Abstract
Telomeres are distinct structures, composed of short, repeated sequences, at the ends of all eukaryotic chromosomes. Telomeres have been shown in yeast to induce late replication in S phase and to silence transcription of neighboring genes. To examine the possibility of similar effects in human chromosomes, we studied cells from a subject with a microdeletion of 130 kb at the end of one copy of chromosome arm 22q, repaired by the addition of telomere repeats. Using fluorescence in situ hybridization of S phase nuclei, a distinct difference was found in the replication timing of the breakpoint region between the intact and truncated copies of chromosome 22. This difference was evident as a shift from middle to late replication time of the breakpoint region adjacent to the repaired telomere. This finding suggests that the human telomere sequence influences activation of adjacent replication origin(s). The difference in replication timing between the two chromosomes was not associated with differences in sensitivity to digestion by DNase I or with methylation of regions immediately adjacent to the breakpoint. Furthermore, both alleles of arylsulfatase A, a gene located at a distance of approximately 54 kb from the breakpoint, were expressed. We conclude that as in yeast, the proximity of telomeric DNA may induce a positional effect that delays the replication of adjacent chromosomal regions in humans.
Telomeres, which constitute the ends of eukaryotic chromosomes, are essential for maintaining normal chromosomal function (1–3). Human telomeric DNA is composed of the hexamer repeat TTAGGG, at a length of ≈4–15 kb (4, 5). Sandwiched between the hexamer repeats and the most distal unique region of the chromosome is the subtelomeric region, whose length varies among different chromosomes from a few tens to a few thousands of kb (6–8) and that contains mainly repetitive sequences of several types (6, 8, 9).
In yeast, proximity to telomeres has been shown to retard the activation of replication origins (10) and also to silence transcription of neighboring genes, a phenomenon known as telomere position effect (refs. 11 and 12; for review, see ref. 13). Similar effects have not yet been described in humans. The occurrence of subtelomeric repetitive sequences, adjacent to human telomeres, renders such analysis more challenging. The only naturally occurring situations in humans wherein telomeric repeats are adjacent to unique sequences at chromosome ends are those that occur in individuals with truncated chromosome ends that have been repaired by the process of telomeric healing (14). Several examples have been reported of individuals in whom idiopathic mental retardation has been associated with such chromosome-end truncations and telomere healing (15–17).
Accordingly, to study position effects of human telomeres, we used lymphoblastoid cells from a child (NT) with mental retardation and a microdeletion of 130 kb at the end of one copy of chromosome arm 22q (18). The broken end of this chromosome has been “repaired” by addition of telomeric repeats. As a consequence, a region of unique DNA that normally is located at a distance of more than 100 kb from the telomere is located adjacent to the telomeric repeats in this truncated chromosome. Here we show that although methylation patterns and DNase I sensitivity show no differences between the normal and truncated chromosomes, a clear difference between the chromosomes was found in the replication timing of the breakpoint region. The replication timing of this region clearly was delayed in the truncated chromosome from middle to late in S phase, thus demonstrating the position effect of a human telomere.
MATERIALS AND METHODS
Cell Lines.
Human lymphoblastoid cell lines NT (15), LAZ (19), and two additional lymphoblastoid cell lines, 3125 and 3133 (kindly provided by Nadine Cohen, Technion, Haifa, Israel), were grown in RPMI supplemented with 15% FCS and antibiotics.
Probes.
The following probes were used for fluorescence in situ hybridization (FISH): cosmids N85A3, N66C4, and N85E7 from the 22q telomeric region (18), cosmid N73A6 (contig 117 from human chromosome 22, http://webace.sanger.ac.uk/cgibin/display?db=acedb22&class=Map&object=Chr_22ctg117&display=graphics), cosmid CL15 from the human muscle glycogen phosphorylase (PYGM) gene region (20), and cosmid cJ21 from the human cystic fibrosis transmembrane region gene (21). Probe MS607, a minisatellite probe from locus D22S163, located near the 22q breakpoint region (22), was used for Southern blots.
Replication-Timing Assay.
Replication timing of genomic regions was determined by using FISH, as described previously (21, 23–25). Briefly, cells from unsynchronized cultures were labeled with 10−5 M BrdUrd for 1 hr before harvesting. BrdUrd containing nuclei were detected by an anti-BrdUrd antibody (PharMingen or Neomarkers Union City, CA). Fluorescence-activated cell sorter analysis was performed on an aliquot of propidium iodide-stained nuclei to confirm the cell cycle status of the culture. At least 100 S phase nuclei were scored for each slide.
In S phase nuclei, nonreplicated sequences are detected as two single hybridization signals (singlets, S.S.) and sequences that have completed replication are detected as two double-hybridization signals (doublets, D.D.). Each such doublet represents a pair of sister chromatids. The earlier the replication of a given sequence occurs, the higher is the expected percentage of S phase nuclei displaying the D.D. pattern (21, 25). A certain background fraction of the nuclei is expected to exhibit a singlet/doublet (S.D.) pattern as a result of incomplete hybridization (20). To test for hybridization efficiency and percentage of S.D. nuclei due to incomplete hybridization, metaphase spreads were examined for the number of hybridization signals at each chromosome copy. In metaphase chromosomes, two hybridization signals are expected at each copy and, thus, provide the expected background of S.D. signals attributed to incomplete hybridization, as well as indicating the relative hybridization efficiency. The cosmid probes that span partially deleted regions in NT cells (Fig. 1a) were examined for hybridization efficiency in approximately 50 metaphase spreads from NT cells, and comparable hybridization signals were found on both copies of chromosome 22. The background level of S.D.s attributed to incomplete hybridization was 10.8%, consistent with previous reports (23).
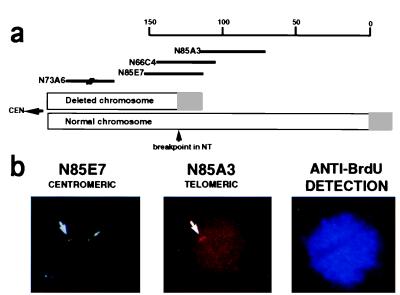
Figure 1.
Replication asynchrony at the 22q breakpoint. (a) A schematic diagram of the ends of both copies of the q arm of chromosome 22 in patient NT. The cosmid contig spanning the region appears above. The position of the distal end of cosmid N73A6 has been determined as indicated. The gray boxes represent telomeric (TTAGGGn) repeats. The small arrowhead points to the breakpoint. CEN, centromeric direction. Scale is in kb. (b) FISH of NT nuclei using two probes. NT nuclei were hybridized simultaneously with digoxigenin-labeled cosmid N85A3 and biotin-labeled cosmid N85E7. Biotin was detected with FITC (green), and digoxigenin was detected with rhodamine (red). Cosmid N85A3 hybridizes only to the normal copy of chromosome 22 in NT cells, thus identifying the source of each signal as arising from the N85E7 probe. Before harvesting, cells were labeled with BrdUrd for detection of S phase nuclei. S phase cells were detected by an antibody to BrdUrd, which, in turn, was detected with AMCA (blue). The triple exposure in this figure represents an NT nucleus in S phase having replicated the N85E7 region in the normal chromosome 22 (green doublet) but not yet in the deleted chromosome (green singlet).
Isolation of Nuclei and DNase I Digestion.
Nuclei were prepared as described (26). Digestion with DNase I was performed at 37°C for 5 min, at several enzyme concentrations. The reaction was terminated by the addition of an equal volume of buffer containing 1% SDS, 0.6 M NaCl, 20 mM Tris (pH 7.9), 10 mM EDTA, and proteinase K (0.4 mg/ml) and incubated for 2 hr at 55°C. Finally, the DNA was purified by phenol-chloroform extractions and precipitated in ethanol.
Southern Blotting and Hybridization.
Purified DNA was subjected to restriction enzyme digestion and gel electrophoresis on 1% agarose gels and was transferred to a Magnacharge nylon transfer membrane (Micron Separations, Westboro, MA) according to the manufacturer’s instructions. Hybridization buffer and conditions were performed as described (27). Filters were washed in 0.1× SSC [standard saline citrate (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)]/0.1% SDS.
Reverse Transcription–PCR (RT-PCR) Heterozygosity Analysis of the Arylsulfatase A (ARSA) Gene.
DNA (100 ng) from the lymphoblastoid cell line, NT, was subjected to PCR amplification of the ARSA gene by using primers 11+12 [shortened versions of published primers (28)]. The sequences of these primers are as follows: primer 11 (a forward primer), 5′-TTGATGGCGAACTGAGTGAC-3′, and primer 12 (a reverse primer), 5′-TTCCTCATTCGTACCACAGG-3′. The PCR was performed with TaKaRa Taq polymerase, manufacturer’s buffer, and 2.5% DMSO under the following conditions: 95°C (7 min), 40 cycles of 50°C (1 min), 72°C (4 min), and 95°C (1 min). To detect a BsrI polymorphism in exon 7 (29), the PCR product was digested with BsrI (MBI Fermentas) and run on a 2% agarose gel.
RNA was prepared from NT cells by extraction with Tri-Reagent (Molecular Research Center, Cincinnati). Five micrograms of total RNA was reverse-transcribed with 200 units of Moloney murine leukemia virus RT (Promega) in a volume of 20 μl. Two microliters from the RT reaction was subjected to PCR by using primers that amplify exon 7 of the ARSA gene: forward primer, no. 14, 5′-AGCCCTCGGCAGTCTCTC-3′, and reverse primer, no. 10, 5′-TTTCAGGGCTTGCAGCAC-3′. Amplification conditions were 95°C (5 min), 40 cycles of 52°C (45 sec), 72°C (45 sec), and 95°C (30 sec). An aliquot of the RT-PCR was run on a 2% agarose gel to ensure that a single product of the correct molecular weight was obtained. The remaining RT-PCR product was purified by using the QIAquick gel extraction kit (Qiagen), digested with BsrI, and run on a 3% MetaPhor agarose gel (FMC). As control for full digestion by BsrI, the PCR product of ARSA, by using primers 11 and 12, was purified and digested under similar conditions.
RESULTS
Replication Timing of the Breakpoint Region.
To analyze the replication timing of the breakpoint region in both the normal and truncated chromosomes 22 in the NT cells, the FISH replication time assay was performed by using cosmids N85E7 and N85A3 (Fig. 1a) (for a more detailed map of the region see ref. 18). The normal replication-timing status of this locus, in comparison with known early and late replicating loci, was determined in a control lymphoblastoid cell line, LAZ, that has no aberrations at the 22q telomeric region (Table 1).
Table 1.
FISH replication-timing patterns of different probes in LAZ cells
| Probe | % D.D. | % S.D. | % S.S. | No. scored | Comments |
|---|---|---|---|---|---|
| CL15 | 60.5 | 10.5 | 29.0 | 104 | Early control |
| cJ21 | 22.0 | 13.5 | 64.5 | 124 | Late control |
| N85E7 | 42.0 | 11.5 | 46.5 | 105 | |
| N85A3 | 18.0 | 13.5 | 68.5 | 105 |
Nuclei from LAZ cells were prepared and hybridized in situ to various cosmid probes: CL15, muscle glycogen phosphorylase (PYGM) gene; cJ21, cystic fibrosis transmembrane region gene; N85E7 and N85A3, probes from the 22q telomeric region (see Fig. 1a). Percentages of nuclei exhibiting each of the three patterns of hybridization (D.D., S.D., S.S.) are indicated together with the total number of nuclei scored for each probe.
The percentage of nuclei from the normal cell line exhibiting the doublet pattern (D.D.) indicates that the proximal N85E7 cosmid region replicates in middle to late S, whereas the distal N85A3 cosmid region, which hybridizes to the most telomeric unique DNA (18), replicates late in S. The difference in the percentage of D.D. signals for the two probes suggests that each region is replicated from an independent origin. Comparison of the percentage of D.D. nuclei by using the N85A3 probe to the corresponding percentage of D.D. signals observed by using control probes to established early and late replication regions indicates that the most distal unique region of 22q replicates at approximately the same time as the gene for the cystic fibrosis transmembrane region, which is known to be late-replicating in cells that do not express the gene (21).
In several cases of monoallelically expressed genes, both imprinted and nonimprinted, asynchronous replication of both alleles was indicated by elevated percentages of S.D. nuclei exceeding 20% of S phase nuclei (23, 30, 31). Initial analysis of the replication timing of the breakpoint region in the NT cell line by using probe N85E7 revealed a relatively high percentage of S.D. nuclei (28%) compared with LAZ cells (11.5%). Because this region shows a pattern of middle–late replication (Table 1), replication-timing delay of one allele would be expected to yield a small but significant increase in percentage of S.D. nuclei. Accordingly, we analyzed multiple slides of several hybridization experiments by using two different probes (N66C4 and N85E7) spanning the breakpoint region, comparing NT with three different normal lymphoblastoid cell lines (LAZ, 3125, and 3133). A significant difference in percentage of nuclei exhibiting the S.D. pattern between NT and the control lymphoblastoid cell lines was observed by using both the N85E7 and N66C4 probes (Table 2). As indicated in Materials and Methods, analysis of metaphase spreads confirmed that higher percentages of S.D. nuclei in NT cells could not be attributed to differences in hybridization efficiency of the probes from the breakpoint region because of reduced target DNA. The incomplete hybridization background rate of 10.8%, evident from metaphase spreads in NT cells, is comparable to that for numerous different probes (23) and indicates that the elevated levels of S.D. nuclei are attributable to asynchronous replication of the region.
Table 2.
Analysis of the S.D. pattern in NT and control cell lines using probes from the breakpoint region
| Probe | N85E7
|
N66C4
|
||||
|---|---|---|---|---|---|---|
| μ | SD | n | μ | SD | n | |
| Control cells | 11.08 | 0.55 | 4 | 10.00 | 2.26 | 2 |
| NT cells | 25.04 | 2.64 | 5 | 23.80 | 0.57 | 2 |
| P value | <.001 | <.01 | ||||
Nuclei from normal control cell lines (LAZ, 3125, and 3133) and from NT were hybridized with probe N85E7 or N66C4. The percentage of S phase nuclei displaying the S.D. pattern was determined in the number (n) of slides indicated, from two different preparations. The data are shown as the mean (μ) and SD.
To determine which of the alleles is delayed in replication time, we took advantage of the ability to distinguish the normal and deleted chromosomes at the N85A3 region (Fig. 1a). The distal N85A3 region is entirely deleted in the truncated chromosome of NT, and, consequently, this probe hybridizes to only one allele in the NT nuclei on the intact chromosome. Therefore, we were able to use this cosmid probe to distinguish between the normal and the truncated chromosomes by performing simultaneous, dual-color FISH with probes N85E7 and N85A3 (Fig. 1b). The results obtained from four slides in two independent preparations showed that in 85.1 ± 6.1% (μ ± SD) of the S.D. nuclei, the normal copy of chromosome 22 was the earlier-replicating copy. Thus, replication of the proximal region that is newly adjacent to a telomere in the truncated chromosome is shifted from an earlier to a later time in S phase.
To examine the possibility that the change in replication timing of the truncated chromosome is due to the loss of an origin of replication in the deleted region, from which the breakpoint region is normally replicated, we determined the direction of replication of the region in normal lymphoblastoid cells. Performing a dual-hybridization experiment with a biotinylated N73A6 probe and a digoxigenin-labeled N85E7 probe on two normal cell lines, we detected 20 S phase nuclei in which one of the two probes displayed a D.D. pattern whereas the other displayed a S.S. pattern. In 16 of these 20 nuclei (80%), the centromeric N73A6 region replicated before the adjacent, more telomeric N85E7 region, which is situated less then 20 kb away (results not shown). This finding indicates that the breakpoint region is replicated from an origin situated centromeric to the region. After establishing the direction of the replication, we determined whether the N73A6 region, situated approximately 40 kb from the breakpoint, is also replicated asynchronously in the NT cells. The results of a dual-FISH experiment by using N73A6 and N85A3 indicated that this region also replicates asynchronously (% S.D. = 21.5), with the normal copy replicating earlier than the copy originating from the truncated chromosome.
Expression of Genes Localized to the Breakpoint Region.
The replication-timing results indicate that the same region on each chromosome 22 replicates at different times during the S phase. Replication timing has been shown to be strongly correlated with gene expression (32), and silencing of gene expression by telomere position effect has been demonstrated in yeast (11). Therefore, we sought potential differences in gene expression between the intact and the abnormal chromosomes at the 22q breakpoint region in NT cells. ARSA is the nearest gene mapped to the breakpoint in NT (18), which is expressed in lymphoblastoid cells (33). By analysis of several sequenced clones, available in electronic databases (accession nos. U62317, Z82245, AC000050), which span the region between the breakpoint and the ARSA locus, the distance is estimated to be between 52.6 and 55.4 kb (detailed analysis available from authors). At least two polymorphic sites have been identified in exons of the ARSA gene (29), therefore enabling distinction of the mRNA products arising from each of the alleles in heterozygous individuals. NT was found to be heterozygous for a site in exon 7, which can be detected by digestion with the BsrI restriction enzyme (Fig. 2 a, b, and d). RNA was prepared from the NT cells and subjected to RT-PCR, by using primers specific to exon 7, which harbors the polymorphic site (Fig. 2c). After digestion by BsrI and separation on a metaphor gel, bands corresponding to transcripts originating from each of the alleles were detected (Fig. 2e), indicating that neither of the alleles of the gene is completely silenced in this cell line. Therefore, if human telomeres do impose a silencing effect, this effect may be incomplete or may be confined to distances shorter than that between the ARSA gene and the NT breakpoint. Additional genes closer to the breakpoint have not been identified to date.
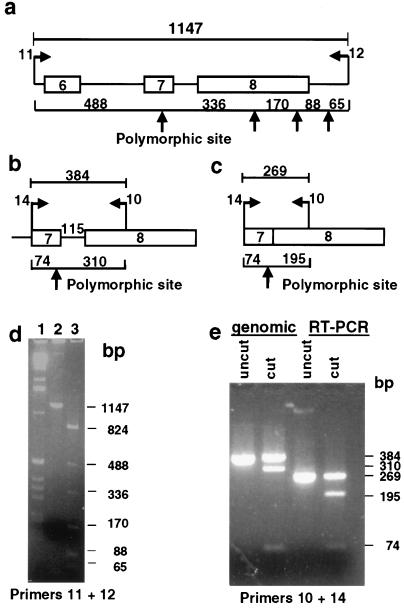
Figure 2.
Expression of ARSA in NT lymphoblastoid cells. (a and b) Partial maps of the genomic region of the ARSA gene harboring the BsrI polymorphism. Boxes represent exons; lines represent introns. Arrows point to BsrI restriction sites. Genomic PCR with primers 11 + 12 or with primers 14 + 10 and subsequent digestion with BsrI produces bands of sizes depicted in the figure (in base pairs). (c) Partial cDNA map of the ARSA gene region harboring the polymorphic BsrI site. RT-PCR with primers 10 + 14 and digestion with BsrI produces bands of sizes depicted in base pairs. (d) Results of PCR of NT DNA with primers 11 and 12, separated on an agarose gel. Lanes: 1, 1-kb ladder (GIBCO/BRL); 2, undigested PCR product; 3, PCR product digested with BsrI. The appearance of bands of sizes 823, 492, and 331 bp indicates that one allele contains a BsrI site and one does not. (e) Products of RT-PCR and PCR of NT cDNA and genomic DNA by using primers 10 and 14, separated on a MetaPhor gel. The appearance of both 269- and 195-bp fragments in the cut RT-PCR product indicates that both alleles are expressed.
DNase I Sensitivity and the Methylation Status at the Breakpoint Region.
In addition to directly assaying gene expression, the potential transcriptional activity of DNA can be evaluated by associated characteristics, one of which is the level of generalized DNase I chromatin sensitivity (34). NT cells provide the opportunity to examine the DNase I sensitivity status near telomeric repeats in direct comparison with the sensitivity of the homologous region on the normal chromosome. The two chromosomes can be distinguished because of the loss of restriction sites from the deleted region and replacement with telomeric repeats, resulting in a restriction fragment length polymorphism (18). To this end, NT nuclei were treated with increasing concentrations of DNase I, and the resulting DNA was analyzed by using the appropriate restriction digestions, followed by Southern blot hybridization with a probe from the breakpoint region (Fig. 3). The digestions with NgoMI and EcoRV detect the sensitivity at distances of up to approximately 6.5 and 15 kb, respectively (Fig. 3a). As seen in Fig. 3b, the EcoRV fragments originating from the two chromosomes disappeared at similar rates as a function of increasing concentrations of DNase I. Similar results were obtained with NgoMI digests (results not shown).
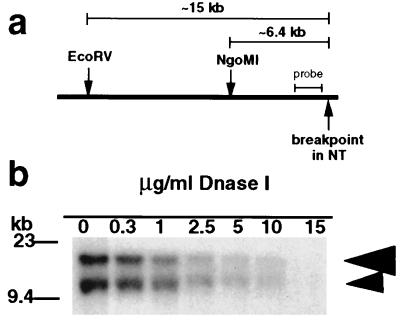
Figure 3.
DNase I sensitivity of the breakpoint region in NT nuclei. (a) A map of the breakpoint region indicating positions of the restriction sites for the enzymes used in this assay (constructed according to the sequence of cosmid N66C4, accession no. AC000050). (b) Southern blot analysis of the DNase I sensitivity of the breakpoint region. Nuclei were digested with increasing concentrations of DNase I (as indicated above each lane). After DNase I treatment, DNA was extracted from nuclei, subjected to digestion with EcoRV, and hybridized with the MS607 probe. Large arrowhead, band originating from the normal chromosome copy; small arrowhead, band originating from the deleted chromosome.
To evaluate the relative potential activity of the breakpoint region by using an additional parameter, we determined the methylation status of several CpG sites in a region encompassing approximately 3 kb upstream to the breakpoint. Several potential methylation sites are present in a unique region of approximately 600 bp upstream to the breakpoint in NT and in the adjacent D22S163 region of variable number of tandem repeats (VNTR) (detected by the MS607 probe) (Fig. 4a). A number of these can be assayed by digestion with methylation-sensitive restriction enzymes followed by Southern blotting and hybridization. The size of the VNTR region was determined at first in NT cells by restriction digestion with MspI, which is insensitive to methylation and digests at both ends of the VNTR (Fig. 4a). The digested product revealed a single band of 2.8 kb after hybridization with the MS607 probe (Fig. 4b), indicating that NT is homozygous at this locus. As a result, the two chromosomes 22 of NT cannot be distinguished by restriction analysis of the VNTR region unless the two chromosomes differ in their methylation patterns. To test this, NT DNA was digested with HpaII, a methylation-sensitive MspI isoschizomer. Southern hybridization with the MS607 probe revealed only a very weak 2.8-kb band (Fig. 4 b and c; seen more clearly in Fig. 4c, at long exposure) but a strong band of approximately 5 kb. This finding indicates that in both chromosomes at least one of the two HpaII sites flanking the VNTR sequence is highly methylated and that only a small fraction of cells are unmethylated at both of these sites. The smear appearing in the HpaII lanes (arrowhead in Fig. 4b) indicates that the most telomeric HpaII site is methylated, because the smear is produced by the fusion at the breakpoint with telomeric repeats that are heterogeneous in length in different cells (4).
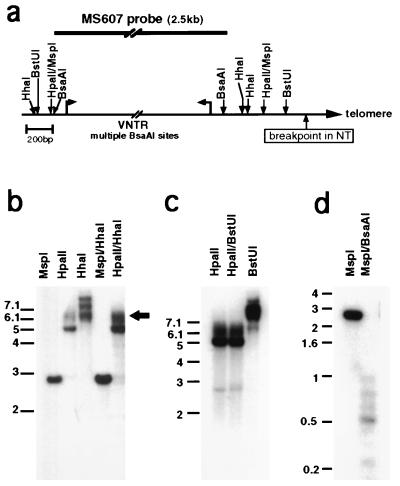
Figure 4.
Methylation patterns in the NT breakpoint region. (a) A map of the breakpoint region indicating the sites for methylation-sensitive enzymes used for digestion, constructed according to the sequence of cosmid N66C4 (see Fig. 1a) (GenBank accession no. AC000050). The position of the MS607 probe is indicated above the map. (b) MspI, HpaII, and HhaI separate and combined digests of NT DNA, probed with MS607. The arrowhead points to the signal originating from the deleted chromosome and contains the region from the most distal HpaII site to the end of the chromosome. The variability of length of telomeres leads to “smearing” of the band. (c) HpaII and BstUI separate and combined digests of NT DNA hybridized with MS607. (d) MspI and combined MspI/BsaAI digest of NT DNA hybridized with MS607 probe.
By digestion with other methylation-sensitive enzymes, additional nearby sites also were found to be methylated on both chromosomes. The HhaI digestion (Fig. 4b) did not produce the approximately 2.75-kb band, which would have been expected if the sites surrounding the VNTR were not methylated. Similarly, the MspI/HhaI double digest did not yield a band that is 150–180 bp smaller than the MspI fragment, as would have been expected if one of the two HhaI sites near the breakpoint had been digested in either of the chromosomes. In addition, the HpaII/HhaI double digest gave a similar pattern of smeared signal as the HpaII digest, again confirming methylation at the HhaI sites near the breakpoint in the broken chromosome. A similar result was obtained with the neighboring BstUI site (Fig. 4 a and c), which also appears to be mostly methylated. In contrast to these methylated sites, the region containing the VNTR appears to be mostly unmethylated. These sites appear in many of the repeat copies in the allele represented by the sequenced N66C4 cosmid (accession no. AC000050), and complete methylation of these sites would be reflected as a band of 2.8 kb in the MspI/BsaAI double digest. The complete absence of a band of this size, both in NT (Fig. 4d) and in the control LAZ cells (result not shown), indicates low methylation levels of the VNTR repeats. These methylation results indicate that in NT both copies of chromosome 22 have similar methylation patterns in the region immediately centromeric to the breakpoint.
DISCUSSION
We have utilized the rare opportunity of a spontaneous chromosomal abnormality to demonstrate a position effect of human telomere sequence on the replication timing of an adjacent DNA region. The effect resulted in a shift of replication timing from middle to late in S phase of a region immediately proximal to a repaired telomere on the truncated q arm of chromosome 22. The change in replication timing was readily apparent from the asynchronous pattern of hybridization of the breakpoint region, visualized by FISH to S phase nuclei, and extends to a distance of at least 40 kb. The influence of the human telomeric sequence on the replication timing of the adjacent region at the NT breakpoint could be caused either by delaying activation of, or by inactivating, nearby origin(s). To understand the mechanism of the telomere effect on origin activation, additional studies will be needed to determine the position of origins of replication in mammalian telomeric regions in general and in the 22q telomere specifically.
In human chromosomes, no individual telomere has been assayed for replication timing. Two studies assayed replication timing of the human telomere hexamer sequence more generally. One study reported replication throughout S phase (35), and the other study found human telomere replication to occur during the first half of the S phase (36). However, the TTAGGG probe hybridizes to interstitial telomere repeats (37, 38), and these may well replicate at different points in S phase compared with the terminal repeats. In the current study, we have shown that the most distal unique region of the 22q arm replicates late in S phase in lymphoblastoid cells, providing data on replication timing of a specific human telomeric region.
Late replication timing often is associated with additional chromatin characteristics (39, 40). Accordingly, our findings of late replication induced by the proximity of a telomere in the NT patient prompted us to investigate the DNase I sensitivity and the methylation status of the breakpoint region. It is noteworthy that although this region is in close proximity to a telomere, it does not contain the repetitive sequences characteristic of normal subtelomeric regions and that may contribute to an inert chromatin structure. We found no difference between the two copies of chromosome 22 in the general DNase I sensitivity of the breakpoint region at distances of up to 15 kb. Therefore, we conclude that the replication-timing difference between the copies is not associated with the differences in chromatin structure that are reflected by DNase I sensitivity.
The methylation status analysis by digestion with several methylation-sensitive enzymes followed by Southern blot analysis of the breakpoint region in NT cells revealed that the region 600 bp upstream of the breakpoint was methylated both in the truncated and normal chromosomes. Both copies of chromosomes 22 in a control cell line, LAZ, were found to be methylated as well, indicating that this area normally is methylated in lymphoblastoid cells. The region of the VNTR adjacent to the breakpoint, in contrast, was found to be unmethylated in both chromosome 22 copies, as well as in the LAZ cells. It has been shown previously that subtelomeric regions of several human chromosomes are highly methylated in somatic tissues (4, 6); therefore, it is exceptional that this VNTR region, which is in close proximity to the telomere of the truncated copy of chromosome 22, escaped de novo methylation during development. The finding of identical methylation patterns at the breakpoint region in both copies of chromosome 22 suggests that as in the case of DNase I sensitivity, so too the replication-timing delay we have observed is not associated with differences in methylation status of the region we assessed.
Based on studies from several organisms, few active genes are located in regions adjacent to telomeres. Subtelomeric regions in yeast contain a low density of ORFs, and those that are present are often members of multigene families (12). In Drosophila, which do not have simple telomeric repeats but, rather, other tandemly repeated sequences, genes located in the subtelomeric regions are subject to variable degrees of repression (reviewed in ref. 41). In humans, no active genes have been reported from analyzed subtelomeric regions, which are those regions normally situated adjacent to telomeric repeats (8, 9). One exception is the RABL2 gene, which is active in spite of its being located in a subtelomeric region but has an additional copy elsewhere in the genome (42). These findings, in addition to the correlation between late replication and gene inactivity (21, 43–46), prompted us to assess possible effects of telomere proximity on transcription of the closest known gene to the breakpoint, the ARSA gene, which is located at a distance of approximately 54 kb from the breakpoint on 22q. Both alleles of this gene are expressed in NT cells, indicating that if a position effect of the repaired telomere on transcription occurs, it does not extend to this distance or is incomplete. Future studies are needed to determine whether the distance over which telomeres effect such an influence on DNA replication and gene expression is a function of telomere length.
In cases of chromosome end breakage and telomere healing, the usual assumption is that the abnormal phenotypes arise as consequence of haploinsufficiency of deleted genes (15). However, if human telomeric repeats do silence genes adjacent to the repaired ends, it is also conceivable that genes residing in closest proximity to the healed chromosome end are inactivated epigenetically and thus may contribute to the phenotype. Such a mechanism may apply to facioscapulohumeral muscular dystrophy (47). It also has been suggested that expression of genes in close vicinity to telomeres may be modulated by age-associated telomere shortening (48).
The rare occurrence of a patient with a specific chromosomal breakage followed by telomere regeneration enabled us to detect the position effect of a human telomere on replication timing. Other such cases of telomere healing may provide us with the opportunity to investigate the potential effect of human or other mammalian telomeres on DNA replication and transcription and promote our understanding of the role of telomere position effects in normal aging and disease.
Acknowledgments
We thank Heather L. Wilson for providing database analysis of the sequence between the NT breakpoint and the ARSA gene. We are grateful to Dr. Jonathan Flint for kindly providing us with the NT cell line. We also wish to thank Howard Cedar, Dale Frank, Daniel Kornitzer, and Maty Tzukerman for critical reading of the manuscript. This work was supported by a grant from the Israeli Ministry of Health.
ABBREVIATIONS
- FISH
fluorescence in situ hybridization
- ARSA
arylsulfatase A
- RT
reverse transcription
- VNTR
variable number of tandem repeats
- S.S.
singlets
- D.D.
doublets
- S.D.
singlet/doublet
Note Added in Proof
Since submission of the manuscript, a similar telomere position effect on replication timing has been reported for chromosome 16p (49).
References
- 1.Harley C B, Villeponteau B. Curr Biol. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E, Bhattcharyya A, Gilley D, Kirk K, Krauskopf A, McEachern M, Prescott J, Ware T. In: Telomeres and Telomerase (Ciba Foundation Symposium 211) Chadwick D J, Cardew G, editors. New York: Wiley; 1997. pp. 2–19. [DOI] [PubMed] [Google Scholar]
- 3.deLange T. Science. 1998;279:334–335. [Google Scholar]
- 4.deLange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Varmus H E. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 6.Cross S, Lindsey J, Fantes J, McKay S, McGill N, Cooke H. Nucleic Acids Res. 1990;18:6649–6657. doi: 10.1093/nar/18.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macina R A, Negorev D G, Spais C, Ruthig L A, Hu X, Riethman H. Hum Mol Genet. 1994;3:1847–1853. doi: 10.1093/hmg/3.10.1847. [DOI] [PubMed] [Google Scholar]
- 8.Flint J, Bates G P, Clark K, Dorman A, Willingham D, Roe B A, Micklem G, Higgs D R, Louis E J. Hum Mol Genet. 1997;6:1305–1314. doi: 10.1093/hmg/6.8.1305. [DOI] [PubMed] [Google Scholar]
- 9.Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett N A, King A, Higgs D R. Nat Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson B M, Fangman W L. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- 11.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 12.Zakian V A. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 13.Shore D. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 139–191. [Google Scholar]
- 14.Cooke H. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 219–245. [Google Scholar]
- 15.Flint J, Wilkie A O M, Buckle V J, Winter R M, Holland A J, McDermid H E. Nat Genet. 1995;9:132–140. doi: 10.1038/ng0295-132. [DOI] [PubMed] [Google Scholar]
- 16.Giraudeau F, Aubert D, Young I, Horsley S, Knight S, Kearney L, Vergnaud G, Flint J. J Med Genet. 1997;34:314–317. doi: 10.1136/jmg.34.4.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horsley S W, Knight S J, Nixon J, Huson S, Fitchett M, Boone R A, Hilton-Jones D, Flint J, Kearney L. J Med Genet. 1998;35:722–726. doi: 10.1136/jmg.35.9.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong A C C, Ning Y, Flint J, Clark K, Dumanski J P, Ledbetter D H, McDermid H E. Am J Hum Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler J T, Krevans J R. Bull Johns Hopkins Hosp. 1961;109:217–233. [PubMed] [Google Scholar]
- 20.Lichter P, Tang C-J C, Call K, Hermanson G, Evans G A, Housman D, Ward D C. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 21.Selig S, Okumura K, Ward D C, Cedar H. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armour J A L, Povey S, Jeremiah S, Jeffreys A J. Genomics. 1990;8:501–512. doi: 10.1016/0888-7543(90)90037-u. [DOI] [PubMed] [Google Scholar]
- 23.Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll D J, Nicholls R D, Cedar H. Nature (London) 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- 24.Ward D C, Boyle A, Haaf T. In: Human Chromosomes: Principles and Techniques. Verma R S, Babu A, editors. New York: McGraw–Hill; 1995. pp. 184–191. [Google Scholar]
- 25.Boggs B A, Chinault A C. Methods Companion Methods Enzymol. 1997;13:259–270. doi: 10.1006/meth.1997.0525. [DOI] [PubMed] [Google Scholar]
- 26.Keshet I, Lieman-Hurwitz J, Cedar H. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 27.Nesslinger N J, Gorski J L, Kurczynski T W, Shapira S K, Siegel-Bartelt J, Dumanski J P, Cullen R F, French B N, McDermid H E. Am J Hum Genet. 1994;54:464–472. [PMC free article] [PubMed] [Google Scholar]
- 28.Polten A, Fluharty A L, Fluharty C B, Kappler J, von Figura K, Gieselmann V. N Engl J Med. 1991;324:18–22. doi: 10.1056/NEJM199101033240104. [DOI] [PubMed] [Google Scholar]
- 29.Zlotogora J, Furman-Shaharabani Y, Goldenfum S, Winchester B, von Figura K, Gieselmann V. Am J Med Genet. 1994;52:146–150. doi: 10.1002/ajmg.1320520205. [DOI] [PubMed] [Google Scholar]
- 30.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 31.Chess A. Science. 1998;279:2067–2068. doi: 10.1126/science.279.5359.2067. [DOI] [PubMed] [Google Scholar]
- 32.Holmquist G P. Am J Hum Genet. 1987;40:151–173. [PMC free article] [PubMed] [Google Scholar]
- 33.Tempesta M-C, Levade T, Salvayre R. Clin Chim Acta. 1991;202:149–166. doi: 10.1016/0009-8981(91)90046-f. [DOI] [PubMed] [Google Scholar]
- 34.Weintraub H, Groudine M. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 35.TenHagen K G, Gilbert D M, Willard H F, Cohen S N. Mol Cell Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hultdin M, Gronlund E, Norrback K-F, Eriksson-Lindstrom E, Just T, Roos G. Nucleic Acids Res. 1998;26:3651–3656. doi: 10.1093/nar/26.16.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber B, Collins C, Robbins C, Magenis R E, Delaney A D, Gray J W, Hayden M R. Nucleic Acids Res. 1990;18:3353–3361. doi: 10.1093/nar/18.11.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells R A, Germino G G, Krishna S, Buckle V J, Reeders S T. Genomics. 1990;8:699–704. doi: 10.1016/0888-7543(90)90257-u. [DOI] [PubMed] [Google Scholar]
- 39.Arapinis C, Elion J, Labie D, Krishnamoorthy R. Eur J Biochem. 1986;156:123–129. doi: 10.1111/j.1432-1033.1986.tb09556.x. [DOI] [PubMed] [Google Scholar]
- 40.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 41.Mason J M, Biessmann H. Trends Genet. 1995;11:58–62. doi: 10.1016/s0168-9525(00)88998-2. [DOI] [PubMed] [Google Scholar]
- 42.Wong A C C, Shkolny D, Dorman A, Willingham D, Roe B A, McDermid H E. Genomics. 1999;59:326–334. doi: 10.1006/geno.1999.5889. [DOI] [PubMed] [Google Scholar]
- 43.Goldman M A. BioEssays. 1988;9:50–55. doi: 10.1002/bies.950090204. [DOI] [PubMed] [Google Scholar]
- 44.Hatton K A, Dhar V, Brown E H, Iqbal M A, Stuart S, Didamo V T, Schildkraut C L. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taljanidisz J, Popowski J, Sarkar N. Mol Cell Biol. 1989;9:2881–2889. doi: 10.1128/mcb.9.7.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon I, Cedar H. In: DNA Replication in Eukaryotic Cells. DePamphilis M L, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 387–408. [Google Scholar]
- 47.Lemmers R J L F, van der Maarel S M, van Deutekom J C T, van der Wielen M J R, Deidda G, Dauwerse H G, Hewitt J, Hofker M, Bakker E, Padberg G W, et al. Hum Mol Genet. 1998;7:1207–1214. doi: 10.1093/hmg/7.8.1207. [DOI] [PubMed] [Google Scholar]
- 48.Wright W E, Shay J W. Trends Cell Biol. 1995;5:293–297. doi: 10.1016/s0962-8924(00)89044-3. [DOI] [PubMed] [Google Scholar]
- 49.Smith Z E, Higgs D R. Hum Mol Genet. 1999;8:1373–1386. doi: 10.1093/hmg/8.8.1373. [DOI] [PubMed] [Google Scholar]