Abstract
Nuclear pore complex (NPC) assembly in interphase cells requires that new NPCs insert into an intact nuclear envelope (NE). Our previous work identified the Ran GTPase as an essential component in this process. We proposed that Ran is required for targeting assembly factors to the cytoplasmic NE face via a novel, vesicular intermediate. Although the molecular target was not identified, Ran is known to function by modulating protein interactions for karyopherin (Kap) β family members. Here we characterize loss-of-function Saccharomyces cerevisiae mutants in KAP95 with blocks in NPC assembly. Similar to defects in Ran cycle mutants, nuclear pore proteins are no longer localized properly to the NE in kap95 mutants. Also like Ran cycle mutants, the kap95-E126K mutant displayed enhanced lethality with nic96 and nup170 mutants. Thus, Kap95 and Ran are likely functioning at the same stage in assembly. However, although Ran cycle mutants accumulate small cytoplasmic vesicles, cells depleted of Kap95 accumulated long stretches of cytoplasmic membranes and had highly distorted NEs. We conclude that Kap95 serves as a key regulator of NPC assembly into intact NEs. Furthermore, both Kap95 and Ran may provide spatial cues necessary for targeting of vesicular intermediates in de novo NPC assembly.
INTRODUCTION
Nuclear pore complexes (NPCs) are large, proteinaceous structures that span the pore formed by fusion of the inner and outer nuclear membranes (INM and ONM). They are required for the passive diffusion of small metabolites and proteins and the selective transport of large proteins and RNAs between the nucleus and cytoplasm (Fahrenkrog and Aebi, 2003; Suntharalingam and Wente, 2003). Structural and proteomic studies of NPCs from several organisms have revealed a highly conserved 40–60-MDa entity with an eightfold axis of rotational symmetry perpendicular to the plane of the nuclear envelope (NE; Yang et al., 1998; Stoffler et al., 2003; Beck et al., 2004). The central core of the complex is formed from a series of spoke and ring-like structures that presumably anchor the NPCs to the NE. Filaments emanate from the cytoplasmic and nuclear NPC faces. Approximately 30 individual proteins, collectively termed nucleoporins or Nups, have been identified (Rout et al., 2000; Cronshaw et al., 2002) and can be isolated in discrete, evolutionarily conserved subcomplexes (reviewed in Vasu and Forbes, 2001; Suntharalingam and Wente, 2003). These Nup subcomplexes are believed to be the building blocks for NPC assembly.
In eukaryotic cells that undergo an open mitosis, both the NE and NPCs are disassembled at the onset of mitosis and must be reassembled after chromosome segregation (Burke and Ellenberg, 2002; Hetzer et al., 2005). Others have elegantly demonstrated that this reassembly is a highly ordered process, with temporal recruitment of individual Nups to the reforming NE and nucleus (Buendia and Courvalin, 1997; Bodoor et al., 1999; Haraguchi et al., 2000; Belgareh et al., 2001; Daigle et al., 2001; Loiodice et al., 2004; Rabut et al., 2004). During reassembly, unique membrane vesicles are rapidly recruited to the chromatin. Specific subsets of Nups also become associated with the chromatin at this time, and failure of these Nups to associate with chromatin before NE closure results in an irreversible block in NPC assembly (Harel et al., 2003b; Walther et al., 2003a; Antonin et al., 2005; Franz et al., 2005). The vesicles then fuse to form a closed NE (Franz et al., 2005). Finally, additional Nups are recruited to complete NPC biogenesis and result in a transport competent nucleus (Macaulay and Forbes, 1996; Goldberg et al., 1997).
Ran, a member of the Ras-like family of small GTPases, regulates key events during mitosis and the subsequent nuclear reassembly process (Dasso, 2002; Quimby and Dasso, 2003). The nucleotide-bound state of Ran is controlled by both a specific GTPase-activating protein (RanGAP, Rna1 in yeast; Bischoff et al., 1995), which stimulates GTP hydrolysis to favor the production of RanGDP, and a guanine nucleotide exchange factor (RanGEF, RCC1 in vertebrates and Schizosaccharomyces pombe; Prp20 in Saccharomyces cerevisiae; Bischoff and Ponstingl, 1991), which promotes nucleotide release to favor RanGTP. During interphase, RanGEF is localized to the nucleus, whereas RanGAP is cytoplasmic. The asymmetric distribution of the RanGEF and RanGAP regulatory factors results in RanGTP predominating in the nucleus, whereas RanGDP is favored in the cytoplasm (Hetzer et al., 2002; Kalab et al., 2002). Because RanGEF is tightly associated with chromatin, the concentration of RanGTP remains high near the chromosomes after nuclear envelope breakdown (Li et al., 2003; Li and Zheng, 2004; Kalab et al., 2006).
A cellular role for Ran was first elucidated in nucleocytoplasmic transport wherein RanGTP interactions with the karyopherin β family of transport receptors (also known as importins, exportins, and transportins) control the directionality of transport (Gorlich and Kutay, 1999; Pemberton and Paschal, 2005). During nuclear import, a karyopherin (Kap) recognizes and binds a nuclear localization sequence (NLS) within the imported protein. Once in the nucleus, RanGTP binds to the Kap causing NLS cargo release. In a parallel process, factors to be exported contain nuclear export sequences (NES), which are recognized by different Kap family members. However, for nuclear export, RanGTP is required to stabilize the Kap-NES cargo interaction. The presence of cytoplasmic RanGAP rapidly converts RanGTP to RanGDP resulting in NES cargo release. Thus, RanGTP acts as the molecular switch controlling Kap-cargo interactions with the asymmetric distribution of its regulatory factors providing the critical spatial information.
The mechanisms for RanGTP action in mitotic events also occurs by modulating the interactions of Kaps with key factors (Harel and Forbes, 2004; Mosammaparast and Pemberton, 2004). For example, importin β blocks spindle assembly by binding and sequestering specific spindle assembly factors (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Tsai et al., 2003). However, when these importin β complexes are near chromatin, the locally high concentration of RanGTP promotes RanGTP-importin β interaction and release of these factors. This localized release ensures that spindle assembly occurs only at chromosomes. Likewise, the balance of importin β and RanGTP controls proper NE assembly after mitosis. Addition of excess importin β to an in vitro nuclear reassembly assay blocks vesicle fusion and inhibits formation of a closed NE (Harel et al., 2003a; Walther et al., 2003b). In contrast, removal of importin β or addition of RanGTP leads to aberrant membrane fusion and the accumulation of cytoplasmic membranes harboring NPC structures (termed annulate lamellae). The molecular target(s) of importin β during NE assembly and membrane fusion are unknown. Interactions between RanGTP and importin β also appear to regulate the assembly of NPCs after mitosis and during subsequent nuclear growth in vitro (Harel et al., 2003a; Walther et al., 2003b; D'Angelo et al., 2006).
Although mitotic NPC reassembly has been extensively studied, there is less known about how NPCs are assembled de novo into an intact NE. In cells undergoing a closed mitosis, all NPCs must insert into a closed NE, presumably by coordinated recruitment of Nups during fusion of the INM and ONM to form the pore. For cells with an open mitosis, NPCs also must assemble into an intact NE during interphase when NPC number increases (Maul et al., 1971). Maintenance of NPC number in postmitotic cells also requires NPC biogenesis into a closed NE. To focus on the mechanism of NPC biogenesis in intact NEs, we have used a genetic strategy in the yeast S. cerevisiae (Ryan and Wente, 2002). We identified Ran as a key regulator of NPC assembly (Ryan et al., 2003). Intriguingly, mutants with perturbations at each stage of the Ran GTPase cycle accumulate small, cytoplasmic vesicles. At least one Nup, Nic96, is associated with these vesicles. This led us to propose a model for NPC assembly in which cytoplasmic Ran is necessary for the fusion of a novel, vesicular intermediate to the ONM and nucleation of new NPC formation.
A cytoplasmic function for the Ran GTPase cycle in de novo NPC assembly into intact NEs presents a paradox due to the predominant localization of RanGAP to the cytoplasm and RanGEF to the nucleus. However, several studies indicate that microenvironments of Ran and its regulatory factors exist within the cytoplasm and the nucleus. For example, RanGTP has been identified at centrosomes where it is involved in regulating duplication (Keryer et al., 2003; Wang et al., 2005). In addition, despite the high levels of RanGTP that are visualized around mitotic chromatin (Kalab et al., 2002, 2006), RanGAP has been localized to kinetochores and its mislocalization results in defects in kinetochore structure (Joseph et al., 2002; Arnaoutov and Dasso, 2003; Arnaoutov et al., 2005). Strikingly, this requirement for RanGAP activity in kinetochore function is also found during closed mitosis in S. cerevisiae (Tanaka et al., 2005), and a temperature-sensitive allele of S. pombe RanGAP (Sprna1ts) has chromosome segregation and centromeric silencing defects (Kusano et al., 2004).
Here, we characterize a mutant allele of KAP95, the yeast orthologue of importin β, that was isolated in our screen for nuclear pore complex assembly (npa) mutants. The mutant allele, kap95-E126K, results in a protein that is unstable at nonpermissive temperatures. Concatenate with the loss of Kap95, cells accumulate extended cytoplasmic membrane sheets and fail to correctly localize Nups to the NE. Taken together, we propose that Kap95 and Ran function together to spatially restrict fusion of a vesicular intermediate to the ONM for productive de novo NPC assembly into an intact interphase NE.
MATERIALS AND METHODS
Yeast Strains and Reagents
Yeast strains used in this study are listed in Table 1. General yeast manipulations were performed according to standard methods. All strains except SWY3189 and SWY3190 (see below) were grown at 23°C in YPD (yeast extract, peptone, 2% glucose) or synthetic complete (SC) media lacking appropriate amino acids and supplemented with 2% glucose unless otherwise indicated. For growth at the nonpermissive temperature, cells were incubated at 34 or 37°C. SWY3189 and SWY3190 (kap95Δ + pGAL-KAP95) were grown in YP + 1% galactose + 1% raffinose. To inhibit KAP95 expression, SWY3189 cells were washed into fresh YPD medium and incubated at 23°C. Cycloheximide (Sigma-Aldrich, St. Louis, MO) was added directly to growing cultures to a final concentration of 10 μg/ml. To compare strain growth at different temperatures, early log phase cultures were pelleted and resuspended in a minimal volume of 10 mM Tris HCl, 1 mM EDTA, pH 8. The number of cells was counted and then diluted to 2 × 107 cells/ml. A series of fivefold dilutions were made, and 5 μl of each was spotted to YPD plates and incubated at the appropriate temperature.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SWY518 | MATa ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | Bucci and Wente (1997) |
| SWY519 | MATα ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | Bucci and Wente (1997) |
| SWY811 | MATα ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ade2-1::ADE2:ura3 nup49ΔGLFG::GFP-S65T-TRP1 | Bucci and Wente (1998) |
| SWY1695 | MATa GFP-nic96:HIS3 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | Bucci and Wente (1998) |
| SWY2089 | MATα GFP-nic96:HIS3 nup170-GFP:URA3 lys2 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | Ryan and Wente (2002) |
| SWY2090 | MATa GFP-nic96:HIS3 nup170-GFP:URA3 trp1-1 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | Ryan and Wente (2002) |
| SWY2119 | MATα nup170-GFP:URA3 ura3-1 his3-11,15 lys2 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | This study |
| SWY3189 | MAT? kap95Δ::HIS3 GFP-nic96:HIS3 nup170-GFP:URA3 trp1-1 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 + pGAL-KAP95-TRP1 (pSW3067) | This study |
| SWY3190 | MATa kap95Δ::HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 + pGAL-KAP95-TRP1 (pSW3067) | This study |
| SWY3299 | MATα kap95-E126K GFP-nic96:HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | This study |
| SWY3300 | MATa kap95-E126K nup170-GFP:URA3 lys2 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | This study |
| SWY3561 | MATα kap95-E126K(npa16) nic96-GFP:HIS3 NUP170-GFP:URA3 lys2 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | This study |
| SWY3562 | MATa kap95-E126K trp1-1 ura3-1 his3-11,15 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | This study |
| SWY3609 | MATa kap95-E126K nup49ΔGLFG::GFP-S65T-TRP1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | This study |
Cloning and Plasmid Construction
Cloning of the wild-type allele of NPA16 was accomplished using a LEU2/CEN yeast genomic library as previously described (Ryan and Wente, 2002). To sequence the npa16 allele of kap95, genomic DNA was isolated from npa16 and used as a template in PCR to amplify the entire open reading frame. The PCR products from two independent reactions were cloned into pRS315 and sequenced. The results of the sequencing were compared with the yeast genome (www.yeastgenome.org) to identify the mutation.
To place KAP95 under the GAL promoter, oligonucleotides 1084 (5′-ACAAGATCTACCGCTGAATTTGCTC-3′) and 1085 (5′-GCGAGATCTTACAGGATCCCTAAGGATAATTGACGCTTC-3′) were used to amplify the KAP95 open reading frame of pSW503 (Iovine and Wente, 1997) using PCR. The resulting product was digested with BglII and cloned into the BamHI site of pSW1328 (Verbsky et al., 2002) to make pGAL-KAP95 (pSW3067). For the plasmid expressing the dominant kap95Δ48 mutant (pSW1037), oligonucleotides 672 (5′-ACCCGGATCCTCGATGAAAATACAAAGCTA-3′) and L15-2 (5′-CGCGGATCCATACATTGACTATTAACGCGCAG-3′) were used to amplify by PCR a fragment from the KAP95 open reading frame (in pSW503) for the sequence from the codon for amino acid 49 through the termination codon. The resulting product was digested with BamHI and cloned in frame with the sequence coding for glutathione S-transferase (GST) under the control of the GAL promoter cassette in pBJ382 (TRP1, 2μ pRS424).
Transport Assays and Microscopy
To assay steady state nuclear import efficiency, strains containing pGAD-GFP (cNLS-GFP; Shulga et al., 1996) or pNS167 (Nab2-NLS-GFP; Shulga et al., 1996) plasmids were grown to early log phase in SC media lacking uracyl at 23°C and then shifted to growth at 37°C for 6 h. Fluorescence and differential interference contrast (DIC) microscopy were performed on an Olympus BX50 microscope (Center Valley, PA) using an UPlan 100×/1.3 objective. Images were captured using a Photometrics CoolSnap HQ camera (Tucson, AZ) with MetaVue software. Indirect immunofluorescence was performed as previously described (Wente et al., 1992) with the mAb118C3 against Pom152 (Strambio-de-Castillia et al., 1995), polyclonal anti-Nup116 C-term antibody (Iovine et al., 1995), or monoclonal anti-Ran antibody (Transduction Laboratories, San Jose, CA) followed by Alexa-594–conjugated anti-mouse or anti-rabbit secondaries (Molecular Probes, Carlsbad, CA). Like-images within a figure were captured for the same exposure time, and all image processing was done in MetaVue. Final figures were assembled in Adobe Photoshop (San Jose, CA). Samples for thin-section electron microscopy were prepared as previously described (Wente and Blobel, 1993). Sections were viewed on a Philips CM-12 electron microscope (Mahwah, NJ), and images were taken using a CCD digital camera.
Western Analysis of Kap95 Protein Levels
Cultures were grown to early log phase and then shifted to 34 or 37°C for the indicated time or were shifted from growth in galactose to glucose media. Crude cell lysates were prepared by a method adapted from Yaffe and Schatz (1984). Briefly, 30 mg of cells was harvested, washed with water, and resuspended in 160 μl of 1.85 M sodium hydroxide, 7.4% β-mercaptoethanol and incubated on ice for 10 min. An equal volume of 50% trichloroacetic acid was added and incubated on ice for an additional 10 min for protein precipitation. Samples were centrifuged at 15K relative centrifugal force in a microcentrifuge for 2 min. Next, pellets were washed with 500 μl 1 M Tris base and resuspended in SDS sample buffer. The equivalents of 1.5 mg of starting cell pellets were separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with affinity-purified anti-Kap95 antibody (1:100; Iovine and Wente, 1997) followed by an alkaline-phosphatase labeled, anti-rabbit secondary antibody.
RESULTS
Identification of a kap95 Mutant in a Genetic Screen for Defective NPC Assembly
In our previous studies, we isolated a collection of nuclear pore complex assembly (npa) mutants in the yeast S. cerevisiae (Ryan and Wente, 2002). The npa mutants are temperature sensitive (ts) and are characterized by their inability to correctly localize GFP-Nic96 and Nup170-GFP to the NE/NPCs at the nonpermissive temperature. Nic96 and Nup170 are both associated with NPC subcomplexes that play critical roles in global NPC structure and assembly (Grandi et al., 1993; Aitchison et al., 1995; Zabel et al., 1996; Marelli et al., 1998). Thus, perturbation of GFP-Nic96 and Nup170-GFP localization at NPCs has proven to be an excellent indicator of NPC structural integrity and defects in NPC biogenesis (Zabel et al., 1996; Ryan and Wente, 2002; Ryan et al., 2003). To further understand the NPC assembly mechanism at the molecular level, we concentrated on the characterization of the npa16 complementation group, which was represented by a single allele in the mutant collection. The wild-type NPA16 gene was identified by complementation of the ts growth phenotype with a yeast genomic library. By DNA sequence analysis and comparison to the yeast genome database, we found that the library plasmid insert contained KAP95. A plasmid harboring only KAP95 was tested and found to rescue both the growth and GFP-Nup mislocalization phenotypes (data not shown). To delineate the precise mutation in the npa16/kap95 strain, the mutant allele was cloned and sequenced. A single point mutation that results in an amino acid substitution of Glu to Lys at amino acid 126 was found. This allele was designated kap95-E126K.
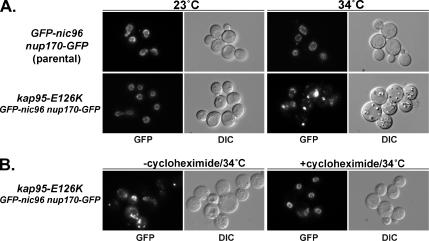
In the wild-type parental cells expressing both GFP-Nic96 and Nup170-GFP, the GFP-Nup signal forms a punctate pattern uniformly distributed around the NE (Figure 1A). This pattern indicates that these Nups are assembled into NPCs. When the kap95-E126K cells were grown at the permissive temperature of 23°C, GFP-Nic96 and Nup170-GFP were also localized to the NE/NPCs. However, after shifting the kap95-E126K cells to the nonpermissive growth temperature of 34°C for 5 h, defects in cell growth and perturbations in GFP-Nup localization were detected. At 23°C, the doubling time of the kap95-E126K cells was the same as the KAP95 parental strain. In contrast, shifting to growth at 34°C inhibited kap95-E126K cell growth, and cell divisions ceased after ∼4 h (data not shown). Coincidentally, the GFP-Nup signal at the NE rim became markedly diminished in the kap95-E126K cells. Most of the GFP-Nup signal was instead mislocalized and often concentrated in bright foci near the nucleus (Figure 1A).
Figure 1.
KAP95 is required for correct assembly of newly synthesized Nups. (A) The parental stain (GFP-nic96 nup170-GFP) and the npa16 mutant (kap95-E126K GFP-nic96 nup170-GFP) were grown at 23°C to early log phase and then shifted to 34°C (nonpermissive temperature) for 5 h. GFP-Nic96 and Nup170-GFP were localized by direct fluorescence microscopy of live cells (GFP columns). DIC images show overall cell morphology. (B) GFP-Nup mislocalization requires new protein synthesis in the npa16-kap95-E126K cells. Cells were grown as in (A) in the absence (−) or presence (+) of cycloheximide. Parental, SWY2089; npa16-kap95-E126K, SWY3561.
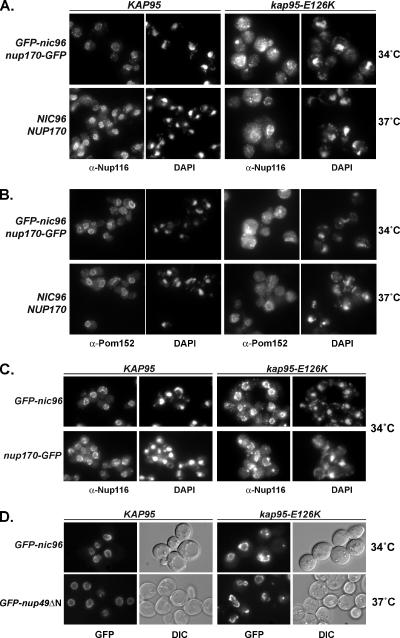
To test if the GFP-Nup mislocalization observed in the kap95-E126K cells at 34°C represented a global defect in Nup localization and NPC structure, the integral membrane pore-associated protein Pom152 and the GLFG-repeat containing nucleoporin Nup116 were localized using indirect immunofluorescence microscopy. Similar to GFP-Nic96 and Nup170-GFP, Pom152, and Nup116 were not correctly localized in the kap95-E126K GFP-nic96 nup170-GFP cells shifted to growth at 34°C (Figure 2, A and B, top rows). Like the GFP-Nups, these proteins appeared concentrated in cytoplasmic foci and in regions associated with the NE.
Figure 2.
Nup localization defects are observed in kap95-E126K cells with multiple combinations of GFP-nup alleles. (A and B) Analysis of Nup116 (A) and Pom152 (B) localization in KAP95 and kap95-E126K strains. Early log phase cells with GFP-nic96 and nup170-GFP (SWY2089, SWY3561) or wild-type alleles of both NUPs (SWY518, SWY3562) were shifted to the nonpermissive temperatures, (as determined in Figure 5) for 5 h and processed for indirect immunofluorescence microscopy with an antibody against Nup116 or Pom152. Note DAPI staining detects both nuclear (large spots) and mitochondrial (small spots) DNA. (C) Nup116 localization in wild-type or the kap95-E126K mutant cells combined with either GFP-nic96 (SWY1695, SWY3299) or nup170-GFP (SWY2119, SWY3300) was determined after shifting to growth at 34°C. Indirect immunofluorescence with anti-Nup116 antibodies was conducted as in A. (D) Localization of GFP-nic96 (SWY1695, SWY3299) or GFP-nup49ΔN (SWY811, SWY3609) in wild-type and kap95-E126K cells was determined after shifting cells at the early log phase of growth to the indicated temperature for 5 and 6 h, respectively. GFP signals were visualized by direct fluorescence microscopy. DIC images show overall cell morphology.
The failure of Nups to localize exclusively in NPCs at the nuclear rim could result from either a block in NPC assembly or an increased turnover of pre-existing NPCs. To distinguish between these two possibilities, new protein synthesis was inhibited by addition of cycloheximide during the shift to the nonpermissive growth temperature. Cycloheximide treatment blocked the mislocalization of the GFP-Nups in kap95-E126K cells grown at 34°C (Figure 1B). Equally significant is the observation that a weak GFP-Nup signal remained associated with the NE in the absence of cycloheximide. We have previously speculated that the fraction of GFP-Nup signal still associated with the NE and forming the weak nuclear ring in npa mutant cells (Figure 1, A and B, −cycloheximide) represents NPCs formed before the temperature shift. Indeed, previous studies of nic96 mutants have shown that if new assembly is blocked then NPC density in the NE decreases by half with every division (Zabel et al., 1996; Gomez-Ospina et al., 2000). Growth of such nic96 NPC assembly mutant cells arrests after two or three cell divisions. This correlates directly with the defects in the kap95-E126K mutant cells. These results strongly suggested that Nup mislocalization in the kap95-E126K cells was the result of a defect in assembling newly synthesized Nups into new NPCs.
The kap95-E126K Mutant Functions as an Effective Null with Complete Loss of Kap95 Protein
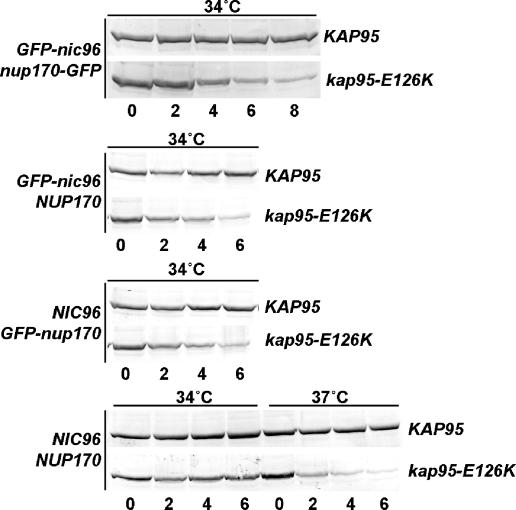
Kap95 is an essential member of the karyopherin-β family and required for nuclear import of cargo proteins containing a basic-type NLS (reviewed in Bednenko et al., 2003; Stewart, 2003). The x-ray crystal structure of Kap95 reveals a framework of 19 HEAT repeats (Lee et al., 2005; Liu and Stewart, 2005). These HEAT repeats form a superhelical structure that provides a large surface area for protein–protein interactions. Like other family members, Kap95 is divided into three functional domains. The N-terminal region binds the small GTPase Ran. The central HEAT repeats form a surface that directly interacts with Nups harboring FG, FxFG, or GLFG domains. The C-terminal domain provides a docking site for the adaptor karyopherin-α (Kap60/Srp1/importinα), which directly binds the NLS cargo. E126 is a highly conserved residue, and based on the crystal structure, lies on the outer surface of the protein (Cingolani et al., 1999; Lee et al., 2005; Liu and Stewart, 2005). However, co-crystal structures also indicate that it is not likely to be involved in binding FG-Nups (Bayliss et al., 2000; Bayliss et al., 2002). To examine the molecular defect caused by the kap95-E126K mutation, the ability of kap95-E126K protein to bind RanGTP, Kap60/Srp1, and GLFG or FxFG nucleoporins was assayed. Using both yeast two-hybrid and in vitro recombinant protein strategies, no qualitative differences in binding were observed between the wild-type Kap95 and kap95-E126K proteins and any of these binding partners (data not shown). Interestingly, Western blot analysis showed that the stability of the kap95-E126K protein in vivo was dramatically altered when cells were shifted to the nonpermissive temperature. In wild-type cells, Kap95 levels remained constant when cells were grown at 34°C. However, the levels of the kap95-E126K protein were dramatically decreased when the kap95-E126K cells were cultured at 34°C (Figure 3). This result suggested that E126 residue contributes to the stability of Kap95, especially at higher temperatures.
Figure 3.
kap95-E126K protein is unstable in cells grown at nonpermissive temperatures. Strains expressing different combinations of NIC96 and NUP170 alleles (left column) with either wild-type KAP95 or mutant kap95-E126K alleles (right column) were grown to early log phase and shifted to the indicated temperatures for various times (in hours). Total cell lysate from each was separated by SDS-PAGE, and immunoblotting with an anti-Kap95 antibody was performed to detect protein levels. Strains, from top to bottom: SWY2089, SWY3561, SWY1695, SWY3299, SWY2119, SWY3300, SWY518, SWY3562.
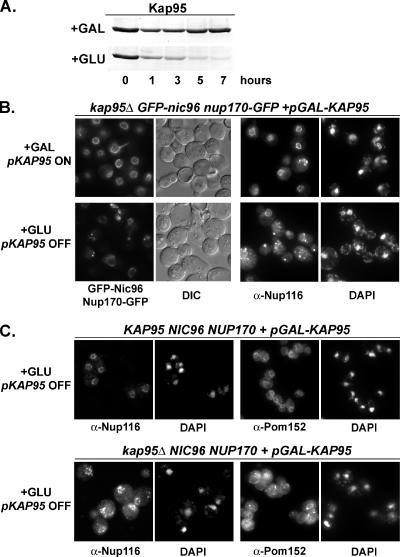
The decrease in protein levels suggested that the kap95-E126K mutant represents a loss of function allele that phenocopies the null allele at the nonpermissive growth temperature. Indeed, the time frame for the protein level decrease was coincident with the time frame for the GFP-Nup mislocalization defect to become apparent. To test if loss of Kap95 was directly responsible for the GFP-Nup mislocalization phenotype, KAP95 was deleted in GFP-nic96 nup170-GFP and wild-type strain backgrounds. A plasmid carrying KAP95 under an inducible promoter was introduced to allow for KAP95 expression and cell growth when the kap95Δ strains were grown in galactose. However, addition of glucose to logarithmically growing cells repressed KAP95 expression and led to the gradual loss of Kap95 protein (Figure 4A). When Kap95 levels decreased in the kap95Δ GFP-nic96 nup170-GFP cells, both GFP-Nup and Nup116 mislocalization was observed (Figure 4B). Likewise, in the kap95Δ cells lacking GFP-Nups, Pom152, and Nup116 were both mislocalized (Figure 4C). In both the kap95Δ and kap95Δ GFP-nic96 nup170-GFP cells, the perturbed Nups showed diminished nuclear rim signal and increased cytoplasmic signal with distinct foci. These phenotypes were indistinguishable from the kap95-E126K mutant phenotypes. Thus, it is the absence of Kap95 in the kap95-E126K mutants that is responsible for the NPC assembly defect.
Figure 4.
A kap95 null (Δ) mutant mimics the kap95-E126K phenotype. A plasmid containing KAP95 under control of a GAL promoter (pSW3067) allowed for KAP95 expression in wild-type KAP95 and kap95Δ strains. (A) Immunoblotting with an anti-Kap95 antibody of crude cell lysates harvested from kap95Δ GFP-nic96 nup170-GFP (SWY3189) cells grown in 1% galactose + 1% raffinose (+GAL) or after shifting to 2% glucose media (+GLU) for various times was performed to detect loss of Kap95 protein. (B) GFP-Nic96 and Nup170-GFP were visualized by direct fluorescence microscopy in the kap95Δ GFP-nic96 nup170-GFP (SWY3189) cells grown in 1% galactose + 1% raffinose (+GAL) or shifted to 2% glucose media (+GLU) for 8 h. In the same cells, indirect immunofluorescence microscopy with an anti-Nup116 antibody was used to detect Nup116 localization. (C) Localization of Pom152 and Nup116 by indirect immunofluorescence microscopy was conducted in the wild-type KAP95 (SWY518, top row) and the kap95Δ strain (SWY3190, bottom row) after shifting the cells from growth in YP 2% galactose to YP 2% glucose.
Functional Connections Revealed by Genetic Interactions between the kap95-E126K and nup Alleles
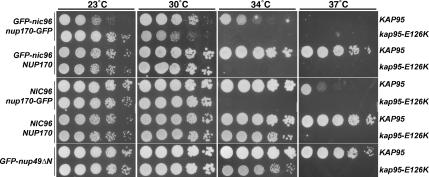
For the npa screen, the GFP sequence was integrated into the genome to generate a parental strain that contained GFP-nic96 and nup170-GFP. The GFP-tagged proteins that result are not fully functional, because the strain expressing both GFP-Nic96 and Nup170-GFP is ts at 37°C but not 34°C (Figure 5, and Ryan and Wente, 2002). In our previous analysis of the Ran GTPase cycle in NPC assembly, we found genetic interactions between the Ran GTPase cycle mutants (e.g., ntf2-H104Y and rna1-S116F) and the GFP-nic96 and nup170-GFP alleles (Ryan et al., 2003). If the Kap95 function in NPC assembly is linked to the role for Ran, we speculated that the kap95-E126K mutant might also have genetic interactions with the nup alleles. As isolated in the npa screen, the kap95-E126K GFP-nic96 nup170-GFP strain (npa16) failed to grow at 34°C. However, when the kap95-E126K mutation was crossed to a strain background containing wild-type alleles of NIC96 and NUP170, the cells were no longer ts at 34°C. The lowest nonpermissive temperature of the kap95-E126K NIC96 NUP170 mutant strain was instead 37°C (Figure 5). To determine which GFP-nup was contributing to the ts phenotype, kap95-E126K was combined independently with the GFP-nic96 and nup170-GFP alleles. Neither the GFP-nic96 nor the nup170-GFP strain was ts, although growth of the nup170-GFP strain was slower at 37°C. However, both the kap95-E126K GFP-nic96 and the kap95-E126K nup170-GFP strains were lethal at 34°C (Figure 5). Thus, the kap95-E126K phenotype was exacerbated by either nup allele.
Figure 5.
The kap95-E126K mutant displays enhanced lethality with GFP-nic96 and nup170-GFP alleles. Serial dilutions of strains with various combinations of NIC96 and NUP170 alleles (left column) in either wild-type KAP95 or kap95-E126K mutant backgrounds (right column) were spotted on YPD for growth at the indicated temperature. Strains, from top to bottom: SWY2090, SWY3561, SWY1695, SWY3299, SWY2119, SWY3300, SWY518, SWY3562, SWY811, SWY3609.
Next, we asked whether the genetic interactions between kap95-E126K and GFP-nic96 or nup170-GFP were specific or if kap95-E126K had enhanced temperature sensitivity with all mutant nup alleles. Unlike GFP-nic96 and nup170-GFP, GFP-nup49ΔN did not enhance the temperature sensitivity of kap95-E126K. The kap95-E126K GFP-nup49ΔN double mutant grew in a similar manner as the kap95-E126K single mutant (Figure 5).
We also tested for protein stability in the kap95-E126K GFP-nup versus NUP strains (Figure 3). Wild-type Kap95 protein levels remained constant in the different GFP-nup strains and at different growth temperatures. In either the GFP-nic96 or nup170-GFP strains, the levels of the kap95-E126K protein decreased over time at 34°C. In contrast, kap95-E126K protein was stable in NIC96 NUP170 cells grown at 34°C. However, when the kap95-E126K NIC96 NUP170 cells were shifted to growth at 37°C (the nonpermissive growth temperature for this strain) the kap95-E126K protein levels decreased. Thus, the temperature sensitivity of the kap95-E126K GFP-nup strains was directly correlated with kap95-E126K protein levels. Moreover, Nic96 and Nup170 both individually contributed to the stabilization of kap95-E126K protein at 34°C but were insufficient at 37°C.
To evaluate whether the GFP-nup alleles also contributed to the Nup mislocalization defect observed in the kap95-E126K GFP-nic96 nup170-GFP mutant, a series of different strains were tested. In the kap95-E126K GFP-nic96 and kap95-E126K nup170-GFP cells, Nup116 localization was altered after growth at the nonpermissive temperature (Figure 2C). Likewise, in kap95-E126K GFP-nic96 and kap95-E126K GFP-nup49ΔN cells, GFP-Nic96 and GFP-nup49ΔN were each independently perturbed (Figure 2D). It is interesting that the GFP-nup49ΔN allele did not shown a genetic interaction with the kap95-E126K mutant, but the GFP-nup49ΔN protein was perturbed in the same relative manner as a GFP-nup from a gene that was genetically linked (nic96 and nup170). This correlates with the above results in the kap95Δ strain where the GFP-tagged Nups were not required for the NPC perturbation (Figure 4C). Thus, the primary defect in NPC assembly in the kap95-E126K mutant is independent of but synergistic with the GFP-nic96 nup170-GFP mutant phenotype.
Cytoplasmic Membrane Accumulation Is Observed in the kap95-E126K Mutant Cells
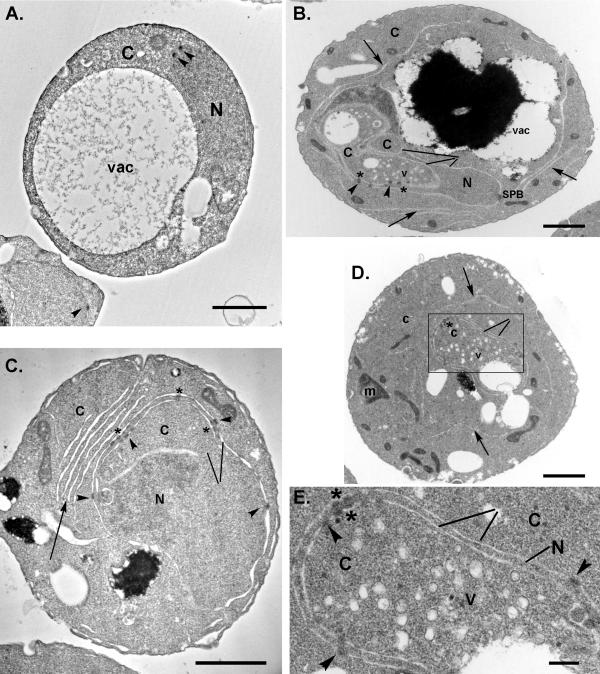
Thin-section electron microscopy was used to assess the ultrastructural consequences associated with the kap95-E126K mutant. At 34°C, cells from the KAP95 GFP-nic96 nup170-GFP parental strain had normal morphology with small amounts of cortical ER and a continuous NE with electron-dense NPCs spanning the INM and ONM (Figure 6A). In contrast, nuclear architecture was severely distorted at the nonpermissive temperature in kap95-E126K GFP-nic96 nup170-GFP cells. We observed several distinct differences compared with wild-type cells. First, large invaginations of the cytoplasm into the nuclear sphere resulted in the nucleus and NE appearing as layered membrane rings (lines, Figure 6, B–E). Often the NPCs aligned between the different juxtaposed membranes rings (Figure 6, B–E). This alignment likely accounts for the bright foci observed by direct fluorescence microscopy of the GFP-Nups (Figures 1 and 4). Second, interestingly, some of the NPC-like structures failed to span the NE and were associated with just the INM (asterisks, Figure 6, B–E). Third, the accumulation of ∼70-nm vesicles was also observed within many of the cytoplasmic invaginations (Figure 6, B, D, and E). Fourth, in addition to nuclear defects, kap95-E126K GFP-nic96 nup170-GFP cells also had long stretches of cytoplasmic membranes (arrow, Figure 6, B–D). Unlike annulate lamellae, no electron-dense NPC structures were observed in these cytoplasmic membrane sheets. All of the electron dense NPC-like structures were consistently associated with membranes linked to the NE.
Figure 6.
Loss of Kap95 leads to disruption of cellular membrane architecture. Cells in early log phase for (A) the parental stain (SWY2089) and (B–E) the corresponding kap95-E126K GFP-nic96 nup170-GFP mutant (SWY3561) were shifted to growth at 34°C for 5 h, then processed for thin-section transmission electron microscopy. Arrowheads denote NPCs that span the NE, asterisks (*) mark areas where NPC-like structures appear restricted to the INM in the kap95-E126K mutant. Lines point to NE; arrows point to stretches of cytoplasmic (nonnuclear) membranes. N, nucleus; C, cytoplasm; v, vesicles; m, mitochondria; SPB, spindle pole body; vac, vacuole. Bars, (A–D) 1 μm; (E) 200 nm.
To confirm that the ultrastructural perturbations were directly linked to the Kap95 defect, we also examined the kap95Δ mutants by thin-section electron microscopy. After growth of the kap95Δ GFP-nic96 nup170-GFP strain under conditions to repress KAP95, membranes structures accumulated in the cytoplasm (Supplementary Figure 1). The morphology was indistinguishable from that in the kap95-E126K GFP-nic96 nup170-GFP cells. Moreover, electron microscopy analysis of kap95Δ NIC96 NUP170 cells again showed the same phenotype (Ryan, unpublished data).
Kap95 Role in NPC Assembly Is Linked to Ran Function in Assembly
Given our prior studies showing an in vivo role for the RanGTPase cycle in NPC assembly (Ryan et al., 2003), we tested for whether the defects in the kap95 mutants were directly linked to Ran function. One possibility was that depleting Kap95 may indirectly alter the cellular levels of RanGTP or Ran localization. By indirect immunofluorescence microscopy, we analyzed the localization of Ran in wild-type and kap95-E126K GFP-nic96 nup170-GFP cells. No dramatic changes were observed (Supplementary Figure 2). We also noted that Nup localization was not altered when wild-type KAP95 was overexpressed (Figure 7B).
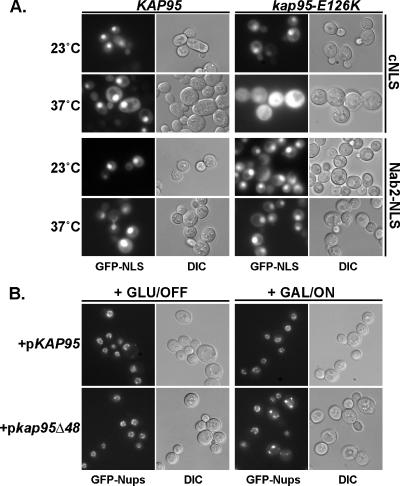
Figure 7.
The role for Kap95 in NPC assembly is linked to a Kap95-RanGTP interaction. (A) Nuclear transport is not globally perturbed in the kap95-E126K mutant. The effects of the kap95-E126K mutant on nuclear import via the Kap95 pathway versus the Kap104 pathway were assayed using GFP reporters in wild-type (SWY518) and kap95-E126K (SWY3562) cells. cNLS-GFP (Kap95 pathway) and Nab2-NLS-GFP (Kap104 pathway) fusions were visualized by direct fluorescence microscopy after shifting to growth at 37°C for 6 h. (B) The kap95Δ48 dominant mutant perturbs GFP-nup localization. Wild-type cells expressing GFP-nic96 and nup170-GFP (SWY2090) transformed with the wild-type pGAL-KAP95 (pSW3067) or dominant pGAL-GST-kap95Δ48 (pSW1307) plasmids were grown to early log phase in SC media lacking tryptophan and supplemented with 2% glucose then shifted to 2% galactose media for 20 h. Direct fluorescence microscopy was used to detect the GFP-Nups. DIC shows overall cell morphology.
A further test of global RanGTP function was conducted by monitoring the steady state nuclear import capacity of two independent Kap pathways. If Ran function was indirectly altered, we would expect all Kap-dependent import pathways to be inhibited. Wild-type and kap95-E126K cells harboring plasmids expressing GFP reporters for a Kap95 cargo (cNLS) and a Kap104 cargo (Nab2-NLS) were examined after growth at the nonpermissive temperature. In wild-type cells, the fluorescence signals for both cNLS-GFP and Nab2-NLS-GFP were concentrated in the nucleus. In kap95-E126K cells, the nuclear cNLS-GFP signal was markedly diminished reflecting decreased nuclear import (Figure 7A, top rows). This was expected as Kap95 import should be abolished as the kap95-E126K protein is degraded. In contrast, the Nab2-NLS-GFP remained localized in the nucleus of temperature-arrested kap95-E126K cells (Figure 7A, bottom row). This indicated that RanGTP function in the Kap104 import pathway was not perturbed.
To directly test for whether Ran interaction with Kap95 was required for NPC assembly, we made use of a dominant mutant wherein the sequence encoding the N-terminal 48 amino acid residues of Kap95 is deleted (kap95Δ48). Others have shown that the N-terminal 44 amino acids of the vertebrate importin β are necessary for RanGTP binding (Kutay et al., 1997). In in vitro import assays, the importin β lacking RanGTP binding acts as a dominant negative, presumably because of the lack of recycling importin α (Kutay et al., 1997). Here, sequence-encoding residues 49–861 of Kap95 was fused in frame with that of GST and placed under the control of the inducible GAL10 promoter in a multicopy plasmid. The cell growth and GFP-nup localization for GFP-nic96 nup170-GFP cells harboring the GAL-GST-kap95Δ48 plasmid were compared with that for cells harboring a wild-type GAL-KAP95 plasmid. Induction of GAL-GST-kap95Δ48 expression resulted in a lethal phenotype when cells were grown on media with galactose as the carbon source (data not shown). Moreover, GFP-Nup localization was markedly altered in the galactose-induced GAL-GST-kap95Δ48 cells (Figure 7B). The GFP-Nup signal at the nuclear rim was diminished and cytoplasmic foci were prevalent. Thus, the kap95Δ48 mutant lacking the RanGTP-binding domain results in dominant negative effects on Nup localization. This links the roles for Kap95 and RanGTP in NPC assembly.
DISCUSSION
Here, we have shown that Kap95, the yeast orthologue of importin β, is required for normal nuclear architecture and cellular membrane dynamics. When Kap95 levels decrease, cells have a highly invaginated NE and accumulate extended sheets of cytoplasmic membranes. These membrane perturbations are distinct from the cytoplasmic honeycomb-like appearance of membranes found in the npa mutants that block ER-to-Golgi transport and indirectly effect Nup localization and NE morphology (Ryan and Wente, 2002). Instead, the membrane defects in kap95-E126K cells are more similar to the annulate lamellae and excess NE observed when RanGTP is added to or importin β is depleted from in vitro Xenopus nuclear reassembly assays (Harel et al., 2003a; Walther et al., 2003b). Our results suggest that, just as in mitotic nuclear reassembly, Kap95 serves to regulate membrane fusion to shape an intact NE. Because these effects are observed in the absence of NE breakdown in yeast, they raise the interesting possibility that importin β may also have a role in NE dynamics during interphase in metazoan cells.
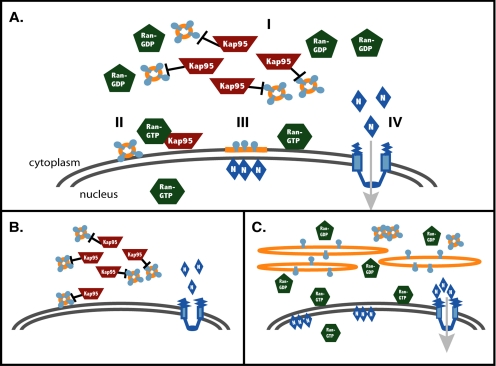
At the molecular level, Ran functions by modulating interactions between Kaps and other proteins. Previously, we have shown that disrupting the Ran GTPase cycle inhibits NPC assembly and results in the cytoplasmic accumulation of distinct Nup containing vesicles (Ryan et al., 2003). We proposed that these vesicles target a subset of Nups and NPC assembly factors to the ONM where RanGTP is required for their fusion to the NE. Significantly, the long stretches of nonnuclear membranes that are observed in the kap95-E126K loss-of-function mutant directly contrast with the cytoplasmic vesicles that accumulate in the Ran GTPase cycle mutants. We conclude that Kap95 is not simply regulating the cellular levels of RanGTP. Instead, the Kap95-RanGTP function in NPC assembly occurs by an independent mechanism. Overall, our results indicate that Kap95 and Ran have antagonistic effects on membrane dynamics. With these results, we have now refined our model for NPC assembly to include an inhibitory role for Kap95 in the fusion of the novel, Nup-containing vesicles (Figure 8).
Figure 8.
Model for the spatial regulation of NPC assembly. (A) In wild-type cells, Kap95 inhibits vesicle intermediates from fusing in the cytoplasm where RanGDP concentrations are high (I). Near the ONM, binding of RanGTP to Kap95 relieves the inhibitory effect and the assembly vesicle can fuse to the ONM (II). This fusion event provides a concentration of NPC assembly factors at the ONM that is able to recruit additional proteins (designated by N-labeled diamonds) to the INM for NPC assembly from both the nuclear and cytoplasmic faces of the NE (III). Such nuclear assembly factors would be imported through existing NPCs (IV). (B) When the Ran GTPase cycle is disrupted, Kap95 continues to block vesicle fusion even at the NE, and the vesicle intermediates accumulate in the cytoplasm. Factors that would normally be transported into the nucleus also remain in the cytoplasm as nucleocytoplasmic transport becomes disrupted with perturbations of the Ran GTPase cycle. (C) The absence of Kap95 allows for the unregulated cytoplasmic fusion of assembly vesicles to form membrane sheets. This results in a failure to target necessary assembly factors to the ONM. Because nuclear import can still occur, nuclear assembly factors concentrate at the INM and are not available for assembly into the cytoplasmic membranes.
We speculate that cytoplasmic Kap95 normally inhibits the fusion of these vesicles until they reach the NE where a highly localized pool of RanGTP binds and sequesters Kap95 (Figure 8A). This would allow specific vesicular fusion to the ONM. In the absence of Kap95, vesicular fusion would not be spatially regulated (Figure 8C). The lack of spatial regulation would result in the indiscriminate fusion of these vesicles into extended sheets of cytoplasmic membranes and the failure to correctly target Nups and other assembly factors to the NE for NPC assembly. Consistent with this hypothesis, both the membrane and Nup mislocalization phenotypes are seen when Kap95 levels decrease in either the kap95-E126K or kap95Δ mutant backgrounds. In addition, the dominant kap95Δ48 mutant also resulted in GFP-nup perturbations. Although necessary, the loss of Kap95 does not appear to be sufficient for fusion, because vesicles still accumulate in the cytoplasmic invaginations. This suggests that additional fusion machinery, which is not trapped in the invaginations, is required for efficient vesicle fusion. The kap95Δ48 mutant could be acting by sequestering such an assembly factor that needs to be released by RanGTP binding to Kap95. Thus, Kap95 could inhibit fusion through interactions with vesicle proteins or cytoplasmic fusion proteins.
Although a vesicle intermediate during interphase NPC assembly may seem counterintuitive, such an intermediate could be important in specifically targeting NPC assembly to the NE. The annulate lamellae found in oocytes, early embryonic cells, and other rapidly proliferating cells indicate that NPC assembly is not restricted to the NE (Kessel, 1992). However, the rapid proliferation of these cell types means NPCs are continually disassembled during the inherent division cycles and can be reformed in the NE or new annulate lamellae during postmitotic nuclear reassembly (Stafstrom and Staehelin, 1984; Cordes et al., 1996). Postmitotic cells or cells undergoing a closed mitosis do not have this alternative assembly opportunity and may require that the location of NPC assembly be more tightly regulated. Assembly vesicles would allow essential factors to be concentrated and then removed from membranes to prevent the nucleation of NPCs at nonnuclear membranes. Interactions between Ran and Kap95 at the ONM would provide the critical spatial information for subsequent vesicle fusion and allow NPCs to assemble only in the NE.
Unlike the annulate lamellae formed in in vitro Xenopus extracts by sequestration of importin β (Harel et al., 2003a; Walther et al., 2003b), the cytoplasmic membranes in kap95-E126K or kap95Δ yeast cells do not contain electron dense NPCs structures. A long-standing model for NPC assembly has been that it occurs from both the nuclear and cytoplasmic faces of the NE (Macaulay and Forbes, 1996), and recent studies have confirmed this model (D'Angelo et al., 2006). During assembly into a closed NE, access of these Nups to the INM could occur by transport through existing NPCs. This has been demonstrated in studies of Nup53 import by Kap121 (Marelli et al., 2001; Lusk et al., 2002). Further, with an intact NE, nuclear import of factors required at INM would make them unavailable for assembly into annulate lamellae in the cytoplasm. This may account for the lack of NPCs structures in the cytoplasmic membranes found in kap95-E126K cells. Moreover, the presence of electron dense plaques at the INM when Kap95 is depleted (Figure 6, asterisks) may reflect the accumulation of such INM/NPC assembly components.
Although nuclear import is most likely required for continued NPC assembly during interphase, defects in nuclear import alone cannot fully account for the perturbations in the kap95-E126K mutant. Indeed, global nuclear import is not perturbed in the kap95-E126K mutant. If defective import were the sole cause, then inhibiting nuclear import by loss of either Ran or Kap95 function should result in the same phenotypes. However, the Ran GTPase cycle mutants and kap95 loss-of-function mutants have opposite effects on membrane morphology, which are not the consequence of altering overall Ran levels. This is compelling evidence for additional, cytoplasmic roles for Ran and Kap95 during NPC assembly into a closed NE. Furthermore, Ran action in the cytoplasm is consistent with published in vitro nuclear assembly and growth results (Harel et al., 2003a; Walther et al., 2003b; D'Angelo et al., 2006). During the assembly process, two distinct steps are inhibited by GTPγS (Macaulay and Forbes, 1996). The first is the fusion of chromatin bound vesicles to form a closed NE, which has been shown to be Ran-dependent (Hetzer et al., 2000). In addition, GTPγS independently blocks the assembly of NPCs into the closed NE (Macaulay and Forbes, 1996), which is a common step in both postmitotic and interphase assembly. This block can be rescued with the addition of fresh extract to the cytoplasmic face of the NE (Macaulay and Forbes, 1996). Addition of importin β also inhibits NPC assembly at this stage (Harel et al., 2003a; Walther et al., 2003b). Continued insertion of NPCs into a growing nucleus was also found to require a balance of importin β and RanGTP at the ONM (D'Angelo et al., 2006). We hypothesize that the kap95-E126K mutant identified in the npa screen is blocked in the assembly mechanism at the stage before fusion of the inner and outer nuclear membranes is triggered (Figure 8C).
Both Nic96 and Nup170 are highly abundant Nups implicated in NPC assembly and structure (Grandi et al., 1993; Aitchison et al., 1995; Zabel et al., 1996; Marelli et al., 1998); however, neither contains a characteristic FG-repeat domain necessary for karyopherin binding during nuclear import or export through the NPC (Bednenko et al., 2003). Therefore, we were surprised to find that the severity of the ts phenotype of kap95-E126K strains was dependent on NIC96 and NUP170 alleles. Both GFP-nic96 and nup170-GFP were able to independently exacerbate the temperature sensitivity of kap95-E126K. In contrast, GFP-nup49ΔN, a mutant allele of a FG-NUP, did not enhance the temperature sensitivity of kap95-E126K. Others have demonstrated that efficient targeting of Nup53 to the NPC requires another karyopherin, Kap121. This targeting is dependent on a Kap121-binding domain in a non-FG region of Nup53 (Lusk et al., 2002). It is also interesting to note that a form of importin β with deceased affinity for FG nup binding, importin β I178D, is still able to inhibit NPC assembly in the Xenopus system (Walther et al., 2003b). These results indicate that Kap95 may interface with a set of factors during NPC assembly that are distinct from those that it interacts with for NPC translocation.
Because of the asymmetric distribution of Ran regulatory factors, accepted models have restricted interactions between RanGTP and Kaps to either the nucleus during interphase or near chromatin during an open mitosis. Thus, the cytoplasmic effects of our Ran GTPase cycle mutants were unexpected (Ryan et al., 2003). However, recent in vitro studies using Xenopus extracts support our model for a cytoplasmic function of RanGTP in NPC assembly into an intact NE (D'Angelo et al., 2006). Furthermore, Hetzer and coworkers found that RanGTP and importin β had opposite effects on the assembly processes in vitro (D'Angelo et al., 2006). Now, our in vivo characterization of the kap95-E126K mutant provides a true parallel to these in vitro results. Exactly how RanGTP is generated at the ONM remains unclear. Because Ran is only 25 kDa in size and within the diffusion limit for the NPC, diffusion of nuclear RanGTP across the NPC may produce a highly localized region of RanGTP at the ONM. Alternatively, localization of Ran regulatory factors to the ONM may establish the correct, local Ran environment. Significantly, these in vitro and in vivo functional requirements for Ran and Kap95 (importin β) during interphase NPC assembly have been discovered in organisms with very distinct nuclear architecture and dynamics (Xenopus and budding yeast). We predict that the mechanism for NPC assembly into intact NEs will be conserved in all eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Ginger Winfrey and Gary Olson for assistance and expertise with the electron microscopy experiments. We thank Kathy Iovine for construction of the plasmid pSW1037 and documentation of the dominant kap95Δ48 growth defect, Michael Rout for providing anti-Pom152 antibodies, David Goldfarb for plasmids, and Wente laboratory members for valuable comments on the manuscript and critical discussions. This work was supported by a grant from the National Institutes of Health, R01 GM-57438 to S.R.W.
Abbreviations used:
- GFP
green fluorescent protein
- INM
inner nuclear membrane
- NE
nuclear envelope
- NES
nuclear export sequence
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- Δ
null
- Nup
nucleoporin
- ONM
outer nuclear membrane
- ts
temperature sensitive.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0525) on December 20, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aitchison J. D., Rout M. P., Marelli M., Blobel G., Wozniak R. W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I. W. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Dasso M. The Ran GTPase regulates kinetochore function. Dev. Cell. 2003;5:99–111. doi: 10.1016/s1534-5807(03)00194-1. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood T., Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood T., Strawn L. A., Wente S. R., Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- Beck M., Forster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Bednenko J., Cingolani G., Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003;4:127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- Belgareh N., et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Krebber H., Kempf T., Hermes I., Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc. Natl. Acad. Sci. USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W. H., Bastos R., Lohka M., Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 1999;112(Pt 13):2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Bucci M., Wente S. R. In vivo dynamics of nuclear pore complexes in yeast. J. Cell Biol. 1997;136:1185–1199. doi: 10.1083/jcb.136.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M., Wente S. R. A novel fluorescence-based genetic strategy identifies mutants of Saccharomyces cerevisiae defective for nuclear pore complex assembly. Mol. Biol. Cell. 1998;9:2439–2461. doi: 10.1091/mbc.9.9.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia B., Courvalin J. C. Domain-specific disassembly and reassembly of nuclear membranes during mitosis. Exp. Cell Res. 1997;230:133–144. doi: 10.1006/excr.1996.3395. [DOI] [PubMed] [Google Scholar]
- Burke B., Ellenberg J. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Cingolani G., Petosa C., Weis K., Muller C. W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Cordes V. C., Reidenbach S., Franke W. W. Cytoplasmic annulate lamellae in cultured cells: composition, distribution, and mitotic behavior. Cell Tissue Res. 1996;284:177–191. doi: 10.1007/s004410050578. [DOI] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M. A., Anderson D. J., Richard E., Hetzer M. W. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. The Ran GTPase: theme and variations. Curr. Biol. 2002;12:R502–R508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B., Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- Franz C., Askjaer P., Antonin W., Iglesias C. L., Haselmann U., Schelder M., de Marco A., Wilm M., Antony C., Mattaj I. W. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. W., Wiese C., Allen T. D., Wilson K. L. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J. Cell Sci. 1997;110(Pt 4):409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- Gomez-Ospina N., Morgan G., Giddings T. H., Jr., Kosova B., Hurt E., Winey M. Yeast nuclear pore complex assembly defects determined by nuclear envelope reconstruction. J. Struct. Biol. 2000;132:1–5. doi: 10.1006/jsbi.2000.4305. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E. C. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Haraguchi T., Koujin T., Hayakawa T., Kaneda T., Tsutsumi C., Imamoto N., Akazawa C., Sukegawa J., Yoneda Y., Hiraoka Y. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 2000;113(Pt 5):779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- Harel A., Chan R. C., Lachish-Zalait A., Zimmerman E., Elbaum M., Forbes D. J. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell. 2003a;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Forbes D. J. Importin beta: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D. J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003b;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Bilbao-Cortes D., Walther T. C., Gruss O. J., Mattaj I. W. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Gruss O. J., Mattaj I. W. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 2002;4:E177–E184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- Hetzer M. W., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Iovine M. K., Watkins J. L., Wente S. R. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine M. K., Wente S. R. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J. Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J., Tan S. H., Karpova T. S., McNally J. G., Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pralle A., Isacoff E. Y., Heald R., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Keryer G., Di Fiore B., Celati C., Lechtreck K. F., Mogensen M., Delouvee A., Lavia P., Bornens M., Tassin A. M. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol. Biol. Cell. 2003;14:4260–4271. doi: 10.1091/mbc.E02-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel R. G. Annulate lamellae: a last frontier in cellular organelles. Int. Rev. Cytol. 1992;133:43–120. doi: 10.1016/s0074-7696(08)61858-6. [DOI] [PubMed] [Google Scholar]
- Kusano A., Yoshioka T., Nishijima H., Nishitani H., Nishimoto T. Schizosaccharomyces pombe RanGAP homolog, SpRna1, is required for centromeric silencing and chromosome segregation. Mol. Biol. Cell. 2004;15:4960–4970. doi: 10.1091/mbc.E04-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Izaurralde E., Bischoff F. R., Mattaj I. W., Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Matsuura Y., Liu S. M., Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- Li H. Y., Wirtz D., Zheng Y. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 2003;160:635–644. doi: 10.1083/jcb.200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Y., Zheng Y. The production and localization of GTP-bound ran in mitotic mammalian tissue culture cells. Cell Cycle. 2004;3:993–995. [PubMed] [Google Scholar]
- Liu S. M., Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Makhnevych T., Marelli M., Aitchison J. D., Wozniak R. W. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 2002;159:267–278. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C., Forbes D. J. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M., Aitchison J. D., Wozniak R. W. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M., Lusk C. P., Chan H., Aitchison J. D., Wozniak R. W. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 2001;153:709–724. doi: 10.1083/jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G. G., Price J. W., Lieberman M. W. Formation and distribution of nuclear pore complexes in interphase. J. Cell Biol. 1971;51:405–418. doi: 10.1083/jcb.51.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Pemberton L. F. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Quimby B. B., Dasso M. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 2003;15:338–344. doi: 10.1016/s0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., McCaffery J. M., Wente S. R. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Wente S. R. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 2002;3:17. doi: 10.1186/1471-2156-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Roberts P., Gu Z., Spitz L., Tabb M. M., Nomura M., Goldfarb D. S. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom J. P., Staehelin L. A. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur. J. Cell Biol. 1984;34:179–189. [PubMed] [Google Scholar]
- Stewart M. Structural biology. Nuclear trafficking. Science. 2003;302:1513–1514. doi: 10.1126/science.1092863. [DOI] [PubMed] [Google Scholar]
- Stoffler D., Feja B., Fahrenkrog B., Walz J., Typke D., Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J. Mol. Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M. P. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S. R. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C., Tanaka T. U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- Vasu S. K., Forbes D. J. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Verbsky J. W., Wilson M. P., Kisseleva M. V., Majerus P. W., Wente S. R. The synthesis of inositol hexakisphosphate Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- Walther T. C., et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003a;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I. W., Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003b;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Wang W., Budhu A., Forgues M., Wang X. W. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 2005;7:823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- Wente S. R., Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Rout M. P., Blobel G. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Rout M. P., Akey C. W. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Zabel U., Doye V., Tekotte H., Wepf R., Grandi P., Hurt E. C. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.