jun and the induction of transactivation by the recruitment of the potent acidic transactivation surface of VP-16 to the minimal E1B promoter. Thus, DACH1 functionally interacts with c-Jun in a mammalian two-hybrid assay (Figure 9B). To determine the effect of DACH1 expression on c-Jun activity, DACH1 was coexpressed with an expression vector encoding Gal4-c-Jun 5-253 and c-Jun activity determined using luciferase reporter activity. DACH1 repressed Gal4-c-Jun activity (Figure 9C). The DACH1 mutant ΔDS, however, was incapable of repressing Gal4-c-Jun activity (Figure 9C). These findings are consistent with the previous studies, in which DACH1 inhibition of AP-1 activity required the DS domain. The activity of Gal4-c-Jun was enhanced by oncogenic Ras, however, deletion of amino acids 1-139 abrogated Ras-induced transactivation (Figure 9D), consistent with previous findings (Baichwal et al., 1991). DACH1 inhibited c-Jun activation by Ras (Figure 9E). Together, these results demonstrate, first, that DACH1 inhibits oncogenic Ras-induced transactivation of c-Jun; second, that DACH1 associates with and inhibits c-Jun activity; third, that physical association of DACH1 with c-Jun is necessary but insufficient for repression; and fourth, that the c-Jun δ domain is required for DACH1 repression.
Figure 9.
DACH1 and c-Jun interact in mammalian two-hybrid system. (A) Schematic representation of c-Jun protein linked to a Gal4 binding domain or DACH1, linked to the VP16 transactivation domain. HEK293T cells were transfected with a multimeric luciferase reporter gene UAS5 E1B TATA-LUC and mammalian expression vectors encoding the Gal4 DNA-binding domain linked to c-Jun with VP16-DACH1 (B) or DACH1 (C). HEK293T cells were transiently transfected with a luciferase reporter UAS5 E1BTATA-LUC and mammalian expression vectors for c-Jun, Ras V12, or DACH1 as shown (D–E). Data are mean ± SEM of n = 7 separate transfections normalized to the β-galactosidase for transfection efficiency.
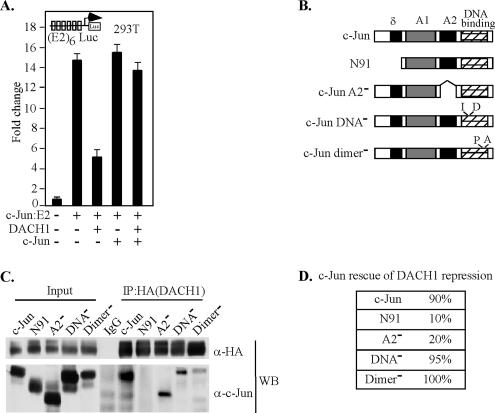
To examine further the mechanisms by which DACH1 represses c-Jun, we compared the properties of DACH1-mediated repression of c-Jun, with the properties of a previously described inhibitor of c-Jun. Previous studies using in vivo competition assays identified an inhibitor of c-Jun activity (Baichwal and Tjian, 1990; Baichwal et al., 1991). In these prior studies, coexpression of the mammalian expression vector encoding either the wild-type or mutant c-Jun was used to identify the domains of c-Jun required for interaction with the δ domain inhibitory factor (Baichwal and Tjian, 1990; Baichwal et al., 1991). In our experiments, the ability of DACH1 to repress activity of c-Jun-E2 was overcome by c-Jun expression in trans (Figure 10A, lane 3 versus lane 5). To determine the domains of c-Jun capable of binding DACH1, immunoprecipitation Western blot analysis were conducted using truncated or mutant c-Jun (Figure 10B). The c-Jun mutants were detectable with a c-Jun antibody directed to the carboxy terminus of c-Jun. Immunoprecipitation of DACH1 with an antibody directed to the HA epitope of DACH1 coprecipitated c-Jun. Deletion of the N-terminal 91 amino acids of c-Jun abolished binding of DACH1 to c-Jun. A c-Jun protein deleted of the A2 domain or mutated of either the DNA binding or dimerization domains remained capable of binding DACH1 (Figure 10C). Mutated of either the DNA binding domain or the dimerization domain, each rescued DACH1 repression. But deletion of the amino-terminal 91 amino acids (N91) and A2 domain resulted in a mutant that failed to overcome DACH1 repression (Figure 10D). Together, these findings are consistent with a model in which DACH1 repression involves an interaction in trans, which requires the c-Jun δ and A2 domains but is independent of the c-Jun dimerization and DNA binding domains. DACH1 binding to c-Jun is insufficient for repression as DACH1 bound a c-Jun mutant deleted of the A2 domain; yet, this mutant failed to rescue DACH1 repression in trans. Together, these findings indicated the c-Jun δ and A2 domains interact in trans to mediate DACH1 repression of c-Jun.
Figure 10.
DACH1 repression of c-Jun activity is rescued in trans by c-Jun requiring the N terminus. (A) HEK293T cells were transiently transfected with a multimeric E2 binding site linked to a luciferase reporter gene (E2)6-LUC and mammalian expression vectors encoding c-Jun-E2, DACH1 or c-Jun. (B) Schematic representation of mammalian c-Jun expression vectors. (C) Western blot analysis of HEK293T cells transiently transfected with c-Jun wild-type or mutant expression vectors with DACH1. HA epitope of DACH1 expression vector was used to immunoprecipitate DACH1. Coprecipitation of c-Jun is shown using a c-Jun–specific antibody. (D) Rescue of DACH1 repression by mammalian c-Jun mutant expression vectors. Rescue is shown as a percentage of wild-type c-Jun for each mutant. Data are mean ± SEM of n = 6.
DISCUSSION
In the current study, DACH1 inhibited DNA synthesis and cellular proliferation in 3T3 cells. Deletion of the c-Jun gene by using c-junfl/fl 3T3 cells and Cre recombinase demonstrated a requirement for c-Jun in DACH1-mediated inhibition of cellular proliferation. c-Jun overexpression rescued DACH1-mediated inhibition of DNA synthesis. c-Jun promotes G1 phase progression and c-Jun is necessary for G0/G1- and S-phase progression (Ryseck et al., 1988; Kovary and Bravo, 1991). The finding that DACH1 inhibited the DNA synthetic phase of the cell cycle is consistent with the known role of c-Jun to promote G1/S-phase transition. The mechanism by which DACH1 inhibited c-Jun expression involved inhibition of AP-1 activity. c-Fos and c-Jun, which contribute to AP-1 activity, were transcriptionally repressed by DACH1. c-Jun abundance is regulated in part by c-Jun gene transcription (Angel et al., 1988; Treier et al., 1994; Fuchs et al., 1996; Musti et al., 1997). The c-Jun promoter, which is autoregulated by AP-1 activity mediated through an AP-1 binding site (Angel et al., 1988; Kayahara et al., 2005) was repressed by DACH1. Thus, c-Jun expression and transcription were both inhibited by DACH1.
The current study identifies DACH1 as a c-Jun repressor protein, extending prior observations that c-Jun binds JDP1 and JDP2 (Piu et al., 2001). Recent studies using genome-wide microarray analysis suggested a potential new mechanism controlling AP-1 activity (Wu et al., 2003). In these studies, inducible expression of the cell fate determination factor DACH1 inhibited expression of the genes governing cellular migration, adhesion, and growth, many of which are AP-1 responsive (Wu et al., 2003). The abundance and activity of c-Jun is a critical determinant of tumor progression and contact independent growth. Compelling evidence for a key role of c-Jun in tumor progression was the finding that disruption of the c-jun gene in murine hepatocytes prevents the emergence of hepatocellular carcinoma (Eferl et al., 2003). DACH1 repression of c-Jun expression provides an additional mechanism controlling c-Jun abundance and function. DACH1 protein is lost during tumor progression, providing a mechanism by which c-Jun activity may be increased, contributing to contact-independent growth. Thus, the current study suggests an additional mechanism by which c-Jun transformation function is kept in check through the endogenous inhibitory protein DACH1.
c-Jun is sufficient for the induction of anchorage-independent growth of Rat1a cells (Schutte et al., 1989; Katabami et al., 2005). Herein, c-Jun–induced contact-independent growth was blocked by the cell fate determination factor DACH1. DACH1 inhibition of c-Jun–induced cellular growth required a conserved domain (DS domain). c-Jun acts in conjunction with Ras-V12 to transform rodent fibroblasts (Alani et al., 1991; Binetruy et al., 1991). c-Jun is required for transformation by several collaborative oncogenes, including Ras, c-Fos, Raf, c-Myc, Mos, and Abl (Rapp et al., 1994; Johnson et al., 1996). The finding that DACH1 blocks c-Jun function raises the possibility that DACH1 may inhibit multiple distinct oncogenic signaling pathways that converge on c-Jun. The mechanism by which c-Jun induces cellular growth and tumor progression is not well understood (Maeda and Karin, 2003). It has been proposed that c-Jun contributes to the onset and progression of tumorigenesis, in part through c-Jun inhibition of apoptosis via a p53-dependent cellular pathway (Eferl et al., 2003). The defect in proliferation of c-jun–deficient cells has been attributed to elevated expression of p53 and p21CIP1, and several downstream targets of c-Jun are required, but not sufficient, for induction of contact-independent growth, including stathmin, HMG 1/Y, and cyclin A (Kinoshita et al., 2003; Hommura et al., 2004; Katabami et al., 2005). DACH1 inhibited the physiological induction by serum of the c-Jun target genes cyclin A and β-PAK. The expression of c-Jun with a doxycycline-inducible stable cell line (Rat1a-J4) induced expression of cyclin A. DACH1 inhibited c-Jun induction of cyclin A expression. DACH1 inhibited c-Jun–mediated induction of several known endogenous c-Jun target genes, including cyclin A and stathmin; and DACH1 was identified within the context of local chromatin of the endogenous AP-1 sites at the known c-Jun–responsive promoter of cyclin A and cyclin D1. DACH1 siRNA reduced DACH1 abundance and induced expression of the c-Jun responsive genes c-Jun and cyclin D1. Collectively, these studies demonstrate that DACH1 is a physiological regulator of endogenous c-Jun function, inhibiting c-Jun expression and c-Jun target gene expression.
The binding and inhibition of c-Jun transactivation by DACH1 but not the structurally related Ski protein, suggests these proteins have evolved to regulate distinct subsets of transcriptional responses. Herein, the interaction of DACH1 with c-Jun was evidenced by mammalian two-hybrid and by immunoprecipitation Western blotting. DACH1 inhibited c-Jun transactivation when c-Jun was linked to a heterologous DNA binding domain. Deletion of the DS domain of DACH1 abrogated interaction with c-Jun. Similarly, deletion of the c-Jun N-terminal amino acids 1-91 abrogated binding of c-Jun to DACH1. Coexpression of c-Jun titrated the inhibitory function of DACH1, consistent with the model in which the relative abundance of c-Jun and DACH1 determined c-Jun transactivation.
In the current studies, DACH1 repression of c-Jun required the c-Jun δ domain. Activity of the v-Jun E2 fusion protein, which deletes the δ domain, was resistant to DACH1 repression. The c-Jun δ domain engages cell-specific inhibitors of c-Jun (Baichwal and Tjian, 1990). Putative inhibitors of this domain include catalytically inactive JNK (Dai et al., 1995; Ljungdahl et al., 1997). JNK binds and phosphorylates c-Jun at serine 63 and 73. The role of the δ domain itself versus JNK phosphorylation within the N terminus of c-Jun for cellular DNA synthesis and transformation remains controversial. C-Jun promotion of G1 phase progression seems to be independent of its phosphorylation (Wisdom et al., 1999). The JNK binding site of c-Jun can also be uncoupled from its transformation capabilities in same studies (Sprowles and Wisdom, 2003). Thus, mutation of the JNK phosphorylation sites did not affect the transformation activity of c-Jun in chicken embryo fibroblasts (Vogt, 2001). Furthermore, JNK-dependent phosphorylation of the c-Jun N terminus may not be required for Ras-induced cellular transformation (Johnson et al., 1996; Kennedy et al., 2003). In contrast with these studies, mice and cells encoding a mutant allele of c-jun in which the JNK phosphorylation site has been mutated demonstrated an important role for these residues in c-jun–dependent transformation (Behrens et al., 2000). Furthermore, although mutation of serine 63 and 73 in v-Jun did not alter transforming activity of v-jun, mutation of these residues in c-jun reduced cooperation with Ha-Ras in oncogenic transformation (Binetruy et al., 1991). The current study is consistent with a role for DACH1 as an inhibitor of the c-Jun δ domain.
The transactivation potential of c-Jun and its oncogenic activity in cooperation with Ras are often correlated (Alani et al., 1991; Smeal et al., 1991), although exceptions exist in avian and yeast systems (Vogt, 2001). In the current study, DACH1 inhibited c-Jun–mediated transactivation. DACH1 binding to the N-terminal domain of c-Jun may repress c-Jun activity by recruiting a corepressor or interfering with coactivator binding. CBP binds to the N-terminal region of c-Jun (Bannister et al., 1991; Mayr and Montminy, 2001) as does RH2/Gu-RNA-helicase (Westermarck et al., 2002). The corecruitment of histone deacetylase proteins HDAC1 and HDAC3 by DACH1, to c-Jun and thereby to an AP-1 binding site, provides one mechanism by which DACH1 may regulate AP-1 signaling. Recent studies suggested that the c-Jun δ domain may interact with a yet unidentified protein that recruits HDAC3 (Weiss et al., 2003). The relative importance of DACH1 in recruitment of histone deacetylase to c-Jun remains to be further explored.
Herein, DACH1 repressed the c-fos promoter. DACH1 transcriptional repression was abolished by point mutation of the SRE site. In the current study, serum-induced c-fos promoter activity was reduced by point mutation in either the TCF or SRE binding site. Extracellular stimuli enhance TCF transcriptional activity through the MAPK family of extracellular regulated kinases (extracellular signal-regulated kinases, JNKs, and p38s), which phosphorylate the TCF transactivation domain (Whitmarsh et al., 1995, 1997). The SRF site of the c-fos promoter is induced by RhoA independently of MAPKs (Sotiropoulos et al., 1999; Miralles et al., 2003). RhoA induction of SRF involves ROCK and phosphorylation of LIM kinase (Hill et al., 1995). Activation of SRF involves a decrease in the cellular pool of monomeric actin (Sotiropoulos et al., 1999). LIM kinase-dependent phosphorylation of cofilin induces the stabilization of polymerized actin (F-actin), which is sensed by SRF to induce its activity (Sotiropoulos et al., 1999; Miralles et al., 2003). DACH1 regulates cytoskeletal proteins and their function as evidenced by genome-wide analysis of molecular genetic targets (Wu et al., 2003). Although speculative, the finding that DACH1 inhibits c-fos promoter activity through the SRF site is consistent with a role for DACH1 in regulating a RhoA/ROCK/LIM kinase cytoskeletal signaling pathway.
ACKNOWLEDGMENTS
We thank Dawn Scardino and Almeta Mathis for manuscript preparation. This work was supported in part by awards from R01CA70896, R01CA75503, and R01CA86072 (to R.G.P.). Work conducted at the Kimmel Cancer Center was supported by the National Institutes of Health Cancer Center Core Grant P30CA56036 (to R.G.P.). This project is funded in part from the Dr. Ralph and Marian C. Falk Medical Research Trust and a grant from the Pennsylvania Department of Health (to R.G.P.). The Department specifically disclaims responsibility for any analysis, interpretations or conclusions. A.C. was supported by National Institutes of Health Grant EY12200, the Research to Prevent Blindness Career Development Award, and American Cancer Society Junior Faculty Institutional Award.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0793) on December 20, 2006.
REFERENCES
- Alani R., Brown P., Binetruy B., Dosaka H., Rosenberg R. K., Angel P., Karin M., Birrer M. J. The transactivating domain of the c-Jun proto-oncoprotein is required for cotransformation of rat embryo cells. Mol. Cell Biol. 1991;11:6286–6295. doi: 10.1128/mcb.11.12.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese C., Johnson J., Watanabe G., Eklund N., Vu D., Arnold A., Pestell R. G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Albanese C., et al. IKKalpha Regulates Mitogenic Signaling through Transcriptional Induction of Cyclin D1 via Tcf. Mol. Biol. Cell. 2003;14:585–599. doi: 10.1091/mbc.02-06-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Hattori K., Smeal T., Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- Angel P., Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- Baichwal V. R., Park A., Tjian R. v-Src and EJ Ras alleviate repression of c-Jun by a cell-specific inhibitor. Nature. 1991;352:165–168. doi: 10.1038/352165a0. [DOI] [PubMed] [Google Scholar]
- Baichwal V. R., Tjian R. Control of c-Jun activity by interaction of a cell-specific inhibitor with regulatory domain delta: differences between v- and c-Jun. Cell. 1990;63:815–825. doi: 10.1016/0092-8674(90)90147-7. [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Cook A., Kouzarides T. In vitro DNA binding activity of Fos/Jun and BZLF1 but not C/EBP is affected by redox changes. Oncogene. 1991;6:1243–1250. [PubMed] [Google Scholar]
- Behrens A., Jochum W., Sibilia M., Wagner E. F. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19:2657–2663. doi: 10.1038/sj.onc.1203603. [DOI] [PubMed] [Google Scholar]
- Binetruy B., Smeal T., Karin M. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature. 1991;351:122–127. doi: 10.1038/351122a0. [DOI] [PubMed] [Google Scholar]
- Bohmann D., Tjian R. Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell. 1989;59:709–717. doi: 10.1016/0092-8674(89)90017-2. [DOI] [PubMed] [Google Scholar]
- Chen R., Amouri M., Zhang Z., Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Dai T., Rubie E., Franklin C. C., Kraft A., Gillespie D. A., Avruch J., Kyriakis J. M., Woodgett J. R. Stress-activated protein kinases bind directly to the delta domain of c-Jun in resting cells: implications for repression of c-Jun function. Oncogene. 1995;10:849–855. [PubMed] [Google Scholar]
- Davis R. J., Shen W., Heanue T. A., Mardon G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev. Genes Evol. 1999;209:526–536. doi: 10.1007/s004270050285. [DOI] [PubMed] [Google Scholar]
- Eferl R., Ricci R., Kenner L., Zenz R., David J. P., Rath M., Wagner E. F. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- Eferl R., Sibilia M., Hilberg F., Fuchsbichler A., Kufferath I., Guertl B., Zenz R., Wagner E. F., Zatloukal K. Functions of c-Jun in liver and heart development. J. Cell Biol. 1999;145:1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M., Rao M., Wu K., Wang C., Zhang X., Hessien M., Yeung Y. G., Gioeli D., Weber M. J., Pestell R. G. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J. Biol. Chem. 2004;279:29436–29449. doi: 10.1074/jbc.M313466200. [DOI] [PubMed] [Google Scholar]
- Fu M., Wang C., Reutens A. T., Wang J., Angeletti R. H., Siconolfi-Baez L., Ogryzko V., Avantaggiati M. L., Pestell R. G. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- Fu M., et al. Acetylation of the androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol. Cell Biol. 2003;23:8563–8575. doi: 10.1128/MCB.23.23.8563-8575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M., et al. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell Biol. 2002;22:3373–3388. doi: 10.1128/MCB.22.10.3373-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S. Y., Dolan L., Davis R. J., Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Wynne J., Treisman R. The Rho family of Rho A, Rac 1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Hommura F., Katabami M., Leaner V. D., Donninger H., Sumter T. F., Resar L. M., Birrer M. J. HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. Mol. Cancer Res. 2004;2:305–314. [PubMed] [Google Scholar]
- Ikeda K., Watanabe Y., Sno H., Ohto I., Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum W., Passegue E., Wagner E. F. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- Johnson R., Spiegelman B., Hanahan D., Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol. Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki P., Jenkinson S., Edelman G. M., Jones F. S. Silencer elements modulate the expression of the gene for the neuron-glia cell adhesion molecule, Ng-CAM. J. Biol. Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- Kallunki T., Deng T., Hibi M., Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- Karin M., Liu Z., Zandi E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Katabami M., Donninger H., Hommura F., Leaner V. D., Kinoshita I., Chick J. F., Birrer M. J. Cyclin A is a c-Jun target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. J. Biol. Chem. 2005;280:16728–16738. doi: 10.1074/jbc.M413892200. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Sato S., Ozaki H., Ikeda K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kayahara M., Wang X., Tournier C. Selective regulation of c-jun gene expression by mitogen-activated protein kinases via the 12-O-tetradecanoylphorbol-13-acetate-responsive element and myocyte enhancer factor 2 binding sites. Mol. Cell Biol. 2005;25:3784–3792. doi: 10.1128/MCB.25.9.3784-3792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N. J., Sluss H. K., Jones S. N., Bar-Sagi D., Flavell R. A., Davis R. J. Suppression of Ras-stimulated transformation by the JNK signal transduction pathway. Genes Dev. 2003;17:629–637. doi: 10.1101/gad.1062903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita I., Leaner V., Katabami M., Manzano R. G., Dent P., Sabichi A., Birrer M. J. Identification of cJun-responsive genes in Rat-1a cells using multiple techniques: increased expression of stathmin is necessary for c-Jun-mediated anchorage-independent growth. Oncogene. 2003;22:2710–2722. doi: 10.1038/sj.onc.1206371. [DOI] [PubMed] [Google Scholar]
- Kleemann R., et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–216. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- Kovary K., Bravo R. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z., Pfeffer P., Kralova J., Paces J., Paces V., Kalousova A., Cvekl A. Molecular cloning and expression of the human and mouse homologues of the Drosophila dachshund gene. Dev. Genes Evol. 1999;209:537–545. doi: 10.1007/s004270050286. [DOI] [PubMed] [Google Scholar]
- Lee J.-S., Collins K. M., Brown A. L., Lee C.-H., Chung J. H. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000a;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- Lee R. J., et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell Biol. 2000b;20:672–683. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Ljungdahl S., Linder S., Sollerbrant K., Svensson C., Shoshan M. C. Signal transduction in fibroblasts stably transformed by [Val12]Ras–the activities of extracellular-signal-regulated kinase and Jun N-terminal kinase are only moderately increased, and the activity of the delta-inhibitor of c-Jun is not alleviated. Eur. J. Biochem. 1997;249:648–656. doi: 10.1111/j.1432-1033.1997.t01-1-00648.x. [DOI] [PubMed] [Google Scholar]
- Maeda S., Karin M. Oncogene at last–c-Jun promotes liver cancer in mice. Cancer Cell. 2003;3:102–104. doi: 10.1016/s1535-6108(03)00025-4. [DOI] [PubMed] [Google Scholar]
- Mayr B., Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A. I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Morton S., Davis R. J., McLaren A., Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musti A. M., Treier M., Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- Nateri A. S., Riera-Sans L., Da Costa C., Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- Piu F., Aronheim A., Katz S., Karin M. AP-1 repressor protein JDP-2, inhibition of UV-mediated apoptosis through p53 down-regulation. Mol. Cell Biol. 2001;21:3012–3024. doi: 10.1128/MCB.21.9.3012-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp U. R., Troppmair J., Beck T., Birrer M. J. Transformation by Raf and other oncogenes renders cells differentially sensitive to growth inhibition by a dominant negative c-jun mutant. Oncogene. 1994;9:3493–3498. [PubMed] [Google Scholar]
- Ryseck R.-P., Hirai S. I., Yaniv M., Bravo R. Transcriptional activation of c-jun during the Go/G1 transition in mouse fibroblasts. Nature. 1988;334:535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- Sage J., Miller A. L., Perez-Mancera P. A., Wysocki J. M., Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Schreiber M., Kolbus A., Piu F., Szabowski A., Mohle-Steinlein U., Tian J., Karin M., Angel P., Wagner E. F. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte J., Viallet J., Nau M., Segal S., Federiko J., Minna J. JunB inhibits and c-fos stimulates the transforming and transactivating activities of c-jun. Cell. 1989;59:987–997. doi: 10.1016/0092-8674(89)90755-1. [DOI] [PubMed] [Google Scholar]
- Smeal T., Binetruy B., Mercola D. A., Birrer M., Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991;354:494–494. doi: 10.1038/354494a0. [DOI] [PubMed] [Google Scholar]
- Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Sprowles A., Wisdom R. Oncogenic effect of delta deletion in v-Jun does not result from uncoupling Jun from JNK signaling. Oncogene. 2003;22:498–506. doi: 10.1038/sj.onc.1206165. [DOI] [PubMed] [Google Scholar]
- Todaro G. J., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-jun degradation in vivo is mediated by the δ domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Vogt P. K. Jun, the oncoprotein. Oncogene. 2001;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- Wang P., Anton M., Graham F. L., Bacchetti S. High frequency recombination between loxP sites in human chromosomes mediated by an adenovirus vector expressing Cre recombinase. Somat. Cell Mol. Genet. 1995;21:429–441. doi: 10.1007/BF02310209. [DOI] [PubMed] [Google Scholar]
- Wang Y., Falasca M., Schlessinger J., Malstrom S., Tsichlis P., Settleman J., Hu W., Lim B., Prywes R. Activation of the c-fos serum response element by phosphatidyl inositol 3-kinase and rho pathways in HeLa cells. Cell Growth Differ. 1998;9:513–522. [PubMed] [Google Scholar]
- Watanabe G., Lee R. J., Albanese C., Rainey W. E., Batlle D., Pestell R. G. Angiotensin II (AII) activation of cyclin D1-dependent kinase activity. J. Biol. Chem. 1996;271:22570–22577. doi: 10.1074/jbc.271.37.22570. [DOI] [PubMed] [Google Scholar]
- Weiss C., Schneider S., Wagner E. F., Zhang X., Seto E., Bohmann D. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 2003;22:3686–3695. doi: 10.1093/emboj/cdg364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermarck J., et al. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 2002;21:451–460. doi: 10.1093/emboj/21.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh A. J., Shore P., Sharrocks A. D., Davis R. J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A. J., Yang S. H., Su M. S., Sharrocks A. D., Davis R. J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol. Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R., Johnson R. S., Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., et al. DACH1 inhibits TGF-beta signaling through binding Smad4. J. Biol. Chem. 2003;278:51673–51684. doi: 10.1074/jbc.M310021200. [DOI] [PubMed] [Google Scholar]
- Zenz R., Scheuch H., Martin P., Frank C., Eferl R., Kenner L., Sibilia M., Wagner E. F. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev. Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]