Abstract
Autophagy is a catabolic membrane-trafficking mechanism conserved in all eukaryotic cells. In addition to the nonselective transport of bulk cytosol, autophagy is responsible for efficient delivery of the vacuolar enzyme Ape1 precursor (prApe1) in the budding yeast Saccharomyces cerevisiae, suggesting the presence of a prApe1 sorting machinery. Sequential interactions between Atg19-Atg11 and Atg19-Atg8 pairs are thought responsible for targeting prApe1 to the vesicle formation site, the preautophagosomal structure (PAS), and loading it into transport vesicles, respectively. However, the different patterns of prApe1 transport defect seen in the atg11Δ and atg19Δ strains seem to be incompatible with this model. Here we report that prApe1 could not be targeted to the PAS and failed to be delivered into the vacuole in atg8Δ atg11Δ double knockout cells regardless of the nutrient conditions. We postulate that Atg19 mediates a dual interaction prApe1-sorting mechanism through independent, instead of sequential, interactions with Atg11 and Atg8. In addition, to efficiently deliver prApe1 to the vacuole, a proper interaction between Atg11 and Atg9 is indispensable. We speculate that Atg11 may elicit a cargo-loading signal and induce Atg9 shuttling to a specific PAS site, where Atg9 relays the signal and recruits other Atg proteins to induce vesicle formation.
INTRODUCTION
Autophagy, as a membrane-trafficking mechanism conserved in all eukaryotic cells, is essential to the survival of unicellular organisms in nutrient starvation conditions. Through a double-membrane–bound vesicle, called an autophagosome, autophagy mediates the degradation of cytosolic components in the lysosome/vacuole and recycles basic building blocks for new macromolecular synthesis or to serve as energy sources (Onodera and Ohsumi, 2005; Yorimitsu and Klionsky, 2005b). Because nutrient starvation stimulates autophagosome formation and the contents of autophagosomes resemble their surrounding cytoplasm, autophagy has long been considered to be a nonselective catabolic pathway for bulk cytosol remodeling. Recent studies, however, have provided evidence for selective cargo transport by autophagosomes. For example, intracellular infectious pathogens are enveloped by autophagosomes and killed upon fusion of the vesicles with lysosomes (Gutierrez et al., 2004; Nakagawa et al., 2004; Ogawa et al., 2005). In autophagy-deficient neuronal cells, intracellular protein aggregates accumulate, which may interfere with neural function and eventually lead to neurodegeneration (Hara et al., 2006; Komatsu et al., 2006). Moreover, autophagy selectively eliminates dysfunctional mitochondria to reduce the production of reactive oxygen species (ROS) and apoptosis (Gu et al., 2004). All these autophagic functions rely on specific signal transduction events and precise cargo-sorting mechanisms to load autophagosomes with appropriate cytosolic materials for elimination. Deciphering these mechanisms in molecular detail is critical to the control of autophagy-related human diseases, but our current knowledge on these issues is still limited. Thus, it is important to study the process of autophagy in an experimentally more amenable model system, such as yeasts.
In the budding yeast Saccharomyces cerevisiae, several examples of selective autophagy to mediate vacuolar delivery of specific cargo have been reported. By switching cells to glucose medium, the numerous peroxisomes induced by culturing cells in lipid-containing medium become superfluous and are targeted to the vacuole for degradation via the pexophagy pathway (Hutchins et al., 1999; Dunn et al., 2005). The vacuolar enzyme aminopeptidase I (Ape1) and α-mannosidase (Ams1) are selectively delivered by starvation-induced autophagosomes (Hutchins and Klionsky, 2001; Huang and Klionsky, 2002). When cells are cultured in nutrient-rich growth conditions, precursors of Ape1 (prApe1) is transported by another mechanism, named the cytoplasm-to-vacuole targeting (Cvt) pathway, a selective trafficking pathway that is topologically similar to autophagy and relies on most of the same molecular machinery. In fact, prApe1 is known to induce Cvt vesicle formation in growing cells (Shintani and Klionsky, 2004). Therefore, the Cvt pathway of budding yeast may better mimic the process of mammalian selective autophagy because cells in a multicellular organism may not face significant nutrient starvation conditions, and mammalian selective autophagy induced by deleterious cytosolic components is likely not associated with starvation signals. Studies on the induction signaling and cargo-sorting molecular mechanisms for the Cvt pathway will provide insight for future work on the mammalian system.
Mutant screens have identified several gene products required for efficient import of prApe1 into the vacuole. Because most of these mutants are also defective in execution of starvation-induced general autophagy, they are named autophagy-related (ATG) genes (Klionsky et al., 2003). Examining the characteristics of prApe1 and the phenotypes of atg mutant strains, has led to our current understanding on prApe1 import mechanism as summarized below. After synthesis in the cytosol, prApe1 quickly forms a homo-dodecamer and then further assembles into a larger Cvt complex (Kim et al., 1997). Atg19, serving as the receptor, binds to the propeptide of prApe1 and mediates targeting of the Cvt complex to the perivacuolar vesicle formation site, the preautophagosomal structure (PAS; Scott et al., 2001; Shintani et al., 2002). Most Atg proteins are then recruited to, and regulate vesicle formation from, the PAS (Suzuki et al., 2001; Kim et al., 2002). Finally, completely assembled vesicles encapsulate and transport prApe1 together with its receptor Atg19 to the vacuole. In growing ape1Δ and atg19Δ cells, Atg proteins are inefficiently recruited to the PAS, indicating that the PAS-targeted prApe1-Atg19 complex likely elicits a signal to regulate the PAS and Cvt vesicle formation. Consequently, identifying the component that interacts with the Cvt complex at the PAS is critical to unveil the molecular mechanism of the Cvt pathway and selective autophagy regulation.
In an atg11Δ strain, the prApe1-Atg19 complex is found away from the perivacuolar PAS, which results in a block of prApe1 transport, whereas general autophagy induction by starvation is not affected (Kim et al., 2001b; Yorimitsu and Klionsky, 2005a). ATG11 encodes a large protein with four potential coiled-coil (CC) domains. The Atg11 C terminus, including its fourth CC domain (CC4), is responsible for interaction with Atg19. Other parts of Atg11 are involved in homo-oligomer formation or interaction with other Atg proteins. The exact timing and subcellular locations of these interactions are not known, but Atg11 and its interacting partners are seen colocalized at the PAS. These Atg11 features and the atg11Δ strain phenotypes indicate its critical role in delivering prApe1-Atg19 to the PAS and subsequently initiating a signal for Cvt vesicle formation. An interaction partner for Atg11 is likely located at the PAS to dock with and transduce signals from the incoming cargo complex, but the identity of such a component is still not clear. Among those PAS-associated Atg proteins, Atg9 is the only one with transmembrane domains (Noda et al., 2000). Atg9 recruits Atg2 and Atg18 to the PAS (Wang et al., 2001; Reggiori et al., 2004). While fulfilling its autophagy regulatory function, Atg9 cycles between subcellular punctate compartments and the PAS. Although the exact role of Atg9 cycling is still a question, this cycling behavior is blocked and Atg9 accumulates at the PAS in atg1Δ and several other atg mutant strains. Interestingly, Atg9 is not restricted to the PAS in growing atg1Δ ape1Δ double knockout cells (Shintani and Klionsky, 2004). These data suggest that Atg9 may participate in an early event for loading prApe1 cargo or integrating signals for control of vesicle formation at the PAS.
In this present study, we have characterized an Atg19-mediated dual interaction prApe1 sorting mechanism and found Atg9 as the partner for interaction with Atg11 to relay the cargo-loading signal at the PAS. The domains required for Atg11-Atg9 interaction have been mapped. Properties of Atg11 and Atg9 variants, which lost interaction with each other, indicate the critical role for a proper Atg11-Atg9 interaction to initiate Cvt vesicle formation. In addition, Atg9 distribution patterns seen in different mutant backgrounds suggest that in the absence of the prApe1 cargo, Atg11 still affects the appearance of multiple perivacuolar PAS sites in starved cells.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae yeast strains used in this study are listed in Table 1. For gene disruptions, the entire coding region was replaced with either the Escherichia coli kanr, Schizosaccharomyces pombe HIS5, or S. cerevisiae TRP1 genes using PCR primers containing ∼40 bases of identity to the regions flanking the open reading frame (ORF). Yeast cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or synthetic medium (SD; 0.67% yeast nitrogen base without amino acids, 2% glucose, auxotrophic amino acids, and vitamins if necessary). For nitrogen starvation, SD-N medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, and 2% glucose) was used.
Table 1.
Yeast strains used in this study
| Strain | Descriptive name | Genotype | Source |
|---|---|---|---|
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | Robinson et al. (1988) | |
| WHY1 | atg1Δ | SEY6210 atg1Δ::HIS5 S.p. | Shintani et al. (2002) |
| WPHYD7 | atg8Δ | SEY6210 atg8Δ::LEU2 | Kim et al. (2001) |
| JKY007 | atg9Δ | SEY6210 atg9Δ::HIS3 | Noda et al. (2000) |
| AHY001 | atg11Δ | SEY6210 atg11Δ::HIS3 | Kim et al. (2001) |
| SSY31 | atg19Δ | SEY6210 atg19Δ::HIS5 S.p. | Scott et al. (2001) |
| WHY2 | atg1Δ ape1Δ | SEY6210 atg1Δ::HIS5 S.p. ape1Δ::LEU2 | Shintani et al. (2002) |
| WHY3 | atg8Δ atg11Δ | SEY6210 atg8Δ::URA3 atg11Δ::HIS3 | Shintani et al. (2002) |
| WHY6 | atg9Δ atg11Δ | SEY6210 atg9Δ::TRP1 atg11Δ::HIS5 S.p. | This study |
| WHY22 | atg11Δ atg19Δ | SEY6210 atg11Δ::HIS5 S.p. atg19Δ::LEU2 | This study |
| TN124 | MATα leu2-3,112 ura3-52 trp1 pho8Δ::PHO8Δ60 pho13Δ::LEU2 | Noda et al. (1995) | |
| WHY28 | atg11Δ | TN124 atg11Δ::TRP1 | This study |
| WHY29 | atg19Δ | TN124 atg19Δ::KAN | This study |
| WHY30 | atg11Δ atg19Δ | TN124 atg11Δ::TRP1 atg19Δ::KAN | This study |
| TN125 | MATa ade2 his3 leu2 lys2 trp1 ura3 pho8Δ::PHO8Δ60 | Noda et al. (1995) | |
| CYY2 | atg9Δ | TN125 atg9Δ::KAN | This study |
| PJ69-4A | MATa his3-200 leu2-3, 112 trp1-901 ura3-52 gal4Δ gal80ΔGAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | James et al. (1996) | |
| WHY10 | atg9Δ | PJ69-4A atg9Δ::TRP1 | This study |
| WHY19 | atg11Δ | PJ69-4A atg11Δ::TRP1 | This study |
Plasmids
For the two-hybrid analysis, we used the system developed by James et al. (1996). The prey plasmid pGAD-ATG9 or its truncated versions were constructed by ligating the corresponding ATG9 ORF fragments into XmaI and PstI sites of pGAD-C3. The bait plasmid pGBDU-ATG11 or truncated versions were constructed by inserting the full-length or the truncated ATG11 ORF into the BamHI and SalI sites of pGBDU-C3. ATG9 or the truncated ATG9 ORF were generated by PCR and ligated into XmaI and XhoI sites of pCuProtA, pCu3xHA, pCuRFP, and pCuGFP. The PCR products of the truncated ATG11 ORF were inserted into the SpeI and KpnI restriction sites of pRS424. For the plasmid expressing GFP-Ape1, the APE1 ORF was amplified by PCR and ligated into XmaI and ClaI sites of pCuGFP. The DNA fragment encoding RFP was inserted into the SpeI and XmaI sites of pCu414-CVT9 to generate a plasmid expressing the RFP-Atg11 fusion protein. Plasmids for expressing GFP-Atg5 and GFP-Atg8 have been described elsewhere (George et al., 2000; Kim et al., 2001a).
Protein A Pulldown
The cells harboring plasmids expressing protein A, ProtA-Atg9, or ProtA-truncated Atg9 and myc-Atg11 or truncated Atg11 were converted into spheroplasts and then lysed with lysis buffer (20 mM HEPES-KOH, pH 6.8, 150 mM KOAc, 5 mM MgOAc, 250 mM sorbitol, 0.5%Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], Complete EDTA-free protease inhibitor; Roche Diagnostics, Alameda, CA). After being centrifuged at 800 rpm for 5 min, the cell lysate was incubated with human IgG–coated Dynabeads (Invitrogen, Carlsbad, CA) at 4°C for 3 h. The resultant protein complexes were eluted by SDS-PAGE sample buffer and analyzed by immunoblotting with anti-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or rabbit PAP (DAKO, Carpinteria, CA).
Alkaline Phosphatase Assay
The system we used was adapted from a previous report (Noda et al., 1995). Briefly, cells starved for 4 h were harvested and lysed with lysis buffer (20 mM PIPES, pH 6.8, 50 mM KCl, 100 mM KOAc, 10 mM MgSO4, 10 μM ZnSO4, 1 mM PMSF, and 0.5% Triton X-100) by glass beads. Autophagic activity was estimated with p-nitrophenol phosphate (p-NPP) as substrate. Protein concentration was measured by Pierce BCA Assay (Pierce Chemical, Rockford, IL).
Analysis of Pexophagy Activity
The degradation of peroxisomes was determined by the loss of thiolase (Fox3) as described previously (Hutchins et al., 1999).
Fluorescence Microscopy
Yeasts cells expressing fluorescent proteins were grown to midlog phase and shifted to SD-N medium as needed. For labeling vacuolar membrane, the cells were incubated with 0.8 μM N-(triethylammoniumpropyl)-4-(p-diethylaminophenylhexatri-enyl) pyridinium (FM 4-64) at 30°C for 20 min. After being washed by medium once, the cells were incubated in YPD at 30°C for 30–45 min and collected for observation.
RESULTS
Atg19 Mediates a Dual Interaction prApe1-sorting Mechanism
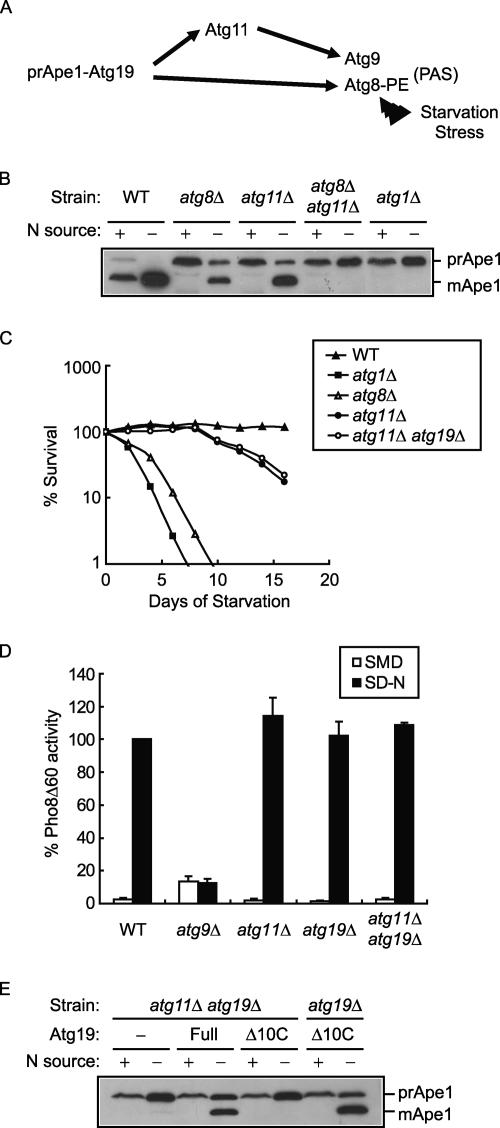
Current information on the molecular mechanism of prApe1 sorting indicates that Atg19 facilitates this process by first associating with prApe1 to form a prApe1-Atg19 Cvt complex, and then sequentially interacting with Atg11 and Atg8 to load the Cvt complex into transport vesicles (Scott et al., 2001; Shintani et al., 2002). Mutations in any one of these components should inevitably result in blockage of prApe1 import. However, paradoxically, this prApe1 import defect is partially reversed in nutrient starvation–treated atg8Δ and atg11Δ cells, whereas an atg19Δ strain does not show this reversal phenotype (Abeliovich et al., 2000; Kim et al., 2001b). These data indicate that only Atg19, but not its two interaction partners, is essential for prApe1 transport by autophagosomes. Because the Atg8 expression level and its conjugation to phosphatidylethanolamine (PE) is increased under starvation stress (Huang et al., 2000; Ichimura et al., 2000), we suspected that interaction between Atg19 and increased levels of Atg8-PE could bypass the Atg11-dependent event and cause the reversal phenotype of an atg11Δ strain. As a consequence, we proposed that by independent, instead of sequential, interaction with Atg11 and Atg8, Atg19 mediates prApe1 sorting by a dual interaction mechanism (Figure 1A).
Figure 1.
Atg19 mediates a dual interaction prApe1 sorting mechanism. (A) Model. Atg19 independently interacts with Atg11 and Atg8 to facilitate prApe1 sorting. Atg11 binds with Atg9 to ensure efficient targeting prApe1 to the PAS in growing cells, whereas starvation stress induces Atg8-PE level and partially bypasses the Atg11-dependent event. (B) Double mutation in atg8Δ and atg11Δ shows synergistic effect in blocking prApe1 transport. Total cell extracts from midlog phase growing cells or overnight nitrogen-starved cells were resolved by SDS-PAGE followed by immunoblotting with anti-Ape1 antiserum. Sample loadings for starved cells were empirically decreased to see similar intensities of Ape1 signals. (C) The atg11Δ atg19Δ double mutant strain survives long-term nitrogen starvation stress. Midlog phase growing cells in YPD medium were collected and shifted to nitrogen starvation SD-N medium for further culture. At the indicated day, viability was determined by plating aliquots of the culture on YPD plates and counting the number of colonies after 2 d growth. (D) The atg11Δ atg19Δ double mutant strain shows normal autophagy induction by short-term starvation. Midlog phase growing cells were shifted from SMD to SD-N medium for 4 h. Autophagy was measured by the levels of PhoΔ60 activity in whole-cell protein extracts. Activity in starved wild-type strain was set to 100%, and activity in the other conditions was normalized accordingly. Error bars, ±SD from three separate experiments. (E) Atg19Δ10C-expressing atg11Δ atg19Δ cells do not reverse prApe1 transport defect after starvation. Corresponding cells were handled as described in B.
To test this dual interaction sorting proposal, we first examined whether atg8Δ atg11Δ double knockout cells showed a synergistic effect in blocking the reversal phenotype of the prApe1 import defect. After overnight incubation the corresponding mutants in nitrogen starvation medium, both atg8Δ and atg11Δ cells generated a significant amount of mApe1, whereas atg8Δ atg11Δ double knockout cells still had a complete block in prApe1 import (Figure 1B). Because atg1Δ cells also did not generate mApe1 by this prolonged starvation treatment, transport by autophagosomes was likely responsible for the reversal phenotype, and this result seemed to support our proposal. However, an atg8Δ strain is known to have limited autophagy activity and assembles abnormally small autophagosomes (Abeliovich et al., 2000). This atg8 defect in combination with the atg11 mutation may account for the synergistic effect we observed instead of indicating a direct involvement of Atg8 in prApe1 sorting by interaction with Atg19. To exclude this possibility, we sought to re-examine this phenomenon in a genetic background with normal autophagy functions.
Our strategy to conduct this experiment relied on the fact that a 10-residue deletion from the carboxy terminus of Atg19 (Atg19Δ10C) is enough to block its interaction with Atg8 (Shintani et al., 2002). To use Atg19Δ10C for our purpose, we generated an atg11Δ atg19Δ double knockout strain. Expressing Atg19Δ10C in this strain should eliminate the Atg19-Atg11 and Atg19-Atg8 interactions without compromising general autophagy execution. The ability of this atg11Δ atg19Δ strain to induce autophagy after long- and short-term starvation was first confirmed by analyzing survival and Pho8Δ60 activity, respectively. The survival of the double knockout cells under nitrogen starvation conditions was similar to that seen with the atg11Δ cells (Figure 1C). Cytosolically accumulated Pho8Δ60 was delivered to, and activated in, the vacuoles of atg11Δ atg19Δ cells as efficiently as wild-type or either single mutant strains (Figure 1D). These two assay results indicated normal autophagy activity for the atg11Δ atg19Δ double knockout strain. Expression of full-length Atg19 complemented the atg19 defect of atg11Δ atg19Δ cells and reversed their prApe1 import defect after starvation treatment (Figure 1E). Atg19Δ10C also supported the reversal phenotype in the atg19Δ background as expected. The atg11Δ atg19Δ cells expressing Atg19Δ10C did not generate any mApe1, in agreement with our proposal of the dual interaction sorting mechanism. To our knowledge, these results were the first to clearly demonstrate the function of Atg8, a Cvt and autophagosomal vesicle membrane component, in facilitating transport cargo selection.
Atg19 Facilitates Targeting prApe1 to the PAS by Interaction with Atg11 and Atg8
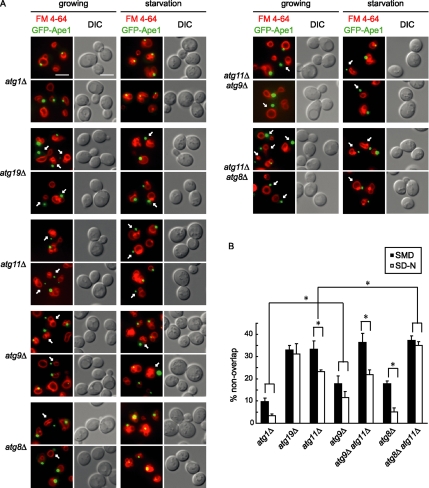
Targeting the prApe1-Atg19 complex to the vesicle formation PAS is a prerequisite for its vacuolar delivery. Despite studies showing that this event is regulated by Atg11, mApe1 is still generated in starved atg11Δ cells, which indicates that partial targeting of prApe1 to the autophagosome formation site in the absence of Atg11 is possible. We next studied the phenotypes of prApe1 PAS targeting to evaluate our proposal of the dual interaction sorting mechanism. A plasmid expressing prApe1 with green fluorescent protein (GFP) tagged to its amino terminus was prepared and the vacuolar delivery of this GFP-Ape1 was confirmed in wild-type cells, suggesting its normal distribution as other researchers observed (Supplementary Figure 1). The plasmid was introduced to different mutant strains to mark their prApe1 cargo complex. The vacuole membrane in those cells was labeled with the lipophilic dye FM 4-64. Because the PAS site is close to the vacuole, overlapping of GFP and FM 4-64 fluorescence signals was proposed as an indication of proper prApe1 PAS targeting. The majority of atg1Δ cells showed efficient targeting of prApe1 to the PAS in either growing or starvation conditions, whereas separated GFP and FM 4-64 signals were detected in a significant percentage of a population of atg19Δ cells, in agreement with its receptor function for prApe1 transport (Figure 2, A and B). For a population of growing atg11Δ cells, the percentage of cells with a targeting error was about the same as that of the atg19Δ strain. Starvation treatment of atg11Δ cells, however, decreased the severity of this defect. Counting the number of cells with nonoverlapping GFP and FM 4-64 signals indicated that the reversal phenotype was statistically significant (Figure 2B). The sorting defect in an atg8Δ background was less severe than in atg11Δ cells, and starvation stress significantly decreased the cells with separated signals. In accord with our proposal, when both the Atg8 and Atg11 branches of the prApe1 sorting pathway were blocked, the percentage of atg8Δ atg11Δ double knockout cells with a sorting error was found to be similar to an atg19Δ strain, and starvation stress did not decrease the sorting error (Figure 2B).
Figure 2.
Both Atg11 and Atg8 contribute to target prApe1 to the PAS. (A) Representative images of prApe1 targeting in different autophagy mutant backgrounds. Expression of GFP-Ape1 was driven by a plasmid. Cells of corresponding strains were grown to midlog phase, labeled with FM 4-64, and immediately examined (SMD) or after additional culturing in nitrogen starvation medium for 2–3 h (SD-N) by fluorescence microscopy. The GFP signals were shown in green and FM 4-64 signals in red. Scale bar, 5 μm. (B) Quantitative results of prApe1-sorting errors. The percentages of cells with separated GFP and FM 4-64 fluorescence signals were counted from at least 200 cells for each condition. Values reported are the means of three independent determinations ± SD. *p < 0.0001 represents statistical differences between groups.
Delivery of the prApe1-Atg19 cargo to the PAS is proposed as an induction signal to stimulate Cvt vesicle formation, but how this induction is achieved is still unknown. Because Atg11 regulates PAS targeting of prApe1-Atg19 and interacts with several Atg proteins, it is a good candidate to elicit and relay a cargo-loading signal and induce vesicle formation (Yorimitsu and Klionsky, 2005a). To mediate these functions, a partner protein localized at the PAS was proposed to interact with this incoming prApe1-Atg19-Atg11 complex. We have found an interaction between Atg11 and Atg9 (see the next section), and proposed that Atg9 served as the counterpart to interact with Atg11 at the PAS. The prApe1 targeting defect in atg9Δ cells was hence examined. Although this strain did not cause as severe a defect as seen with the atg11 mutation, cells with both atg9 and atg11 mutations (atg9Δ atg11Δ) still lessened the prApe1-sorting error after starvation treatment (Figure 2, A and B), in agreement with the idea that they are in the same branch of Atg19-Atg11 mediated prApe1 sorting.
Atg9 Physically Interacts with Atg11
Despite the fact that starvation-induced Atg8 contributes to prApe1 sorting, the defect in atg8Δ cells was much weaker than that seen in atg19Δ and atg11Δ strains (Figure 2, A and B), indicating the critical role of Atg11 in this process. We next sought to identify the Atg11 interaction partner at the PAS to evaluate its involvement in targeting the prApe1-Atg19-Atg11 complex. Because Atg9 is an integral membrane protein and its shuttling to the PAS requires the presence of properly targeted prApe1, we considered it a good candidate to accept or relay cargo-loading signals at the PAS (Noda et al., 2000; Shintani and Klionsky, 2004). To test its possible interaction with Atg11, we applied human IgG–coated magnetic beads to affinity purify bacterial protein A fragment–tagged Atg9 fusion proteins (ProtA-Atg9) from atg1Δ cells. Proteins associated with the PAS were subjected to immunoblotting analyses with antisera against autophagy regulatory proteins. Atg11 was indeed found coisolated with ProtA-Atg9 (Figure 3, B and D). The interaction between Atg9 and Atg11 is likely to be direct because Atg11 was seen in the pulldown sample from detergent Triton X-100–treated protein extract, whereas prApe1 and several other Atg proteins were not (data not shown). These results exclude the possibility of indirect coisolation of Atg11 with the Atg9-localized PAS as a whole entity. Previous studies have found four potential CC domains of Atg11, which are involved in homo-oligomerization or interaction with other autophagy regulatory proteins, including Atg1, Atg17, and Atg20 (Yorimitsu and Klionsky, 2005a). Atg9 is also known to interact with Atg2 and Atg18 (Wang et al., 2001; Reggiori et al., 2004). We have confirmed that the coisolation of Atg11 with ProtA-Atg9 is not affected by deletion of atg2, atg17, atg18, or atg20 (Supplementary Figure 2). Our pulldown experimental results identified another unreported Atg11 interaction partner, Atg9.
Figure 3.
Atg11 interacts with Atg9 and mapping of the binding domains. (A and B) The CC1 and CC2 domains of Atg11 are required for interaction with Atg9. (A) A schematic of Atg11 indicates the location of the CC domains and the exact cloning sites for the different Atg11 variant constructs. The two-hybrid atg11Δ strain was transformed with plasmids to express AD-Atg9 and the indicated BD-Atg11 variants. Transformants were grown 2 d at 30°C on plates lacking histidine with 3 mM of 3-amino-1,2,4-triazole (3-AT). Those transformants grew on test plates were indicated with + signals. (B) Protein extracts of atg1Δ cells expressing ProtA-Atg9 and the indicated myc-tagged Atg11 variants were incubated with human IgG–coated Dynabeads. Coisolated proteins were subjected to SDS-PAGE followed by immunoblotting with anti-myc antibody and PAP reagents for detecting protein A–tagged Atg9. Degradation products of myc-Atg11 were labeled with an asterisk (*). (C and D) The Atg9 residues 154-201 are required for interaction with Atg11. (C) A schematic of Atg9 indicates the location of the cloning sites for the different Atg9 variant constructs. An atg9Δ two-hybrid test strain was used for this study. The two-hybrid assay was conducted as described in A. (D) The affinity pulldown assay confirms the Atg11-binding domain in Atg9. Cells of an atg1Δ strain harboring an empty protein A vector (vector) or different protein A–tagged Atg9 variants were prepared for this study. Experimental procedures were as described in B.
To map the Atg11 domain required for interaction with Atg9, we conducted a yeast two-hybrid analysis. Atg11 fused with the Gal4 DNA binding domain (BD-Atg11) and Gal4 activation domain fused Atg9 (AD-Atg9) recapitulated the specific interaction in an atg11Δ two-hybrid test strain despite the fact that Atg9 is a membrane protein with multiple transmembrane domains (Figure 3, A and C). The atg11Δ background was chosen for this study to prevent any possible interference caused by endogenous full-length Atg11. To further characterize the Atg11 domain responsible for this interaction, several truncated variants of Atg11 were prepared (Figure 3A). The constructs were confirmed to express the desired Atg11 variants by an immunoblotting analysis probed with antiserum against the Gal4 BD domain (data not shown). The amino terminal half of Atg11 (BD-Atg11ΔCC3-4) retained the ability to interact with AD-Atg9, but the carboxy terminus of Atg11 (BD-Atg11ΔCC1-3) did not support cell growth on a test plate without histidine (Figure 3 A). Our construct of the Atg11 amino terminus contained the first and second CC domains (CC1 and CC2). Deletion of either one of them (BD-Atg11ΔCC1 and BD-Atg11ΔCC2) inhibited cell growth on a test plate, indicating that both the CC1 and CC2 of Atg11 are required for interaction with Atg9.
The two-hybrid results were further confirmed by affinity pulldown experiments. ProtA-Atg9 was coexpressed with amino terminus myc-tagged full-length or truncated Atg11 proteins by introducing corresponding low-copy plasmids to atg1Δ cells. Cell extracts were subjected to affinity purify ProtA-Atg9 together with its associated proteins. Consistent with the two-hybrid results, both full-length and the amino terminus of myc-tagged Atg11 were coisolated with ProtA-Atg9, whereas truncation of either the CC1 or CC2 domain compromised Atg11 and Atg9 interaction (Figure 3B).
The Atg9 protein is predicted to have six to eight central-located membrane-spanning segments with large hydrophilic domains at both termini (Noda et al., 2000). We have constructed a series of truncated ATG9 plasmids for a two-hybrid assay to identify the Atg11-interacting domain (Figure 3C). All the constructs supported more or less similar expression levels of the corresponding AD-Atg9 variants (data not shown). Deletion of 200 residues from the carboxy terminus (AD-Atg9Δ200C) did not affect its interaction with BD-Atg11 (Figure 3C). Neither did 152 residues removed from the Atg9 amino terminus (AD-Atg9Δ152N) inhibit Atg9-Atg11 interaction. Further removing an additional 49 residues from the amino terminus (AD-Atg9Δ201N), however, blocked interaction. A plasmid expressing AD-Atg9 with residues 154-201 truncated (AD-Atg9Δ154-201) also prevented cell growth on a test plate, suggesting that this fragment is critical for recognition with Atg11.
The conclusion drawn from the two-hybrid assay was then confirmed by affinity purification experiments. Low-copy plasmids that express protein A fragment–tagged full-length or truncated Atg9 proteins were individually introduced into atg1Δ cells for evaluating their interaction with myc-Atg11. All of the ProtA-Atg9 proteins were first demonstrated to be efficiently isolated by human IgG-coated magnetic beads (Figure 3D). Unexpectedly, ProtA-Atg9Δ154N and -Atg9Δ201N variants were found migrating faster than ProtA-Atg9Δ200C in a gel, which seemed to not correlate with the number of residues deleted in the corresponding variants. To ensure that the plasmids drive the expression of the desired Atg9 proteins, their ATG9 coding regions were confirmed by sequencing. We suspected that this unusual mobility associated with different Atg9 variants was due to different amino acid compositions at the two termini. Full-length (ProtA-Atg9) and the Atg9 variant with 200 residues truncated from the carboxy terminus (ProtA-Atg9Δ200C) efficiently coisolated myc-Atg11 (Figure 3D). Atg9 with residues 154-201 truncated (ProtA-Atg9Δ154-201) lost the ability to coprecipitate myc-Atg11, which was in agreement with the two-hybrid results indicating the involvement of this region for interaction with Atg11. However, deletion of 152 residues from the amino terminus of Atg9 (ProtA-Atg9Δ152N) also strongly obstructed coisolation of myc-Atg11, which was contradictory to the two-hybrid data. This inconsistency may result from different stringency inherent in the two assays.
Deletion of the CC1 and CC2 of Atg11 Affects the Vacuolar Transport of prApe1
Mapping of the binding domains has identified the requirement of Atg11 CC1 and CC2 for interaction with Atg9. We rationalized that Atg9 could function as a partner at the PAS to relay a cargo-loading signal and regulate subsequent vesicle formation. If our hypothesis is correct, preventing the Atg9-Atg11 interaction should also inhibit prApe1 transport by the Cvt pathway, and starvation could reverse the defect due to the Atg19-Atg8 interaction. An immunoblotting analysis against Ape1 showed that this was indeed the case (Figure 4A). Expression of Atg11ΔCC1 or Atg11ΔCC2 in atg11Δ cells did not rescue the prApe1 import defect in growing conditions, but starvation treatment generated mApe1 in these cells, which was also seen in control atg11Δ cells with an empty plasmid. Furthermore, we examined prApe1-sorting phenotypes for cells expressing these two Atg11 variants. In populations of Atg11ΔCC1- and Atg11ΔCC2-expressing cells, a significant percentage of cells had GFP-Ape1 separated from FM 4-64–labeled vacuoles (Figure 4B).
Figure 4.
The CC1 and CC2 domain of Atg11 is required for its proper function in control of prApe1 vacuolar transport. (A) Loss interaction with Atg9 also prevents Atg11ΔCC1 and ΔCC2 to mediate the transport of prApe1 by the Cvt pathway. Total cell extracts of midlog phase growing cells or 4 h nitrogen-starved cells were resolved by SDS-PAGE followed by immunoblotting with anti-Ape1 antiserum. (B) Atg11ΔCC1 and ΔCC2 are defective in targeting prApe1 to the PAS. Cells coexpressing GFP-Ape1 and the indicated Atg11 variants were grown to midlog phase, labeled with FM 4-64, and examined immediately (SMD) or after additional culturing in nitrogen-starvation medium for 2–3 h (SD-N) by fluorescence microscopy. (C) Restriction of GFP-Atg9 to the PAS is blocked in cells expressing Atg11ΔCC1 and ΔCC2. Cells of atg1Δ atg11Δ strains were transformed with plasmids to coexpress GFP-Atg9 and the indicated Atg11 variants. GFP-Atg9 fluorescence signals were observed from 4-h nitrogen-starved cells. Scale bar, 5 μm.
Next, we also examined the Cvt and autophagy functions of cells expressing Atg9Δ154-201, a truncated Atg9 that lost interaction with Atg11. Transport efficiency for prApe1 was first evaluated. As reported before, atg9Δ cells were not able to deliver prApe1 regardless of the nutrient conditions (Figure 5A). Introducing a plasmid to express full-length Atg9 complemented this defect. Interestingly, expressing Atg9Δ154-201 did not support prApe1 transport in nutrient-rich growing cells but starvation stress reversed this transport defect, which is a phenotype similar to atg11Δ cells (Figures 1A and 5A). This result again suggests that a proper Atg9-Atg11 interaction is critical for the Cvt pathway regulation. Next, the autophagy activity induced by short-term starvation was evaluated by the Pho8Δ60 alkaline phosphatase assay. Atg9Δ154-201 supported delivery of cytosolically accumulated Pho8Δ60 to, and subsequent activation in, the vacuoles to a level of ∼75% of that seen with full-length Atg9 (Figure 5B). A survival curve assay was then conducted to test the viability of cells expressing Atg9Δ154-201 in nitrogen starvation medium. Cells of an atg9Δ strain died within 7 d of starvation treatment (Figure 5C). Expression of full-length Atg9 complemented the atg9Δ phenotype and maintained cell viability similar to the wild-type cell level. Atg9Δ154-201–expressing cells survived a long duration of starvation stress but eventually died at the end of the assay period of more than 2 wk. Taken together, Atg9Δ154-201 seems to completely block the Cvt pathway but only mildly affects autophagy regulation. Finally, in addition to facilitate prApe1 transport via the Cvt pathway, Atg11 is also required for efficient elimination of superfluous peroxisomes during pexophagy (Hutchins et al., 1999). We tested if Atg9Δ154-201 also inhibited pexophagy due to its defect in interacting with Atg11. After switching lipid-grown cells to glucose-containing medium, peroxisomes were efficiently degraded in cells expressing either full-length Atg9 or Atg9Δ154-201 (Figure 5D), suggesting that pexophagy execution does not require direct interaction between Atg9 and Atg11.
Figure 5.
Atg9Δ154-201 is defective in the Cvt transport of prApe1, but only mildly affects autophagy, and does not interfere pexophagy. (A) Import of prApe1 in growing cells expressing Atg9Δ154-201 is blocked. Total cell extracts from midlog phase growing cells or 4-h nitrogen-starved cells were resolved by SDS-PAGE followed by immunoblotting with anti-Ape1 antiserum. (B and C) Compared with cells expressing full-length Atg9, Atg9Δ154-201 only mildly affects autophagy. (B) Midlog phase growing cells harboring an empty plasmid or plasmids driving expression of the indicated Atg9 variants were shifted from SMD to SD-N media for 4 h. Autophagy was measured by the levels of PhoΔ60 activity in whole-cell protein extracts. Activity in starved cells expressing full-length HA-tagged Atg9 was set to 100% and activity in the other conditions normalized, accordingly. Error bars, ±SD from three separate experiments. (C) Midlog phase growing cells in SMD medium were collected and shifted to nitrogen starvation SD-N medium for further culture. At the indicated day, viability was determined by plating aliquots of the culture on YPD plates and counting the number of colonies after 2 d growth. (D) Atg9Δ154-201 supports pexophagy. Cells harboring an empty plasmid (atg9Δ) or plasmids driving expression of the indicated Atg9 variants were grown under peroxisome-inducing conditions and shifted to SD-N for 24 h. At the indicated times, proteins extracts were prepared and subjected to immunoblotting against anti-Fox3 antiserum.
Cargo Loading Induces the Assembly of the PAS
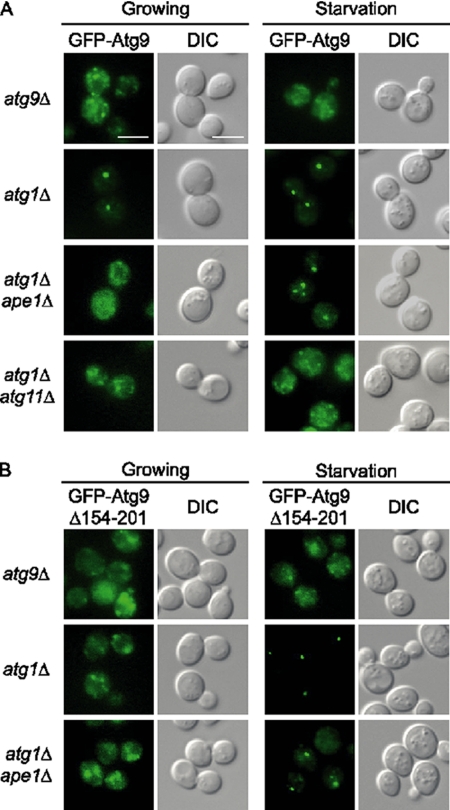
Atg9 shuttles between mitochondria and the PAS to regulate Cvt vesicle and autophagosome formation (Reggiori et al., 2004; Reggiori and Klionsky, 2006). Although the exact function of this cycling behavior is still elusive, a wild-type Atg9 distribution pattern is not observed in several autophagy mutant strains, including atg1 and atg2. In atg1Δ cells, the majority of GFP-Atg9 was restricted at the PAS in either exponentially growing or starved cells (Figure 6A). In growing cells, this PAS-restricted Atg9 is likely stuck in the process of regulating Cvt vesicle formation in order to deliver prApe1 to the vacuole. Therefore, in the absence of prApe1 cargo, GFP-Atg9 restriction did not occur and a diffuse fluorescence signal pattern was detected in growing atg1Δ ape1Δ double knockout cells (Shintani and Klionsky, 2004; Figure 6A). Starvation treatment caused GFP-Atg9 to be restricted in these cells, which was likely due to the induction of autophagosome formation. Interestingly, most cells in this condition showed more than one GFP-Atg9 dot. In atg1Δ atg11Δ double knockout cells, GFP-Atg9 showed yet another distribution pattern. Dispersed GFP-Atg9 signals in the cytosol were detected in most cells regardless of the nutrient conditions, although starvation seemed to have the tendency to concentrate GFP-Atg9 signals in this genetic background. Taken together, these distinctive GFP-Atg9 signal patterns observed in different strains clearly indicate that rather than merely facilitating transit of the prApe1-Atg19 complex to the PAS, Atg11 also affect the appearance of the PAS.
Figure 6.
The Cvt complex and starvation treatment induce Atg9 restriction at the PAS. (A) Transport cargo, such as the prApe1-formed Cvt complex, together with Atg11 induce Atg9 restriction. Midlog phase growing cells or 4-h nitrogen-starved cells of the indicated genetic backgrounds were prepared for detecting GFP-Atg9 signals by fluorescence microscopy. Scale bar, 5 μm. (B) GFP-Atg9Δ154-201 does not concentrate to the PAS in growing atg1Δ cells. GFP-Atg9Δ154-201 was expressed in the corresponding strains. Images were acquired as described in A.
To find out if this role of Atg11 also relies on its direct interaction with Atg9, we examined GFP-Atg9Δ154-201 fluorescence signals. Unlike full-length GFP-Atg9, GFP-Atg9Δ154-201 was dispersed in the cytosol in growing atg1Δ cells (Figure 6B), which is consistent with its defect in the regulation of Cvt vesicle formation through interaction with Atg11. Starvation treatment restricted GFP-Atg9Δ154-201 to a single PAS in most atg1Δ cells, a pattern shared with GFP-Atg9. Interestingly, GFP-Atg9Δ154-201 behaved exactly the same as GFP-Atg9 in both growing and starved atg1Δ ape1Δ cells (Figure 6, A and B). These results indicate that Atg9Δ154-201 retains the ability to cooperate with Atg11 to control the appearance of a single or multiple autophagosome assembly PAS sites depending on the presence of prApe1 or not, and the cooperation does not require direct Atg11-Atg9 interaction.
The multiple GFP-Atg9 dots induced by starvation treatment of atg1Δ ape1Δ double knockout cells might represent nucleated sites of autophagosome assembly. Because the PAS is usually found close to the vacuole in wild-type and atg1Δ strains, we examined whether those multiple GFP-Atg9 dots in double knockout cells also associated with the vacuole. No matter whether or not prApe1 was present, GFP-Atg9 signals were found colocalized with FM 4-64–labeled vacuoles in starved cells of the atg1Δ genetic background (Figure 7A). Next, a functional PAS recruits many Atg proteins to regulate vesicle formation, and we applied this as a criterion to evaluate the GFP-Atg9 dots as sites of autophagosome formation. To colocalize Atg9 with other Atg proteins, a functional RFP-Atg9 was prepared and its ability to complement atg9Δ defects was confirmed (data not shown). Like GFP-Atg9, RFP-Atg9 was also restricted to multiple dots in starved atg1Δ ape1Δ cells and these dots colocalized with GFP-tagged Atg8 (Figure 7B), a marker of autophagosomes, and Atg5 (Figure 7C), a component of the autophagy-specific ubiquitin-like conjugation system (Mizushima et al., 1998; Kirisako et al., 1999; Huang et al., 2000). Furthermore, because GFP-Atg9 did not form clear puncta in the absence of a functional Atg11 (Figures 4C and 6B), we rationalized that the appearance of those Atg9-positive multiple PAS sites require the function of Atg11. As a consequence, Atg11 should also associate with Atg9 dots. Our colocalization data showed that this was indeed the case (Figure 7D). Finally, Atg11 facilitates not only targeting of the prApe1-Atg19 complex to the PAS but also selective peroxisome degradation by pexophagy, which prompted us to speculate that the Atg9 dots formed in the absence of prApe1 were stimulated by an Atg11-dependent selective autophagic cargo. We have tried to colocalize these GFP-Atg9 dots with several fluorescent marker–labeled organelles, including peroxisomes, mitochondria, and endosomes, but none of them were found to colocalize (Supplementary Figure 3 and data not shown). Whether these multiple GFP-Atg9 dots are assembling around particulate selective autophagic cargo, equivalent to the prApe1-formed Cvt complex, remains to be tested.
Figure 7.
The multiple GFP-Atg9 puncta in atg1Δ ape1Δ cells are likely structures for autophagosome formation. Cells of corresponding strains harboring plasmids to express the indicated Atg proteins were grown to midlog phase, labeled with FM 4-64, shifted to starvation medium for 2–3 h, and then examined by fluorescence microscopy. (A) Regardless of prApe1 present or not, GFP-Atg9 is restricted to perivacuolar sites in strains of atg1Δ background. Scale bar, 5 μm. (B–D) The multiple Atg9 dots colocalize with Atg8 (B), Atg5 (C), and Atg11 (D).
DISCUSSION
Right after being synthesized by cytosolic free ribosomes, prApe1 assembles into a particulate Cvt complex and triggers the Cvt pathway to mediate its own efficient import into the vacuole in growing yeast cells. When cells experience starvation stress, the now highly induced prApe1 is delivered together with nonspecific cytosolic components by autophagosomes. Compared with the nonspecific autophagy transport maker Pho8Δ60, however, prApe1 delivery by either pathway is much more efficient indicating the presence of a prApe1 sorting mechanism.
The early event of prApe1 sorting involves the Atg11-mediated targeting of the prApe1-Atg19 cargo-receptor complex to the vesicle formation PAS. Atg8, through its covalent linkage to the vesicle membrane, assists the subsequent incorporation of prApe1 into transport vesicles (Huang et al., 2000). By means of direct interaction between Atg19 and Atg8, the prApe1-Atg19 complex may actually facilitate vesicle assembly by serving as a scaffold. In accordance with this current understanding, targeting prApe1 to the PAS is largely unaffected but further transportation is blocked due to the impairment of vesicle formation in atg8Δ cells (Kirisako et al., 1999; Huang et al., 2000). It is known that starvation stress induces atg8Δ cells to generate small and/or aberrant vesicles, which could account for the ability of this strain to reverse its prApe1 import defect (Abeliovich et al., 2000). However, what seems to be incompatible with this working model of the prApe1 sorting mechanism is that starvation only induces maturation of prApe1 in atg11Δ but not atg19Δ strains, whereas a direct interaction between Atg11 and Atg19 was proposed as a crucial step for linking the prApe1 cargo to the vesicle-forming machinery (Yorimitsu and Klionsky, 2005a). Therefore, we suspected that Atg19 achieved its prApe1 sorting functions via independent, instead of sequential, interactions with Atg11 and Atg8.
Here we found that a strain with both atg8 and atg11 mutations lost the starvation-induced prApe1 maturation phenotype. Autophagy was confirmed to be induced to full-strength by starvation in atg11Δ atg19Δ double knockout cells, but expression of Atg19Δ10C, which could not interact with Atg8, did not restore the ability to deliver prApe1 under starvation conditions in this strain (Figure 1). Therefore, we concluded that Atg19 might mediate a dual interaction mechanism to support prApe1 sorting, one by interaction with Atg11 and the other with Atg8. Quantifying the percentage of cells with a severe prApe1 sorting defect seemed to support this proposal (Figure 2). Starvation decreased the percentage of cells with separated GFP-Ape1 and FM 4-64 fluorescence signals in atg8Δ and atg11Δ strains, consistent with their abilities to reverse prApe1 import defect. In an atg8Δ atg11Δ double knockout strain, starvation did not diminish the sorting defect, and the number of cells with separated fluorescence signals also reached the level shown in atg19Δ strain, again in agreement with our proposal.
However, regardless of the sorting function contributed by Atg8, Atg11 indeed plays an essential role to target the prApe1 cargo. In addition to Atg19, previous studies have identified multiple interacting partners of Atg11, but none of these proteins were confirmed responsible for targeting the whole prApe1-Atg19-Atg11 complex to the PAS (Kim et al., 2001b; Yorimitsu and Klionsky, 2005a). Here we reported a previously unidentified interaction between Atg11 and Atg9 (Figure 3), and their proper interaction was required for not only prApe1 import by the Cvt pathway but also normal patterns of Atg9 subcellular distribution (Figures 4–6). Interestingly, PAS-targeting of GFP-Ape1 was affected by inhibiting the Atg11 and Atg9 interaction (Figures 2 and 4). In the atg9Δ atg11Δ double knockout strain, the GFP-Ape1 targeting defect was partially relieved by starvation, which suggests that mutations in atg11 and atg9 affect the same prApe1 sorting event. Taken together, these results seem to indicate that Atg9 plays an important role for targeting prApe1 cargo to the vesicle-forming PAS by interaction with Atg11. However, the GFP-Ape1 targeting defect in the atg9Δ strain was not as severe as in the atg11Δ strain (Figure 2). Besides, Atg9 is known to cycle between mitochondria and the PAS (Reggiori et al., 2005; Reggiori and Klionsky, 2006). It is necessary to distinguish the two populations of Atg9 in order to correctly target prApe1 cargo to the PAS. Therefore, we suspect that additional components participate in regulation of the Atg11-mediated targeting process, and Atg9 is responsible for relaying cargo-loading signals to induce vesicle assembly. This postulate is also consistent with our observation that localization of GFP-Atg11 to the perivacuolar PAS is not significantly affected in atg9Δ cells (Supplementary Figure 4). It is worth noting that, unlike a previous report, we have found cells expressing Atg11ΔCC1 defective in prApe1 transport (Yorimitsu and Klionsky, 2005a). The reason for this inconsistency is not clear, but our Atg11Δ CC1 construct has four more residues deleted from the CC1 region (Δ 272-325 of Atg11) than the one used in earlier studies (Δ 272-321 of Atg11). It remains to be confirmed whether this accounts for the different observation.
As Atg9 fulfills its function in regulation of vesicle formation at the PAS, it is retrieved and returned to peripheral mitochondria surface by the actions of several autophagy regulatory proteins, including the Atg1-Atg13 complex (Reggiori et al., 2004). Hence, Atg9 is restricted at the PAS in atg1Δ cells, and studies of the distribution pattern of Atg9 in this genetic background have led to the conclusion that cargo proteins facilitate vesicle formation at least in vegetative growth conditions (Shintani and Klionsky, 2004). Inspired by these results, we examined the fluorescence signals of GFP-tagged Atg9Δ154-201, an Atg9 variant unable to interact with ATG11, in several mutant strains and found interesting results (Figure 6). First, GFP-Atg9Δ154-201 was not restricted to the PAS in growing atg1Δ cells, a result supporting the idea that a proper interaction between Atg11 and Atg9 is critical for the Cvt pathway regulation. Second, starvation induced restriction of GFP-Atg9Δ154-201 to a single PAS, whereas both GFP-Atg9 and GFP-Atg9Δ154-201 did not restrict to a major structure in the absence of Atg11. Because Atg9Δ154-201 cannot directly interact with Atg11 and yet its restriction is under Atg11 control, we propose that other than targeting cargo proteins, Atg11 also affects PAS function and that does not rely on its direct interaction with Atg9. This conclusion is in agreement with the data that Atg11ΔCC1 or Atg11ΔCC2 did not restore the GFP-Atg9 restriction phenomenon in atg1Δ atg11Δ double knockout cells (Figure 4). Finally, in the absence of prApe1, starvation treatment induced the recruitment of the vesicle-forming machinery to several perivacuolar dots under the control of Atg11 (Figures 6 and 7). Because Atg11 also regulates other specific cargo sorting for transportation by autophagosomes, such as peroxisome degradation, we postulate whether unidentified specific cargo are nucleating vesicle formation in this situation (Hutchins et al., 1999). Our efforts to colocalize these dots with organelle markers were failed, and this hypothesis remains to be examined. Alternatively, the PAS may become unstable and tend to break off without prApe1 as a concrete cargo, which would also show this PAS signal pattern. Whatever the reason is, these Atg9 puncta are clearly separated from mitochondria indicating their involvement in regulation of vesicle formation (Supplementary Figure 3).
Although the vesicle formation mechanism for the Cvt pathway and autophagy is quite distinctive from those for trafficking between the endomembrane system, efficient cargo sorting is equally important to all events (Reggiori and Klionsky, 2005). It is not unprecedented to find cargo-sorting machineries with components generating multiple interactions. Taking the endosomal sorting complexes required for transport (ESCRTs) for example, all the three complexes contain multiple ubiquitin-binding motifs, which may compensate for the low affinity between each motif and ubiquitin, and facilitate subsequent cargo delivery at the endosomes (Hurley and Emr, 2006; Slagsvold et al., 2006). In the case of prApe1 sorting, its assembly into a Cvt complex, association with Atg19 and Atg11, and Atg11 oligomerization eventually lead to a huge structure with multiple sites to interact with downstream sorting components, Atg8 and possibly Atg9. Maybe only with these multiple interactions it becomes possible to load such a huge protein inclusion into a forming Cvt vesicle or autophagosome. Excitingly, recent studies in mammalian systems have provided parallel comparisons. The polyubiquitin-binding protein p62 was found forming aggregates, associating with LC3, the mammalian homolog of yeast Atg8, and facilitating mutant huntingtin degradation by autophagosomes (Kabeya et al., 2000; Bjorkoy et al., 2005). These data provide striking similarity between prApe1 sorting and p62-mediated huntingtin aggregates elimination. It will be interesting to test if an Atg11-equivalent component links huntingtin-p62 complex to the mammalian autophagy machinery, say mAtg9, in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Daniel Klionsky, Marks Longtine, Yoshinori Ohsumi, and Benedikt Westermann for supplying plasmids and strains and Chia-Wei Ku for technical assistant. This work was supported by Grants NSC94-2311-B-002-026 and NSC95-2311-B-002-019-MY3 from the National Science Council of Taiwan (W.-P.H.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0683) on December 27, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abeliovich H., Dunn W. A., Jr., Kim J., Klionsky D. J. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. A., Jr., Cregg J. M., Kiel J. A., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- George M. D., Baba M., Scott S. V., Mizushima N., Garrison B. S., Ohsumi Y., Klionsky D. J. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Wang C., Cohen A. Effect of IGF-1 on the balance between autophagy of dysfunctional mitochondria and apoptosis. FEBS Lett. 2004;577:357–360. doi: 10.1016/j.febslet.2004.10.040. [DOI] [PubMed] [Google Scholar]
- Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Huang W.-P., Klionsky D. J. Autophagy in yeast: a review of the molecular machinery. Cell Struct. Funct. 2002;27:409–420. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- Huang W.-P., Scott S. V., Kim J., Klionsky D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins M. U., Klionsky D. J. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins M. U., Veenhuis M., Klionsky D. J. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112(Pt 22):4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Klionsky D. J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P. E., Klionsky D. J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr., Klionsky D. J. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Scott S. V., Oda M. N., Klionsky D. J. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komatsu M., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Nakagawa I., et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D. J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Onodera J., Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J. Cell Sci. 2006;119:2903–2911. doi: 10.1242/jcs.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D. J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W., Kim J., Huang W.-P., Abeliovich H., Stromhaug P. E., Dunn W. A., Jr., Klionsky D. J. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005a;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005b;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.