Abstract
Cdc42-associated tyrosine kinase 1 (ACK1) is a specific down-stream effector of Cdc42, a Rho family small G-protein. Previous studies have shown that ACK1 interacts with clathrin heavy chain and is involved in clathrin-coated vesicle endocytosis. Here we report that ACK1 interacted with epidermal growth factor receptor (EGFR) upon EGF stimulation via a region at carboxy terminus that is highly homologous to Gene-33/Mig-6/RALT. The interaction of ACK1 with EGFR was dependent on the kinase activity or tyrosine phosphorylation of EGFR. Immunofluorescent staining using anti-EGFR and GFP-ACK1 indicates that ACK1 was colocalized with EGFR on EEA-1 positive vesicles upon EGF stimulation. Suppression of the expression of ACK1 by ACK-RNAi inhibited ligand-induced degradation of EGFR upon EGF stimulation, suggesting that ACK1 plays an important role in regulation of EGFR degradation in cells. Furthermore, we identified ACK1 as an ubiquitin-binding protein. Through an ubiquitin-association (Uba) domain at the carboxy terminus, ACK1 binds to both poly- and mono-ubiquitin. Overexpression of the Uba domain-deletion mutant of ACK1 blocked the ligand-dependent degradation of EGFR, suggesting that ACK1 regulates EGFR degradation via its Uba domain. Taken together, our studies suggest that ACK1 senses signal of EGF and regulates ligand-induced degradation of EGFR.
INTRODUCTION
The Cdc42-associated tyrosine kinase (ACK) family contains five members: ACK1, ACK2, Tnk1, Kos1, and Gene33 (also Mig-6 or RALT). Among the members, Gene33 is the only protein that does not have tyrosine kinase domain. ACK1, ACK2, and Gene33 are specific effectors of Rho family small GTPase Cdc42 (Manser et al., 1993, Yang and Cerione, 1997; Makkinje et al., 2000). ACK, including both ACK1 and ACK2, is activated by signaling of epidermal growth factor receptor (EGFR), M3 muscarinic receptor, and cell adhesion mediated by integrin and proteoglycan (Satoh et al., 1996; Yang and Cerione, 1997; Eisenmann et al., 1999; Yang et al., 1999; Linseman et al., 2001). In Drosophila, DACK mediates function of Cdc42 in dorsal closure during the embryo development (Sem et al., 2002). An ACK homolog, Ark-1, in Caenorhabditis elegans negatively regulates EGF signaling (Hopper et al., 2000).
The members of the ACK family function in regulation of cell growth. Ark-1 has shown to inhibit EGFR signals and suppress cell division in embryo development (Hopper et al., 2000). Overexpression of ACK2 in NIH3T3 cells severely impairs cell growth (Yang et al., 2001a). Kos-1, the homologue to the N-terminus portion of Tnk1 and ACK, suppresses Ras-mediated cellular transformation (Hoare et al., 2003). Gene33, the homologue to the carboxy terminus of ACK that includes Cdc42-binding and proline-rich domains, inhibits tyrosine phosphorylation and signaling of EGFR and ErbB2, thus blocking the mitogenic effect of EGFR and ErbB2 (Fiorentino et al., 2000; Xu et al., 2005). However, studies on the effect of ACK1 on Ras-GRF activity found that ACK1 activates Ras and is required for Ras-mediated cellular transformation in NIH3T3 cells (Nur-E-Kamal et al., 2005). Recent studies have shown that ACK1 enhances tumor metastasis by mediating integrin signaling (van der Horst et al., 2005) and promotes tumorigenesis by inhibiting tumor-suppression activity of WWOX (Mahajan et al., 2005). The variety of these studies suggests a complexity of the effects of members of ACK family on cell growth and tumorigenesis.
Both ACK1 and ACK2 possess a highly conserved clathrin-binding motif and interact with clathrin (Teo et al., 2001; Yang et al., 2001b). Overexpression of ACK2 severely impairs transferrin receptor endocytosis, causes aberrant localization of AP-2, and induces changes in clathrin assembly. Furthermore, ACK2 interacts with SH3PX1, a member of sorting nexin family, via its proline-rich domain 1 and phosphorylates SH3PX1 to facilitate the degradation of EGF receptors (Lin et al., 2002). In C. elegans, Ark-1 genetically interacts with UNC101, the homologue of mammalian clathrin-associated protein AP47, and SLI-1, the homologue of mammalian Cbl that is an E3 ubiquitin ligase for ubiquitination of EGFR, and negatively regulates EGFR signaling (Hopper et al., 2000). These data suggest a role of ACK in EGFR degradation.
Here we show that ACK1 is an ubiquitin-binding protein and interacts with EGFR through a conserved EGFR-binding domain (EBD) at the carboxy terminus. The interaction of ACK1 with EGFR is dependent on EGF stimulation and kinase activity of EGFR. ACK1 colocalizes with EGFR during the internalization of EGFR. We have demonstrated that ACK1 plays a role in EGFR degradation by using ACK-RNA interference (RNAi) knockdown of endogenous ACK1 and overexpression of the ubiquitin-binding defective mutant. Our studies suggest that ACK1 is a component of EGFR signaling and regulates ligand-induced EGFR degradation.
MATERIALS AND METHODS
Plasmids, Antibodies, and Chemicals
The mouse ACK1 and ubiquitin C cDNAs were purchased from IMAGE (the Integrated Molecular Analysis of Genomes and their Expression) Consortium (ACK1: IMAGE 5702405; Ubiquitin C: IMAGE 5123918). The ACK1 cDNA was subcloned into pcDNA3 with an amino terminal Myc-tag. The ubiquitin C contains the same eight ubiquitin repeats. To obtain mono-ubiquitin and different poly-ubiquitin, we performed PCR with primers that match the sequences at the start codon and the stop codon of ubiquitin. The PCR yielded multiple products representing different number of ubiquitin repeats. These PCR products were subcloned into the glutathione S-transferase (GST)-fusion protein vector pGEX4T-3. The EGFR/ErbB2 chimera, which contains amino acid residues 1–680 of human EGFR and amino acid residues 689-1255 of human ErbB2, was made by PCR-directed mutagenesis and subcloned in the pcDNA3 mammalian expression vector. The point mutations were performed using the mutagenesis kits purchased from Stratagene (La Jolla, CA) according the instructions by the manufacturer. The DNA sequences of mutants and new constructs were verified by DNA sequencing. Anti-EGFR (1005), -ACK1(C-20), -ACK1(A-11), and -c-Cbl antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti-PY (4G10) from UBI (Lake Placid, NY); anti-ubiquitin from Covance (Madison, WI); anti-proteasome S10B from Affinity BioReagents (Golden, CO). Anti-EEA1 from BD Transduction Laboratories (Lexington, KY). Anti-EGFR Mab528 was obtained from culture medium of hybridoma cell line 528 (ATCC, Manassas, VA). The rabbit anti-ACK (anti-ACKPCC) antibody was made at the animal facility of Cornell Veterinary School (Ithaca, NY) using the first 100 amino acid residues of ACK2 as an antigen that are conserved in both ACK1 and ACK2. The proteasome inhibitor MG-132 was purchased from Sigma (St. Louis, MO). The recombinant human EGF was purchased from Invitrogen (Carlsbad, CA).
Cell Culture and Transfection
COS7, 293, HeLa, CHO, breast cancer MDA-MB-231, nonsmall cell lung cancer H-358, the neuroblastoma Neuro-2a, SK-N-DZ, SH-SY5Y, and BE-2C cells were cultured in DMEM plus 10% fetal bovine serum. All cells were maintained in 5 or 10% CO2 at 37°C. For transfection, the cells were cultured overnight to 90% confluency. The transfection was performed with the LipofectAmine transfection kit according to the manufacturer's instructions (Invitrogen). For EGF treatment, the cells were serum-starved overnight (12–16 h) and then treated with EGF (100 ng/ml) for the indicated time. After 48 h of transfection, the cells were lysed with precold mammlian cell lysis buffer (40 mM HEPES, pH 7.4, 100 mM NaCl, 1% Triton X-100, 25 mM glycerol phosphate, 1 mM sodium orthovanadate, 1 mM EDTA, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) by rocking the plates at 4°C for 30 min. The cell lysates were cleared by centrifugation at 14,000 rpm in a microfuge for 4 min at 4°C before use.
Immunoprecipitation and Immunoblot
For immunoprecipitation, the precleared cell lysate was incubated with primary antibody on ice for 30 min, then protein A beads were added, and the mixture was incubated at 4°C for 2 h with rotation. The beads were washed with lysis buffer three times, and the immunoprecipitation complexes were ready either for enzymatic assays or directly dissolved in SDS-PAGE sample buffer for SDS-PAGE. The immunoblot was performed as instructed by ECL immunoblot kits (Amersham Pharmacia, Piscataway, NJ).
Expression and Purification of GST-Fusion Protein
GST-fusion proteins were expressed in Escherichia coli (JM109) and purified by affinity purification with glutathione-agarose beads as described previously (Yang et al., 2001b).
The Pulldown Assays with GST-Fusion Proteins
The GST-fusion protein beads containing 20–60 μg of GST-fusion protein (60 μg for microsequencing or 20 μg for immunoblotting) were incubated with the mammalian cell lysates (10–20 mg for microsequencing or 1 mg for immunoblotting) at 4°C for 3 h with rotation. The beads were washed three times with the mammalian cell lysis buffer and resuspended with 2× SDS-PAGE sample buffer. For microsequencing, after separated by SDS-PAGE, the proteins that are precipitated by the GST-fusion protein were visualized by staining with 0.5–1% Coomassie Blue.
Immunofluorescence Staining
The cells were cultured in the glass coverslip-bottomed culture dishes (MatTek, Ashland, MA) to 50–80% confluence. For EGF stimulation, the cells were serum-starved for 12 h before the treatment. After the culture medium was removed, the cells were rinsed with PBS twice, fixed with 3.7% paraformaldehyde at 25°C for 10 min, and permeabilized with 0.2% Triton X-100 in PBS at 25°C for 10 min. After washing with PBS, the cells were incubated with primary antibody at 37°C for 30 min. Then the cells were washed with PBS three times and incubated with secondary antibody that is conjugated with a fluorescent dye at 37°C for 30 min. Finally, the cells were washed with PBS three times (for 10 min each), and the immunofluorescence staining was visualized under a Zeiss inverted fluorescent microscope (Thornwood, NY).
RNAi
The ACK-RNAi experiment was performed according to the method described by Elbashir et al. (2001). The 21-nucleotide small interference RNA (siRNA) sequence (AAGAUGGUGACAGAGCUGGCA) corresponding to the coding region of the tyrosine kinase domain of ACK1 was selected. This nucleotide sequence was conserved in both ACK1 and ACK2 from human, mouse, and bovine. The short interfering RNA (siRNA) oligos were chemically synthesized (Dharmacon, Lafayette, CO). The negative control was set up using the 21-nucleotide RNA oligos (AAGUUCAGGUCGAUAUGUGCA), which does not match any DNA sequence in GenBank, as determined by NCBI Blast search. Transfection of the siRNA (final concentration 40 nM) into HEK293 or COS7 cells was carried out using LipofectAmine transfection kits (Invitrogen). The suppression of the expression of endogenous ACK1 by the siRNAs was determined by immunoblotting the anti-ACKPCC-immunoprecipitated ACK1 with anti-ACKPCC.
RESULTS
Identification of Endogenous ACK
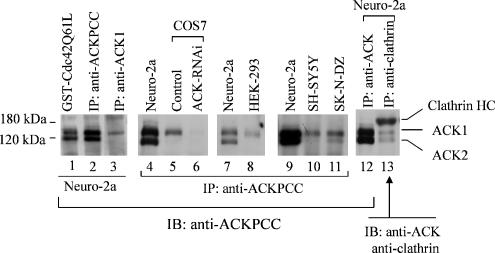
To study the function of endogenous ACK, we raised an anti-ACK antibody in rabbits that is against the first 100 amino acid residues of bovine ACK2. We designated this antibody as anti-ACKPCC. These 100-amino acid residues of bovine ACK2 are highly conserved in ACK1. Therefore, anti-ACKPCC reacts with both ACK1 and ACK2. To identify endogenous ACKs that are detected by anti-ACKPCC, we also performed GST-Cdc42Q61L pulldown to cross-examine the results. As shown in lanes 1–3 of Figure 1, immunoprecipitation of mouse neuroblastoma Neuro-2a cell lysates with anti-ACKPCC yielded two bands at 140 and 120 kDa that cross-react with anti-ACKPCC (Figure 1, lane 2). The pulldown by GST-Cdc42Q61L yielded the same two bands (Figure 1, lane 1), indicating that these two proteins are ACKs. The coimmunoprecipitation of Neuro-2a cell lysates with anti-clathrin light-chain antibody CON.1 also yielded the 140- and 120-kDa proteins that cross-reacted with anti-ACKPCC (Figure 1, lane 13), confirming that these two proteins are ACKs. To identify ACK1 from these two ACKs, we used the anti-ACK1 antibody (Santa Cruz Biotechnology) that is against the C-terminus of ACK1, which is not conserved in ACK2, to immunoprecipitate ACK1 from Neuro-2a cell lysates and to immunoblot the immunoprecipitated proteins with anti-ACKPCC. As shown in Figure 1, lane 3, only the 140-kDa protein was immunoprecipitated by anti-ACK1 antibody, indicating that the 140-kDa protein is ACK1. Comparing with exogenous Myc-tagged ACK1 also confirmed that the 140-kDa protein band is ACK1 (data not shown). The 120-kDa ACK was not immunoprecipitated by anti-ACK1; therefore, we designated the 120-kDa ACK as p120ACK2 in order to distinguish it from the 97-kDa ACK2 that we previously identified from bovine brain (Yang and Cerione, 1997). Considering that only one ACK gene is found in the human and mouse genome, we speculate that p120ACK2 in Neusro-2a cells is an alternative splicing form of ACK that is structurally similar to bovine ACK2. ACK1 is expressed in every cell line we have examined (Figure 1). However, p120ACK2 is only expressed in mouse neuroblastoma cell lines Neuro-2a and human neuroblastoma SK-N-DZ (Figure 1), suggesting that p120ACK2 may exert cellular function different from that of ACK1.
Figure 1.
Identification of endogenous ACK. ACKs in Neuro-2a, COS7, HEK-293, SH-SY5Y, and SK-N-DZ cell lysates were immunoprecipitated with anti-ACKPCC or anti-ACK1 (Santa Cruz) antibody (lane 3) or by pulldown with GST-Cdc42Q61L (40 μg of the GST-fusion protein in 1 ml lysates) and detected by immunoblotting with anti-ACKPCC. For immunoprecipitation of clathrin, anti-clathrin light chain antibody (CON.1) was incubated with 1 ml of the lysates. The coimmunoprecipitated ACK was detected by immunoblotting with anti-ACKPCC and the clathrin heavy chain was detected by immunoblotting with anti-clathrin heavy chain antibody (lane 13).
ACK1 Interacts with EGFR upon EGF Stimulation
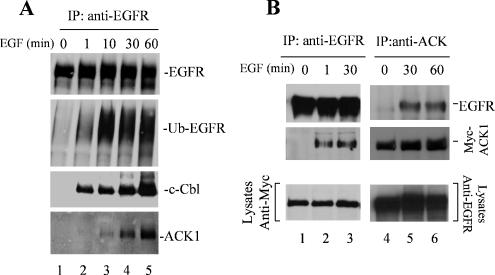
Tyrosine phosphorylation of ACK1 was enhanced by EGF stimulation in previous studies (Satoh et al., 1996). To examine that ACK1 is involved in EGF signaling, we determined the interaction of ACK1 with EGFR. We stimulated serum-starved COS7 cells with EGF for 1, 10, 30, and 60 min, subsequently immunoprecipitated EGFR with an anti-EGFR antibody (Mab528) that is against the extracellular domain of EGFR, and detected endogenous ACK1 in the EGFR complex by immunoblotting with anti-ACKPCC. As shown in Figure 2A, ACK1 was coimmunoprecipitated with EGFR upon the stimulation of EGF (the bottom panel), indicating that ACK1 is in the EGFR complex. The amount of coprecipitated ACK1 with EGFR increased along with the time course of EGF stimulation, corresponding to the ubiquitination of EGFR (the second panel from top). Cbl, an E3-ubiquitin ligase that specifically ubiquitinates EGFR, was also coimmunoprecipitated with EGFR upon the EGF stimulation (the third panel). The timing of the binding of ACK1 to EGFR upon EGF stimulation lagged behind the ubiquitination of EGFR and the binding of Cbl to EGFR (the bottom three panels of Figure 2A), suggesting that association of ACK1 with EGFR may require Cbl-mediated ubiquitination of EGFR. We also examined the binding of exogenous ACK1 to EGFR with coimmunoprecipitation. As shown in Figure 2B, immunoprecipitation of either EGFR (lanes 1–3) or ACK1 (lanes 4–6) confirmed the binding of exogenous Myc-tagged ACK1 to EGFR upon EGF stimulation. However, the binding of exogenous ACK1 with EGFR after EGF stimulation was much faster than the endogenous ACK1 (Figure 2B, lanes 2 and 3). Our explanation for the difference in the EGFR-interaction timing between endogenous and exogenous ACK1 is that the endogenous ACK1 may be restricted in cells; thus the interaction of endogenous ACK1 with EGFR may have a transition process that is involved in dissociation of ACK1 from other protein complexes or intracellular compartments, whereas overexpression of exogenous ACK1 produces free ACK1 available for interaction with EGFR. Nevertheless, all the data indicate that interaction of ACK1 with EGFR requires activation of EGFR, suggesting that interaction of ACK1 with EGFR is downstream of EGFR activation and in response to EGFR signaling.
Figure 2.
Interaction of ACK1 with EGFR is dependent on EGF stimulation. (A) The interaction of endogenous ACK1 with EGFR. The COS7 cells were serum-starved overnight and then stimulated with EGF (100 ng/ml) for the indicated time. EGFR was immunoprecipitated with anti-EGFR Mab528. The ubiquitinated EGFR was detected by an anti-ubiquitin antibody (Covance), and the coprecipitated c-Cbl or ACK1 was detected by immunoblotting the immunoprecipitation complex with an anti-c-Cbl (third panel from top) or anti-ACKPCC (bottom panel). (B) The interaction of exogenous ACK1 with EGFR. After transfection with Myc-tagged ACK1 for 36 h, the COS7 cells were serum-starved overnight and subsequently stimulated with EGF (100 ng/ml) for the indicated time. Endogenous EGFR was immunoprecipitated with an anti-EGFR antibody (Mab528; lanes 1–3). Myc-tagged ACK1 was immunoprecipitated with anti-ACK1 (A-11, Santa Cruz Biotechnology; lanes 4–6). The coimmunoprecipitated ACK1 with EGFR or immunoprecipitated ACK1 was detected by immunoblotting with an anti-Myc antibody (middle panels). The coimmunoprecipitated EGFR with ACK1 or immunoprecipitated EGFR was detected with anti-EGFR (1005, Santa Cruz Biotechnology; top panels). The bottom panels show the expression level of Myc-ACK or EGFR in the lysates used for immunoprecipitation.
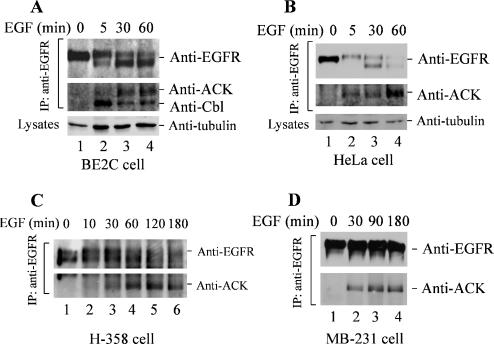
We further characterized ligand-induced degradation of EGFR and the interaction of endogenous ACK1 with EGFR upon EGF stimulation by coimmunoprecipitation assays in multiple cancer cell lines. As shown in Figure 3, EGFR in the neuroblastoma BE2C cells and the adenocarcinoma HeLa cells was promptly degraded upon 60-min stimulation with EGF (Figure 3, A and B), whereas EGFR in the nonsmall cell lung cancer H-358 cells was degraded after 120-min stimulation with EGF (Figure 3C), and EGFR in the breast cancer MDA-MB-231 cells did not show significant degradation over 180-min stimulation with EGF (Figure 3D). These data indicate that the rate of ligand-induced EGFR degradation varies in different cell lines. This variation in rate of ligand-induced EGFR degradation reflects a complexity in regulation of EGFR degradation.
Figure 3.
Interaction of ACK1 with EGFR occurs in cancer cells. (A–D) The adenocarcinoma HeLa cells, neuroblastoma BE2C cells, NSCLC H-358 cells, and breast cancer MDA-MB-231 cells were serum-starved overnight and stimulated with EGF (100 ng/ml) for the indicated time. EGFR was immunoprecipitated by anti-EGFR Mab528 and immunoblotted by anti-EGFR (1005) (top panels). Coimmunoprecipitated ACK1 or Cbl was detected by immunoblotting with anti-ACKPCC (A–D) or anti-c-Cbl (middle panels in A and B and bottom panels in C and D). In A and B, the amount of tubulin in whole cell lysates is shown in the bottom panels as a lysate-loading control.
Similar to that in COS7 cells, the interaction of ACK1 with EGFR is dependent on EGF stimulation, and the maximal level of the interaction occurred at about 60-min stimulation with EGF in all four cancer cell lines (Figure 3), which lagged behind the interaction of c-Cbl with EGFR (Figure 3A). Surprisingly, the amount of coimmunoprecipitated ACK1 was not proportional to the amount of immunoprecipitated full-length EGFR in BE2C, HeLa, and H-358 cells (Figure 3, A–C). Because the anti-EGFR Mab528 used for EGFR immunoprecipitation is against extracellular domain, whereas the anti-EGFR (1005; Santa Cruz Biotechnology) for immunoblotting is against the very carboxy terminus, it is possible that ACK1 is able to coimmunoprecipitate with degraded EGFR fragments that could be immunoprecipitated by Mab528 but not detected by immunoblotting with the anti-EGFR (1005). However, the interaction of ACK1 with EGFR is not dependent on degradation of EGFR because ACK1 was coprecipitated with EGFR in MDA-MB-231 equally well to the other three cell lines, even though EGFR in MDA-MB-231 did not have significant degradation (Figure 3D). These data further confirm that the in vivo interaction of ACK1 with EGFR is dependent on activation of EGFR.
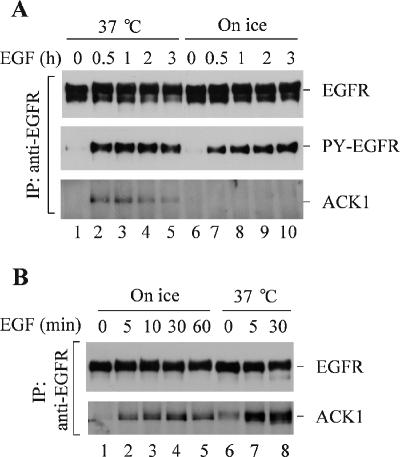
To determine whether the interaction of ACK1 with EGFR occurs on the plasma membrane, the cells were preincubated on ice and subsequently stimulated with EGF on ice, which prevents endocytosis of EGFR and restricts the activated EGFR on the plasma membrane. As shown in Figure 4A, although EGFR activation by EGF on ice was comparable to that at 37°C (the middle panel), endogenous ACK1 was not coimmunoprecipitated with EGFR upon EGF stimulation on ice (lanes 7–10), implying that the interaction of endogenous ACK1 with EGFR does not occur on plasma membrane. These data also suggest that activation of EGFR is necessary but not sufficient for the interaction of endogenous ACK1 with EGFR. However, when overexpression of exogenous ACK1 in cells, as shown in Figure 4B, the interaction of ACK1 with EGFR occurs upon ice incubation (lanes 2–5), which may result from the interaction of EGFR with excessive free ACK1. These data are consistent with the results shown in Figure 2 and suggest that endogenous ACK1 might be restrained in intracellular compartments. Interaction of endogenous ACK1 with EGFR may require transport of EGFR to endosomes or a subcellular location, which is dependent on activation of EGFR.
Figure 4.
Determination of the location for the interaction of ACK1 with EGFR. (A) Interaction of endogenous ACK1 with EGFR does not occur on the plasma membrane. The COS7 cells were starved in serum-free medium for 12 h and then placed on ice for 30 min. EGF (100 ng/ml) was added to the cells on ice for the indicated time. The control cells were preincubated and stimulated with EGF (100 ng/ml) at 37°C (lanes 1–5). EGFR was immunoprecipitated with anti-EGFR Mab528 from the cell lysates and immunoblotted with anti-EGFR (1005; top panel). Coimmunoprecipitated ACK1 was detected by immunoblotting with anti-ACKPCC (bottom panel). Tyrosine phosphorylation of EGFR was detected by immunoblotting with anti-PY (4G10; middle panel). (B) Interaction of exogenous ACK1 with EGFR may occur on the plasma membrane. The Myc-tagged ACK1 was transfected into COS7 cells for 36 h followed by serum starvation for 12 h. The experimental procedures, including ice incubation, EGF stimulation, immunoprecipitation, and immunoblotting, were the same as described in A. Top panel, immunoprecipitated EGFR detected by immunoblotting with anti-EGFR (1005); bottom panel, coimmunoprecipitated ACK1 detected by immunoblotting with anti-ACKPCC.
The Interaction of ACK1 with EGFR Is Dependent on Tyrosine Kinase Activity of EGFR
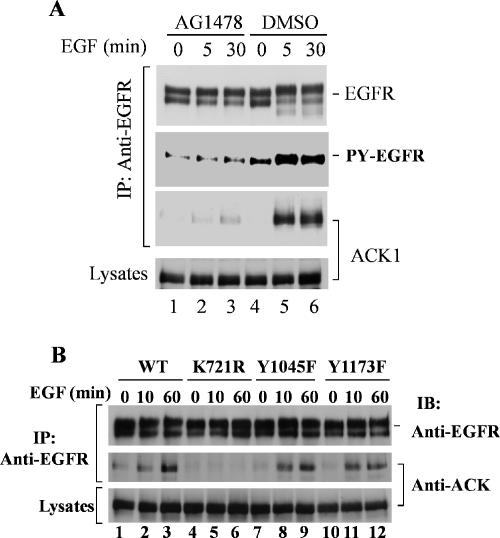
Because ACK1 interacts only with activated EGFR, we suspect that the interaction of ACK1 with EGFR requires tyrosine phosphorylation or tyrosine kinase activity of EGFR. To examine the effect of EGFR kinase activity on the interaction, we used a specific EGFR kinase inhibitor AG1478 to block tyrosine kinase activity of EGFR and determined the binding of ACK1 to EGFR by coimmunoprecipitation assays. As shown in Figure 5A, the treatment of the cells with AG1478 inhibited the tyrosine kinase activity of EGFR (the second panel from top) and eliminated the interaction of ACK1 with EGFR (third panel from top, lanes 1–3), indicating that the tyrosine phosphorylation or the kinase activity of EGFR is required for the binding of ACK1 to EGFR.
Figure 5.
Interaction of ACK1 with EGFR is dependent on EGFR kinase activity. (A) EGFR kinase inhibitor AG1478 blocks the interaction of ACK1 with EGFR. ACK1-transfected COS7 cells were starved with serum-free medium overnight and then treated with AG1478 (100 nM) for 30 min at 37°C before EGF stimulation. DMSO (the solvent for AG1478) was added in the control cells (lanes 4–6). EGF (100 ng/ml) was added to the medium for indicated time. EGFR was immunoprecipitated from the cell lysates with anti-EGFR Mab528 and immunoblotted with anti-EGFR (1005; top panel). The tyrosine phosphorylation of EGFR was detected by immunoblotting with anti-phosphotyrosine (4G10; second panel from the top). Coimmunoprecipitated ACK1 was detected by immunoblotting with anti-ACKPCC (third panel from the top). The expression level of ACK1 was determined by immunoblotting of the cell lysates with anti-ACKPCC (bottom panel). (B) ACK1 does not interact with kinase-dead EGFR. ACK1 was cotransfected with EGFR wild type, the Cbl-binding defective mutant Y1045F, the SHP-1–binding defective mutant Y1173F, or the kinase-dead mutant K721R into CHO cells. The cells were starved in serum-free medium overnight and then stimulated by EGF (100 ng/ml) for the indicated time. EGFR and its mutants were immunoprecipitated from the cell lysates with anti-EGFR Mab528 and immunoblotted with anti-EGFR (1005; top panel). Coimmunoprecipitated ACK1 was detected by immunoblotting with anti-ACKPCC (middle panel). The expression level of ACK1 is shown in the bottom panel by immunoblotting the cell lysates with anti-ACKPCC.
To further confirm that the tyrosine kinase activity is essential for the interaction of ACK1 with EGFR, we made EGFR kinase-dead mutant K721R and performed the binding assay in Chinese hamster ovary (CHO) cells. In addition, we also examined the interaction of ACK1 with c-Cbl–binding defective EGFR mutant Y1045F and SHP-1–binding defective mutant Y1173F. After cotransfection of EGFR or the mutants with ACK1, the CHO cells were stimulated with EGF for 0, 10, and 60 min. The EGFR or its mutants were immunoprecipitated with anti-EGFR Mab528, and the coimmunoprecipitated ACK1 was detected by immunoblotting with anti-ACKPCC. Consistent with the results obtained by treatment of EGFR kinase inhibitor AG1478 as shown in Figure 5A, the kinase-dead mutant K721R became ACK1-binding defective (Figure 5B, middle panel, lanes 4–6), while wild-type EGFR, the Cbl-binding defective mutant Y1045F, and the SHP-1–binding defective mutant Y1173F were capable of interacting with ACK1 after EGF stimulation (Figure 5B, middle panel, lanes 1–3 and 7–12). These data further support the conclusion that the interaction of ACK1 with EGFR requires tyrosine phosphorylation or kinase activity of EGFR and suggest that the interaction of ACK1 with EGFR is independent on pY1045 or pY1173 of EGFR.
ACK1 Interacts with EGFR through a Domain That Is Conserved in Gene-33/Mig-6/RALT
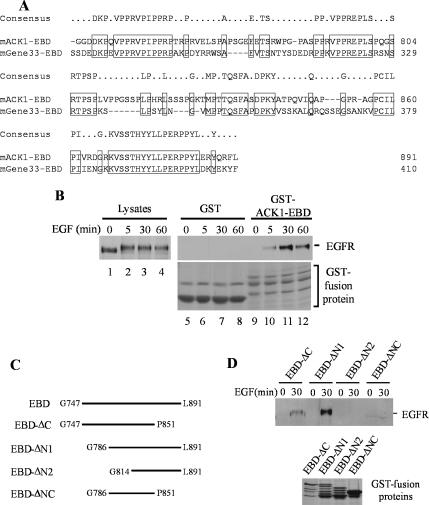
Gene-33/Mig-6/RALT, the nonkinase member of ACK family, has been shown to interact with EGFR and ErbB2 through a domain at its carboxy terminus (Fiorentino et al., 2000; Xu et al., 2005). This domain is conserved in ACK1 (Figure 6A). We designate this domain as the EBD. We subcloned the ACK1 EBD domain into a GST-fusion protein vector and performed the GST-ACK1-EBD pulldown assay with EGF-stimulated COS7 cell lysates. As expected, GST-ACK1-EBD precipitated EGFR from EGF-stimulated COS7 cell lysates (Figure 6B, lanes 10–12) but not from unstimulated cell lysates (Figure 6B, lane 9), whereas GST alone did not precipitate EGFR from either EGF stimulated or unstimulated cell lysates (lanes 5–8). These data indicate that the EBD domain is the domain responsible for interaction with EGFR in ACK1.
Figure 6.
The EGFR-binding domain (EBD) of ACK1 is located in the carboxy terminus region between G747 and L891. (A) Alignment of the EBD of ACK1 with the EBD of Gene-33/Mig-6/RALT. The identical residues are boxed. (B) Binding of GST-ACK1EBD to EGFR is dependent on EGF stimulation. The region that contains G747-L891 of ACK1 was cloned in pGEX-4T3, expressed in E. coli, and affinity-purified by glutathione-conjugated agarose beads. The purified GST-ACK1EBD was immobilized on the beads and incubated with EGF-stimulated COS7 cell lysates (0.5 mg cell lysates/20 μg GST-ACK1EBD). The GST-ACK1EBD beads were precipitated by centrifugation. The coprecipitated EGFR was detected by immunoblotting with anti-EGFR (1005; right top panel, lanes 9–12). The immobilized GST beads were used as a control (lanes 5–8). The input of EGFR in cell lysates was shown in lanes 1–4. The GST-fusion proteins used in the pulldown assays were stained with 1% Coomassie Blue and are shown in lanes 5–12 of the right bottom panel. The top band in lanes 9–12 is the full length of GST-ACK1EBD. The lower two bands are the degradation products of GST-ACK1EBD. (C) Schematic representation of the truncation mutants of the EBD domain. The truncation mutants were generated by PCR, subcloned into GST-fusion protein vector pGEX4T3, expressed in E. coli, and affinity-purified with and immobilized on glutathione-conjugated agarose beads. (D) Determination of the minimal length in the EBD domain that is capable of interacting with EGFR. The EGFR pulldown by the truncation mutants of GST-ACK1EBD was performed the same as the procedures described in B. The GST truncation mutant proteins of the EBD domain used in the experiment were shown in the bottom panel by Coomassie blue staining. The multiple bands under the top band in lanes 1–3 are degradation products of the GST-fusion proteins.
To determine the minimum length in the EBD domain that is capable of binding to EGFR, we made a number of truncation mutants of the EBD domain (Figure 6C), expressed the mutants as GST-fusion proteins in E. coli, and used the GSH-bead–bound GST-mutants for EGFR pulldown assays. As shown in Figure 6D, truncation of ∼40 amino acid residues at either N-terminus (EBD-ΔN1) or C-terminus (EBD-ΔC) of the EBD domain did not eliminate the binding capacity of the EBD domain (Figure 6D, top panel, lanes 1–4). However, truncation of ∼40 amino acid residues at both N- and C- termini (EBD-ΔNC) resulted in elimination of the interaction of the EBD domain with EGFR. These data suggest that at least two regions, one is between G786 and P851 and the other is in either N-terminus or C-terminus of the EBD domain, are required for interaction with EGFR. Further truncation of 28 amino acid residues at the N-terminus of EBD-ΔN1 (EBD-ΔN2) also eliminated the EGFR binding capacity of the EBD domain (Figure 6D), implicating that one of the regions required for interaction with EGFR may locate between G786-G814. Interestingly, there are three visible proline-rich regions, two at each terminus and one between G786-G814, in the EBD domain (Figure 6A). Regarding this structure feature and the EGFR binding of the truncation mutants shown in Figure 6D, it is possible that these three proline-rich regions are critical for the EBD domain to interact with active EGFR, and two of the three proline-rich regions are required for the minimal interaction with EGFR. Thus, we suspect that the interaction of ACK1 with EGFR is indirect, which may be mediated by an SH3 domain-containing EGFR-adaptor protein. Further investigation will be performed to confirm this hypothesis by identification of EGFR-adaptor proteins that interact with the EBD domain.
ACK1 Functions in EGFR Degradation
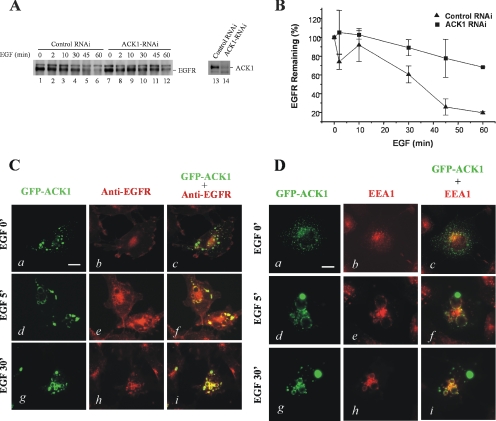
The next question is the function of ACK1 in EGFR signaling. Previous studies have shown that ACK1 interacts with clathrin heavy chain and regulates receptor-mediated endocytosis (Teo et al., 2001). Ligand-induced endocytosis and degradation of EGFR play an important role in down-regulation of the EGFR signal (Yarden, 2001). In C. elegans, the ACK homologue Ark-1 interacts with clathrin-binding protein AP-47 and ubiquitin-ligase Cbl and suppresses EGFR signaling. We suspect that ACK1 may play a role in regulation of EGFR degradation in mammalian cells. In HEK293 cells, ACK1 is the only endogenous ACK kinase, and the ligand-induced degradation rate of EGFR is faster than that in COS7 cells (data not shown). It is anticipated that effect of ACK1-knockdown with ACK1-RNAi on EGFR degradation in HEK293 cells is more visible than in COS7 cells. Thus we used HEK293 cells for ACK1-knockdown experiments. As shown in Figure 7A, the transfection of ACK-RNAi oligos in HEK293 cells effectively knocked down the expression of ACK1 (right panel). The degradation of EGFR after EGF stimulation upon the ACK1-RNAi transfection was significantly inhibited (Figure 7A). We quantified the effect of ACK1-RNAi on EGFR degradation in Figure 7A, as shown in Figure 7B. The maximum ligand-induced degradation of EGFR within 1 h was ∼80% of total EGFR. The maximum degradation of EGFR upon ACK-RNAi dropped to ∼30% of total EGFR. Compare to the control oligos, ACK-RNAi inhibited ∼60% of the total degradation of EGFR. These data suggest that ACK1 plays an important role in ligand-induced EGFR degradation in HEK293 cells.
Figure 7.
ACK1 functions in EGFR degradation. (A and B) Knockdown of ACK1 blocks EGFR degradation. The HEK293 cells were transfected with ACK RNAi (final concentration 40 nM) or the control RNA oligos. The cells were serum-starved for 12 h and subsequently stimulated with EGF (100 ng/ml) for 2, 10, 30, 45, and 60 min. The EGFR in whole lysates was detected by immunoblotting with an anti-EGFR antibody (left panel in A) and quantified by Kodak EDAS290 system and plotted with Origin (B). To assess the suppression of ACK1 expression by ACK RNAi, the endogenous ACK1 in the cell lysates that from either ACK RNAi- or the control HEK293 cells was immunoprecipitated and immunoblotted with anti-ACKPCC (right panel in A). The data in B are the average from two independent experiments. (C and D) ACK1 is colocalized with EGFR on EEA1-positive vesicles during the internalization of EGFR. The GFP-ACK1 was transfected into COS7 cells. After serum-starvation for 12 h, the cells were stimulated by EGF (100 ng/ml) at 37°C for 5 and 30 min. The cells were then fixed by 3.7% paraformaldehyde in PBS for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. For immunofluorescent staining, the cells were incubated with anti-EGFR (Mab528; 1:100) or anti-EEA1 (1:100) for 1–2 h at 22°C followed by incubation with Texas red–conjugated anti-mouse IgG (1:150) for 1–2 h at 22°C. The immunofluorescent staining was visualized under a Zeiss fluorescence microscope. Bar, 10 μm.
To characterize the participation of ACK1 in EGFR endocytosis and degradation in cells, we performed fluorescent microscopic studies with green fluorescent protein (GFP)-tagged ACK1. After transfected with GFP-ACK1, the cells were stimulated with EGF for 5 and 30 min and then immunostained with anti-EGFR to visualize the endocytosis of EGFR. As shown in Figure 7C, ACK1 and EGFR were not colocalized in the cells before the stimulation by EGF (top three panels). GFP-ACK1 was shown as a punctate structure in cells, whereas EGFR was on the plasma membrane. Upon EGF stimulation for 5 min, EGFR was endocytosed and accumulated along the large vesicles, and ACK1 was colocalized with EGFR on the same structure in the cells (middle three panels). Upon 30-min EGF stimulation, EGFR and ACK1 were still colocalized on the large vesicles (bottom three panels). EEA1, an early endosome marker, is positively stained on these vesicles (Figure 7D), suggesting that these vesicles are endosomes. Upon EGF stimulation, ACK1 is colocalized with EEA1 on these EEA1-positive vesicles (Figure 7D). These data suggest that ACK1 is colocalized with EGFR on EEA1-positive endosomes during EGFR endocytosis upon EGF stimulation and support the idea that ACK1 functions in regulation of EGFR degradation.
ACK1 Is a Ubiquitin-binding Protein and Regulates EGFR Degradation via Its Uba Domain
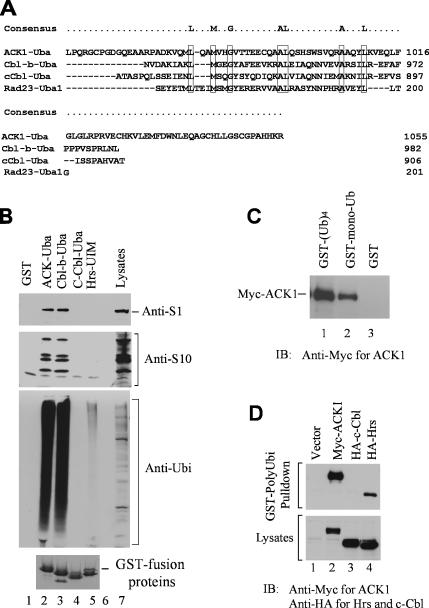
To define the molecular mechanism by which ACK1 regulates EGFR degradation, we have searched the functional domain in ACK1 that potentially mediates the effect of ACK1 on EGFR degradation. Analysis of the primary sequence of ACK1 indicates a putative ubiquitin association (Uba) domain localized at the very carboxy terminus (amino acid residues 970-1055; Figure 8A). Alignment of the putative Uba domain of ACK1 with the Uba domains of Cbl-b, c-Cbl, and Rad23 shows the conserved ubiquitin-binding residues in ACK1 (Figure 8A). It is known that ubiquitination and clathrin-coated vesicle endocytosis are required for the degradation of EGFR (Sorkin and Carpenter, 1993; Levkowitz et al., 1998; Waterman and Yarden 2001; Haglund et al., 2003a, 2003b). To determine the interaction of the putative Uba domain of ACK1 with ubiquitinated proteins, we performed the GST-fusion protein pulldown experiments. The putative ACK1-Uba domain was fused with GST, expressed in E. coli, and purified with the glutathione (GSH)-conjugated agarose beads. The immobilized GST-ACK1-Uba was incubated with cell lysates to precipitate associated proteins. As controls, we also did the pulldown assays with the GST-Cbl-b-Uba, GST-c-Cbl-Uba, and GST-Hrs-UIM domains along with the GST-ACK1-Uba domain. The coprecipitated proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. There were seven bands, migrating at 42, 45, 50, 70, 58, 62, and 100 kDa, that specifically associated with the putative ACK1 Uba domain (data not shown). The microsequencing of these bands at the Harvard Microchemistry Facility indicates that they are 26S proteasome subunits (data not shown). The immunoblot of the GST-ACK1 Uba-associated proteins in COS7 cells with anti-proteasome S1 and S10B antibodies that recognizes proteasome subunits S1, S10B, S4, and S8 has confirmed the microsequencing data (Figure 8B, top and middle panels, lane 2). We also observed that the Uba domain of Cbl-b precipitated proteasome subunits from COS7 cell lysates (Figure 8B, top and middle panels, lane 3). Because the Uba domain is a known domain for binding to ubiquitin, we speculated that binding of the ACK1 and Cbl-b Uba domains to the proteasome subunits might be indirect and mediated by ubiquitinated proteins that were associated with proteasomes. Thus, we immunoblotted the Uba domain precipitated proteins with anti-ubiquitin antibody. As shown in Figure 8B, large amount of ubiquitinated proteins in COS7 cell lysates was precipitated by the ACK1 Uba domain (the third panel, lane 2), suggesting that ACK1 Uba domain is the ubiquitin-binding domain. Cbl-b Uba domain had the same capacity as the ACK1-Uba domain to bind to ubiquitinated proteins (the third panel, lane 3), whereas the Hrs-UIM domain had much weak binding affinity to ubiquitinated proteins (the third panel, lane 5). Furthermore, Hrs-UIM domain did not pull down 26S proteasome subunits (lane 5, the top and the middle panels), suggesting that association of the Uba domains of ACK1 and Cbl-b with proteasome subunits might be specific. Thus, it is possible that the ACK1 and Cbl-b Uba domains can directly interact with proteasomes. It has been shown that the Uba domain of hPLIC-2 directly interacts with the 26S proteasomes (Kleijnen et al., 2003). Further investigation is needed to determine the direct association of the Uba domain with proteasomes. To our surprise, the Uba domain of c-Cbl was defective in binding to ubiquitinated proteins (the third panel, lane 4), which is consistent with the observation from Davies et al. (2004). To further verify the binding of ACK1 to ubiquitin, we used GSH-agarose bead-bound GST-mono-ubiquitin and GST-poly-ubiquitin to incubate with myc-ACK1– expressed COS7 cell lysates and determined the coprecipitation of ACK1 with mono- and poly-ubiquitin. As shown in Figure 8C, both GST-mono-ubiquitin and poly-ubiquitin precipitated myc-ACK1 from the cell lysates, indicating that ACK1 interacts with both mono- and poly-ubiquitin. We compared the binding of poly-ubiquitin to ACK1, Hrs, and c-Cbl and found that polyubiquitin bound to ACK1 with a higher affinity than to Hrs (Figure 8D, lanes 2 and 4) and had no binding to c-Cbl (Figure 8D, lane 3), which is consistent with results from the GST-Uba domain pulldown experiments in Figure 8B.
Figure 8.
ACK1 is an ubiquitin-binding protein. (A) Alignment of the putative ACK1-Uba domain with the Uba domains of Cbl-b, c-Cbl, and Rad23. The conserved residues are boxed. (B) The Uba domain of ACK1 interacts with proteasomes and ubiquitinated proteins. The GST-ACK1-Uba, GST-Cbl-b-Uba, GST-c-Cbl-Uba, or GST-Hrs-UIM domain was expressed in E. coli and purified by affinity pulldown with glutathione-agarose beads. The bead-bound GST-fusion proteins (∼20 μg) were incubated with COS7 cell lysates (∼0.5 mg). The coprecipitated proteins were subjected to SDS-PAGE electrophoresis and immunoblotting with anti-proteasome S1 (top panel), anti-proteasome S10B (second panel), or anti-ubiquitin antibody (third panel). The loaded GST-ACK1-Uba, GST-Cbl-b-Uba, GST-c-Cbl-Uba, or GST-Hrs-UIM proteins were shown in the bottom panel by Coomassie blue staining. (C) Both mono-ubiquitin and poly-ubiquitin bind to ACK1. The bead-bound GST mono-ubiquitin or poly-ubiquitin (Ubi)4 was incubated with Myc-tagged ACK1-overexpressed COS7 cell lysates. The coprecipitated Myc-ACK1 was detected by immunoblotting with an anti-Myc antibody (top panel). (D) ACK1 has a high binding affinity to poly-ubiquitin. Myc-tagged ACK1, HA-tagged c-Cbl, or Hrs was transfected into COS7 cells for expression. The cell lysates were incubated with bead-bound GST-(Ubi)4. The GST-(Ubi)4 beads were precipitated by centrifugation. The coprecipitated ACK1, c-Cbl, or Hrs was detected by immunoblotting with anti-Myc or anti-HA antibodies (top panel). The input amount of ACK1, c-Cbl, or Hrs in lysates was shown in the bottom panel.
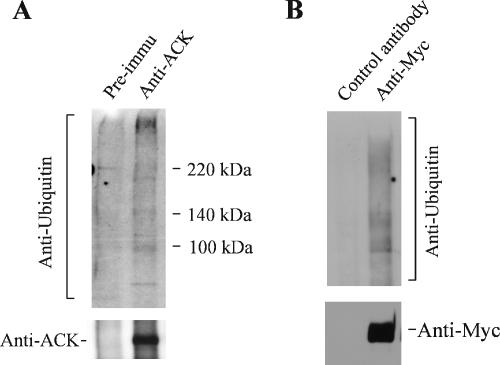
To determine the interaction of ACK1 with ubiquitinated proteins in vivo, we immunoprecipitated both endogenous and overexpressed ACK1 and detected the coimmunoprecipitated ubiquitinated proteins with anti-ubiquitin antibody (Figure 9, A and B). Multiple ubiquitinated proteins were detected in the immunoprecipitation of endogenous ACK1 with anti-ACKPCC and of Myc-tagged ACK1-expressed COS7 cell lysates with anti-Myc antibody (Figure 9), whereas no ubiquitinated protein was detected in the immunoprecipitation with the preimmune serum or control antibody. Taken together, we conclude that interaction of ACK1 with ubiqitinated proteins occurs in vivo.
Figure 9.
ACK1 interacts with ubiquitinated proteins in vivo. (A) The interaction of endogenous ACK1 with ubiquitinated proteins. The endogenous ACK1 was immunoprecipitated from COS7 cell lysates with an anti-ACKPCC. As a control, we used the preimmune serum to incubate with the cell lysates. The precipitated endogenous ACK1 was detected with the anti-ACKPCC (bottom panel). The coprecipitated ubiquitinated proteins were detected with an anti-ubiquitin antibody (top panel). (B) The interaction of overexpressed ACK1 with ubiquitinated proteins. The COS7 cells were transfected with Myc-tagged ACK1. The overexpressed myc-ACK1 was immunoprecipitated with an anti-Myc antibody (bottom panel, lane 2). The ubiquitinated proteins in the immunoprecipitation complex were detected by immunoblotting with an anti-ubiquitin antibody (top panel). The control antibody for the nonspecific binding to ubiquitinated proteins during the immunoprecipitation was using an anti-HA antibody (lane 1).
Ubiquitination is a key event for EGFR endocytosis and degradation (Levkowitz et al., 1998; Haglund et al., 2003). Recent studies indicate that mono-ubiquitination is a sorting signal for EGFR degradation (Haglund et al., 2003; Mosesson et al., 2003). We wonder whether the Uba domain confers the binding of ACK1 to EGFR. We used GST-ACK1-Uba to incubate EGF stimulated COS7 cell lysates and detected the coprecipitated EGFR by immunoblotting. However, we observed a very weak pulldown of EGFR by ACK1-Uba (data not shown). An explanation for the result is that the ubiquitin-binding domains, including the Uba, UIM, and CUE domains, recognize only ubiquitin moiety, not ubiquitinated proteins. This feature of the interaction between ubiquitin and ubiquitin-binding domains may be determined by the fact that ubiquitin is a polypeptide, not a single residue. Therefore, ubiquitin-binding is unlikely to confer the specificity in protein–protein interaction. The Uba domain may not determine the interaction of ACK1 with EGFR but aid the EBD domain to bind to ubiquitinated EGFR.
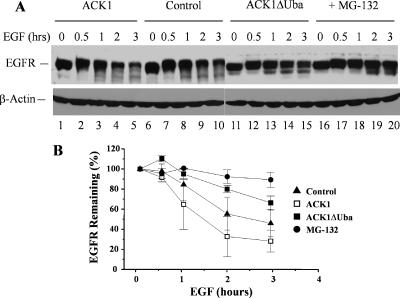
The ubiquitin-binding has been shown to play important roles of endocytic adaptor proteins, such as Hrs and Eps15, in regulation of EGFR endocytosis and degradation (Bache et al., 2003; de Melker et al., 2004). To define the role of the Uba domain of ACK1 in EGFR degradation, we transfected the wild type and the Uba domain-deletion mutant of ACK1 into COS7 cells and then determined ligand-dependent degradation of EGFR by immunoblotting the cell lysates with an anti-EGFR antibody (Figure 10A). The degradation of EGFR in the control (vector transfected) was ∼50% of the total EGFR in the cells after 3-h EGF stimulation (Figure 10B). Overexpression of ACK1 facilitated the degradation of EGFR up to ∼70% of the total EGFR in the cells (Figure 10B), confirming that ACK1 is an important regulator in EGFR degradation. However, overexpression of the Uba domain-deletion mutant ACK1ΔUba decreased the maximum level of ligand-induced degradation of EGFR down to ∼30% of the total EGFR (Figure 10B). Consider the transfection efficiency (∼80% in COS7 cells), we estimate that ACK1ΔUba mutant blocked ∼60% of ligand-induced EGFR degradation in cells, which is consistent with the results in ACK-RNAi experiments (Figure 7). These data suggest that the ACK1 Uba domain plays an important role in regulating ligand-induced EGFR degradation. In addition, MG-132, a proteasome inhibitor, markedly inhibited ligand-induced EGFR degradation in COS7 cells (Figure 10A, lanes 16–20, and B), raising a possibility that proteasome activity is required for EGFR degradation in COS7 cells. However, we did not observe a significant effect of MG-132 on ligand-induced degradation of EGFR in HeLa cells (data not shown), suggesting that proteasome-mediated EGFR degradation is limited in certain cell lines.
Figure 10.
The ACK1-Uba domain is required for EGFR degradation. The COS7 cells were transfected with Myc-tagged ACK1 and the ACK1 Uba domain deletion mutant ACK1ΔUba. The controls were transfected with the vector. The cells were serum-starved for 12 h and subsequently stimulated with EGF (100 ng/ml) for 0.5, 1, 2, and 3 h. For the negative controls of EGFR degradation, the cells were treated with the 26S proteasome inhibitor MG-132 (10 μM) for 30 min before the EGF stimulation. The EGFR degradation was assessed by immunoblotting the whole-cell lysates with an anti-EGFR antibody and quantified by Kodak EDAS290 system (Eastmann Kodak, Rochester, NY). Two independent experiments were performed, and data from both experiments were consistent. (A) The immunoblot of the cell lysates with anti-EGFR antibody (top panel). The lysate loading is shown by immunoblotting with anti-β-actin (bottom panel). (B) The plot of the quantification of the immunoblot from the average of two independent experiments by Kodak EDAS290 system.
DISCUSSION
ACK is a specific downstream effector of Cdc42, a member of Rho family small G-proteins. While Cdc42 functions in cytoskeletal organization, membrane trafficking and mitogenesis, the signal transduction pathway of ACK has not been defined. Previous studies have shown that tyrosine phosphorylation of ACK1 is enhanced in response to the EGF signal (Satoh et al., 1996), and ACK1 possesses a clathrin-binding motif, interacts with clathrin, and participates in clathrin-mediated endocytosis (Teo et al., 2001). Here we report that ACK1 is an ubiquitin-binding protein and interacts with EGFR upon EGF stimulation. Knockdown of ACK1 by ACK1-RNAi or overexpression of the Uba domain-deletion mutant of ACK1 inhibits ligand-induced degradation of EGFR. Our data point to a function for ACK1 to be a regulator in ligand-induced degradation of EGFR.
The interaction of ACK1 with EGFR is in a way that is very similar to that of Gene-33/Mig-6/RALT, a protein homologous to the carboxyl portion of ACK1, with ErbB2 (Fiorentino et al., 2000; Xu et al., 2005). For example, Gene-33 interacts only with activated ErbB2 and the ErbB2-interactive domain of Gene-33 is the conserved EBD domain (Fiorentino et al., 2000). Gene-33 also interacts with EGFR through the EBD domain, however, independent of stimulation of EGF (Xu et al., 2005). It is not clear why Gene-33 binds to ErbB2 dependent on tyrosine phosphorylation of the receptor, whereas to EGFR independent on tyrosine phosphorylation of the receptor. Because those binding assays were performed with transfection of exogenous EGFR and Gene-33, the overexpression level of the receptor and Gene-33 in the assays may be a cause for diversified receptor-binding properties of Gene-33. Nevertheless, it seems that Gene-33 and ACK1 share a conserved EGFR/ErbB2-interactive mechanism. It has been shown that overexpression of Gene-33 inhibited tyrosine phosphorylation of EGFR and ErbB2-initiated cell proliferation and Erk activation (Fiorentino et al., 2000; Xu et al., 2005), suggesting that Gene-33 is a negative regulator of EGFR or ErbB2 signaling. There is a research report showing that ACK1 is required for Ras-mediated cellular transformation (Nur-E-Kamal et al., 2005). However, genetic analysis of ACK function in C. elegans has indicated a negative role of ACK in EGFR signaling (Hopper et al., 2000). Further investigation of the role of ACK1 in EGFR-mediated cell proliferation is necessary to define the role of ACK1 in cellular mitogenesis.
We observed a striking difference in interaction with EGFR between endogenous ACK1 and exogenous overexpressed ACK1 (Figures 2–4): interaction of exogenous ACK1 with EGFR occurred as quickly as 1 min after EGF stimulation (Figure 2B), and consistent with this the interaction took place on plasma membrane (Figure 4), whereas interaction of endogenous ACK1 with EGFR occurred relatively slowly (maximal binding at 1 h after EGF stimulation; Figures 2A and 3), and consistent with this the interaction did not take place on plasma membrane (Figure 4). These data suggest a possibility that endogenous ACK1 is restrained in cells. The interaction of endogenous ACK1 with EGFR may require transport of EGFR to an ACK1-containing intracellular compartment (EEA1 positive endosomes?) or release of ACK1 from the restrained sites by EGF signaling. Overexpression eliminates the restraint of ACK1 and yields free ACK1 that is capable of binding to active EGFR. Because endogenous ACK1 interacts with EGFR at late stage of ligand-induced EGFR activation, ACK1 is likely to function at late phase of EGFR endocytosis on EEA1-positive endosomes.
Cbl, the E3 ubiquitin ligase for the ubiquitination of EGFR, the HGF-regulated tyrosine phosphorylation substrate (Hrs), and the tumor-suppressor gene 101 (Tsg101) have been reported as the key regulators in ubiquitination-mediated EGFR degradation (Levkowitz et al., 1999; Yarden, 2001; Bishop et al., 2002; Lu et al., 2003). Cbl directly interacts with EGFR phosphotyrosine residue (tyrosine 1045) via its SH2 domain upon the activation of EGFR and subsequently catalyzes the ubiquitination. Hrs recognizes the mono-ubiqitinated EGFR and brings EGFR-loaded CCVs to multivesicular bodies (MVBs) by interaction with Tsg101 in the endosomal sorting complex required for transport I (ESCRT-I). The EGFR in MVBs eventually transports to lysosomes for degradation. In C. elegans, the ACK homologue, Ark-1, is in the same pathway as that of Cbl for regulation of EGFR signaling (Hopper et al., 2000). However, we did not observe an interaction between ACK1 and Cbl with coimmunoprecipitation assays in COS7 cells (data not shown). The results in Figures 2A, 3A, and 5B also suggest that ACK1 and c-Cbl interact with EGFR at different steps during ligand-induced endocytosis and degradation of EGFR. To dissect the signaling events of ACK1 in regulation of EGFR degradation, we need to determine the exact timing and location of the interaction of ACK1 with EGFR in future studies. Furthermore, we will address the biological consequence of ACK1-regulated EGFR degradation in future investigations. Because EGFR has a pivotal role in regulation of cell growth and overexpression of EGFR has been found in many types of solid tumors, the function of ACK1 in tumorigenesis related to EGFR degradation should be another interesting area to explore.
ACKNOWLEDGMENTS
We thank Dr. Yosef Yarden at the Weizmann Institute of Science and Dr. Mark I. Greene at University of Pennsylvania for sending us the human EGFR cDNA mammalian expression plasmids, Dr. Inger Helene Madshus at University of Oslo for the c-Cbl cDNA mammalian expression plasmids, and Dr. Stan Lipkowitz at National Institutes of Health for GST-Cbl-b-Uba bacterial expression plasmids. This work was supported in part by a grant to W.Y. from the American Cancer Society (ACS-RSG TBE-110602).
Abbreviations used:
- ACK1
activated Cdc42-associated tyrosine kinase 1
- EBD
EGFR-binding domain
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- RNAi
RNA interference
- SHP-1
SH2-containing phosphatase-1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0142) on December 20, 2006.
REFERENCES
- Bache K. G., Raiborg C., Mehlum A., Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 2003;278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- Bishop N., Horman A., Woodman P. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 2002;157:91–101. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. C., Ettenberg S. A., Coats A. O., Mussante M., Ravichandran S., Collins J., Nau M. M., Lipkowitz S. Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene. 2004;23:7104–7115. doi: 10.1038/sj.onc.1207952. [DOI] [PubMed] [Google Scholar]
- de Melker A. A., van der Horst G., Borst J. c-Cbl directs EGF receptors into an endocytic pathway that involves the ubiquitin-interacting motif of Eps15. J. Cell Sci. 2004;117:5001–5012. doi: 10.1242/jcs.01354. [DOI] [PubMed] [Google Scholar]
- Eisenmann K. M., McCarthy J. B., Simpson M. A., Keely P. J., Guan J. L., Tachibana K., Lim L., Manser E., Furcht L. T., Iida J. Melanoma chondroitin sulphate proteoglycan regulates cell spreading through Cdc42, Ack-1 and p130cas. Nat. Cell Biol. 1999;1:507–513. doi: 10.1038/70302. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fiorentino L., Pertica C., Fiorini M., Talora C., Crescenzi M., Castellani L., Alema S., Benedetti P., Segatto O. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol. Cell. Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Di Fiore P. P., Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003a;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003b;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Hoare K., Hoare S., Smith O. M., Kalmaz G., Small D., Stratford May W. Kos1, a nonreceptor tyrosine kinase that suppresses Ras signaling. Oncogene. 2003;22:3562–3577. doi: 10.1038/sj.onc.1206480. [DOI] [PubMed] [Google Scholar]
- Hopper N. A., Lee J., Sternberg P. W. ARK-1 inhibits EGFR signaling in C. elegans. Mol. Cell. 2000;6:65–75. [PubMed] [Google Scholar]
- Kleijnen M. F., Alarcon R. M., Howley P. M. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol. Biol. Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G., et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Lo C. G., Cerione R. A., Yang W. The Cdc42 target ACK2 interacts with sorting nexin 9 (SH3PX1) to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 2002;277:10134–10138. doi: 10.1074/jbc.M110329200. [DOI] [PubMed] [Google Scholar]
- Linseman D. A., Heidenreich K. A., Fisher S. K. Stimulation of M3 muscarinic receptors induces phosphorylation of the Cdc42 effector activated Cdc42Hs-associated kinase-1 via a Fyn tyrosine kinase signaling pathway. J. Biol. Chem. 2001;276:5622–5628. doi: 10.1074/jbc.M006812200. [DOI] [PubMed] [Google Scholar]
- Lu Q., Hope L. W., Brasch M., Reinhard C., Cohen S. N. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan N. P., Whang Y. E., Mohler J. L., Earp H. S. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 2005;65:10514–10523. doi: 10.1158/0008-5472.CAN-05-1127. [DOI] [PubMed] [Google Scholar]
- Makkinje A., Quinn D. A., Chen A., Cadilla C. L., Force T., Bonventre J. V., Kyriakis J. M. Gene 33/Mig-6, a transcriptionally inducible adapter protein that binds GTP-Cdc42 and activates SAPK/JNK. A potential marker transcript for chronic pathologic conditions, such as diabetic nephropathy. Possible role in the response to persistent stress. J. Biol. Chem. 2000;275:17838–17847. doi: 10.1074/jbc.M909735199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E., Leung T., Salihuddin H., Tan L., Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Nur-E-Kamal A., Zhang A., Keenan S. M., Wang X. I., Seraj J., Satoh T, Meiners S., Welsh W. J. Requirement of activated Cdc42-associated kinase for survival of v-Ras-transformed mammalian cells. Mol. Cancer Res. 2005;3:297–305. doi: 10.1158/1541-7786.MCR-04-0152. [DOI] [PubMed] [Google Scholar]
- Satoh T., Kato J., Nishida K., Kaziro Y. Tyrosine phosphorylation of ACK in response to temperature shift-down, hyperosmotic shock, and epidermal growth factor stimulation. FEBS Lett. 1996;386:230–234. doi: 10.1016/0014-5793(96)00449-8. [DOI] [PubMed] [Google Scholar]
- Sem K. P., Zahedi B., Tan I., Deak M., Lim L., Harden N. ACK family tyrosine kinase activity is a component of Dcdc42 signaling during dorsal closure in Drosophila melanogaster. Mol. Cell. Biol. 2002;22:3685–3697. doi: 10.1128/MCB.22.11.3685-3697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A., Carpenter G. Interaction of activated EGF receptors with coated pit adaptins. Science. 1993;261:612–615. doi: 10.1126/science.8342026. [DOI] [PubMed] [Google Scholar]
- Teo M., Tan L., Lim L., Manser E. The tyrosine kinase ACK1 associates with clathrin-coated vesicles through a binding motif shared by arrestin and other adaptors. J. Biol. Chem. 2001;276:18392–18398. doi: 10.1074/jbc.M008795200. [DOI] [PubMed] [Google Scholar]
- van der Horst E. H., et al. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proc. Natl. Acad. Sci. USA. 2005;102:15901–15906. doi: 10.1073/pnas.0508014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman H., Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- Xu D., Makkinje A., Kyriakis J. M. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J. Biol. Chem. 2005;280:2924–2933. doi: 10.1074/jbc.M408907200. [DOI] [PubMed] [Google Scholar]
- Yang W., Cerione R. A. Cloning and characterization of a novel Cdc42-associated tyrosine kinase, ACK-2, from bovine brain. J. Biol. Chem. 1997;272:24819–24824. doi: 10.1074/jbc.272.40.24819. [DOI] [PubMed] [Google Scholar]
- Yang W., Lin Q., Guan J. L., Cerione R. A. Activation of the Cdc42-associated tyrosine kinase-2 (ACK-2) by cell adhesion via integrin beta1. J. Biol. Chem. 1999;274:8524–8530. doi: 10.1074/jbc.274.13.8524. [DOI] [PubMed] [Google Scholar]
- Yang W., Lin Q., Zhao J., Guan J. L., Cerione R. A. The nonreceptor tyrosine kinase ACK2, a specific target for Cdc42 and a negative regulator of cell growth and focal adhesion complexes. J. Biol. Chem. 2001a;276:43987–43993. doi: 10.1074/jbc.M104819200. [DOI] [PubMed] [Google Scholar]
- Yang W., Lo C. G., Despenza T., Cerione R. A. The Cdc42 target ACK2 directly interacts with clathrin and influences clathrin assembly. J. Biol. Chem. 2001b;276:17468–17473. doi: 10.1074/jbc.M010893200. [DOI] [PubMed] [Google Scholar]
- Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur. J. Cancer. 2001;37(Suppl 4):3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]