Abstract
The antitumor agent camptothecin targets DNA topoisomerase I by reversibly stabilizing a covalent enzyme-DNA intermediate. The subsequent collision of DNA replication forks with these drug-enzyme-DNA complexes produces the cytotoxic DNA lesions that signal cell cycle arrest and ultimately lead to cell death. Despite intense investigation, the character of the lesions produced and the repair processes that resolve the damage remain poorly defined. A yeast genetic screen was implemented to isolate conditional mutants with enhanced sensitivity to DNA topoisomerase I-mediated DNA damage. Cells exhibiting temperature-sensitive growth in the presence of the DNA topoisomerase I mutant, Top1T722Ap, were selected. Substitution of Ala for Thr722 increases the stability of the covalent Top1T722Ap-DNA intermediate, mimicking the cytotoxic action of camptothecin. Two mutants isolated, cdc45-10 and dpb11-10, exhibited specific defects in DNA replication and a synthetic lethal phenotype in the absence of DNA damaging agents. The accumulation of Okazaki fragments under nonpermissive conditions suggests a common function in promoting processive DNA replication through polymerase switching. These results provide a mechanistic basis for understanding the cellular processes involved in the resolution of DNA damage induced by camptothecin and DNA topoisomerase I.
The helical structure of double-stranded DNA allows for the faithful replication of chromosomes, where each strand serves as a template for complementary strand synthesis. However, the intertwining of these template strands imposes topological constraints such that unwinding of the duplex is required for replication fork progression (reviewed in refs. 1–3). Eukaryotic DNA topoisomerase I catalyzes changes in DNA topology via the transient cleavage and religation of a single strand (1, 4). A covalent phosphotyrosyl linkage is formed between the 3′ phosphate of the cleaved DNA strand and the active site tyrosine of the enzyme. This presumably allows for the noncovalently bound end of the nicked DNA to rotate around the intact complementary strand to effect changes in the linkage of the DNA strands. A second transesterification completes the catalytic cycle; the covalent enzyme-DNA intermediate is resolved, and the phosphodiester bond in the DNA resealed.
The antitumor agent camptothecin (Cpt) targets DNA topoisomerase I (Top1p) by reversibly stabilizing the covalent enzyme-DNA intermediate (reviewed in refs. 1, 3, 5, and 6). In the yeast Saccharomyces cerevisiae, DNA topoisomerase I is nonessential, and top1-null strains or cells expressing a catalytically inactive Top1p are resistant to Cpt (3, 7). Cpt cytotoxicity further depends on DNA synthesis as aphidicolin blocks cell sensitivity to the drug (3, 7–9). Yeast rad52 mutants, deficient in homologous recombination and DNA double strand break repair, are hypersensitive to Cpt (3, 7). Based on such data, a model was proposed in which collision of replication forks with the Cpt-stabilized covalent Top1p-DNA intermediates generates double strand breaks, cell cycle arrest in G2, and cell death (8, 10). Consistent with such a mechanism, the abrogation of DNA damage checkpoints, either by mutation in yeast rad9 strains or by treatment with UCN-01 in mammalian cells, abolishes cell cycle arrest in G2 and enhances Cpt cytotoxicity (3, 11, 12).
Despite intense investigation, the character of the DNA lesions produced by the collision of the advancing replication fork with the Cpt-enzyme-DNA complexes remains poorly characterized, as do the specific repair processes required for their resolution. Cpt-induced breakage of SV40 DNA replication forks has been examined in infected cells and in cell-free replication reactions (10, 13). These data suggest a dependence on replication fork polarity in which irreparable DNA lesions are more likely when Cpt-induced breaks reside on the leading, rather than lagging, strand template (6, 10). The mechanistic basis for this is unclear. Further, SV40 DNA replication differs from that of genomic DNA in several respects. It is driven by T antigen helicase and does not depend on DNA polymerase ɛ (Pol ɛ), which is required for cellular DNA replication and a replication checkpoint (reviewed in refs. 14 and 15).
To address events downstream of the ternary Cpt-enzyme-DNA complex, we performed a genetic screen in S. cerevisiae to identify conditional mutants with enhanced sensitivity to DNA damage induced by DNA topoisomerase I. Because the pleiotropic drug resistance network can modulate yeast cell sensitivity to Cpt (16), the top1T722A mutant was used as a source of DNA damage. Substitution of Ala for Thr722 increases the stability of the covalent enzyme-DNA intermediate in the absence of Cpt (12). Overexpression of Top1T722Ap is toxic to wild-type cells whereas constitutive low level expression induces a hyper-recombination phenotype (12). The homologous Thr718-to-Ala substitution in human Top1p induces similar alterations in enzyme function and cell viability (37).
Here, we present the characterization of two mutants that exhibit temperature-sensitive (ts) lethality in the presence of low levels of Top1T722Ap. Complementation analysis and sequencing defined mutations in cdc45 and dpb11, both essential genes involved in DNA replication. We show that these mutants are synthetically lethal with each other and provide evidence suggesting a common function in promoting processive DNA replication through polymerase switching. These findings are also discussed in terms of the cytotoxic action of Cpt.
MATERIALS AND METHODS
Materials, Yeast Strains, and Plasmids.
Ethyl methane sulfonate and methyl methanesulfonate (MMS) were from Kodak. α mating hormone, hydroxyurea (HU) and Cpt were from Sigma. 5-fluoroorotic acid was from American Biorganics (Niagra Falls, NY).
Plasmids YCpSctop1T722A and YCpScTOP1 and strains EKY2 (MATa, ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1∷HIS3), EKY3 (MATα, ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1∷TRP1), and EKY4 (MATa, ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1∷TRP1) were as described (17). RRY71 (EKY4, dpb11-10), RRY72 (EKY4, cdc45-10), MSY32 (EKY4, dpb11-10, cdcd45-10), RRY113 (EKY4, rhoo), RRY114 (RRY71, rhoo), RRY115 (RRY72, rhoo), and RRY116 (MSY32, rhoo) were made by standard techniques.
A yeast genomic DNA library in vector pRS416 was made as described (16). FY250 genomic DNA, partially digested with Sau 3A, was size-fractionated in agarose gels and was ligated into the dephosphorylated BamHI ends of pRS416. After transformation into Escherichia coli, the amplified YCp-FY250 library plasmids were purified by CsCl gradient centrifugation.
tah Mutant Isolation.
In brief, top1T722A hypersensitive (tah) mutants were isolated as follows: EKY3 cells, transformed with plasmid YCpSctop1T722A, were ethyl methane sulfonate-mutagenized (17) and screened for colonies showing ts growth at 35°C. Potential tah mutants were plated at 26°C on 5-fluoroorotic acid media to select against the URA3 marked top1T722A plasmid and were screened for loss of the ts phenotype. tah strains were repeatedly backcrossed to isogenic wild-type cells, and spore products were tested for ts hypersensitivity to top1T722A to identify strains showing 2:2 segregation of single gene mutations.
Cloning CDC45 and DPB11 by Complementation.
tah1-1 and tah2-1 strains, transformed with YCp-FY250 library DNA, were grown at 35°C to isolate genomic DNA inserts that complement hypersensitivity to 5 mg/ml HU. Ends of overlapping clones were sequenced and compared with the Saccharomyces genome database. TAH1 clone 4-4 contained 7.4 kilobases of chromosome X including YJL091c, DPB11, and a 5′ deletion of HPR5. TAH2 clone 2-A contained 7.9 kilobases of chromosome XII encompassing five complete ORFs, YLR099-YLR102, and CDC45. Subcloning showed DPB11 and CDC45, respectively, complemented the HU and top1T722A hypersensitivity of tah1 and tah2.
The genetic identity of TAH1 and TAH2 was established by integrating URA3 into sequences flanking wild-type DPB11 and CDC45, respectively. These strains were mated to tah1 or tah2 mutants, and the meiotic products were assessed for segregation of the mutant phenotype and uracil prototrophy. Integration constructs were as follows: a 3.2-kilobase ClaI fragment excised from TAH1 clone 4-4 was ligated into pRS416 (18), from which a SacI-KpnI fragment was subcloned into pRS306 to make YIpDPB11. Transformation of EKY2 cells with AvrII-digested DNA targeted integration to sequences 5′ to DPB11. A 2.9-kilobase SpeI-BglII fragment containing sequences flanking CDC45 from TAH2 clone 2-A was ligated into the SpeI and BamHI sites of pRS416. A ClaI-SacI fragment was subcloned to yield YIpCDC45. Integration was targeted to CDC45 flanking sequences by restriction with SphI.
Mutant dpb11-10 and cdc45-10 alleles were recovered after targeted integration of the YIp vectors to the mutant loci. Purified yeast genomic DNA (19) was restricted with EcoRI (dpb11-10) or Nsi I (cdc45-10). Size-selected DNA was ligated and transformed into E. coli. Amino acid substitutions were defined by DNA sequencing.
Cell Viability Assays.
Wild-type, dpb11-10, and cdc 45-10 strains, transformed with YCpScTOP1, YCpSctop1T722A, or pRS416, were serially 10-fold diluted. Five-microliter aliquots were spotted onto SC-uracil plates supplemented with 25 mM Hepes (pH 7.2) and 5 μg/ml Cpt in a final 4% DMSO, or DMSO alone. To assess MMS and HU sensitivity, serial dilutions of wild-type and mutant strains were spotted onto yeast extract/peptone/dextrose (YPD) plates containing 0.0125 or 0.025% MMS or 5 mg/ml HU. UV sensitivity was tested by spotting cells onto YPD plates and irradiating with 0, 10, or 20 μJ/M2 UV. Cell viability was assayed at 26 and 35°C.
α Factor Arrest and Flow Cytometry.
Exponential cultures were diluted in YPD, were treated with 5 μg/ml alpha factor for 2 hours, and were visually monitored for G1 arrest (20). An additional 2 μg/ml α factor was added, and the cells were shifted to 35°C. After 2 hours, cells were released into the cell cycle by the addition of 100 μg/ml Streptomyces griseus proteinase E (Sigma) and were prepared for flow cytometry as described (20).
Checkpoint Assays.
Cells grown in YPD were treated with 15 mg/ml HU and were incubated at 26°C or 35°C. Every 2 hours, aliquots were plated to assay cell viability or fixed for microscopy. As in ref. 21, S-phase progression also was assessed in the presence of DNA damage. Cells arrested with 5 μg/ml α factor at 26°C were shifted to 35°C and were released into YPD with or without 0.03% MMS. At 20-minute intervals, aliquots were fixed for flow cytometry.
Analysis of Okazaki Fragments by Alkaline Sucrose Gradient Sedimentation.
As described (22), yeast cells grown at 26°C in YPD were diluted to ≈2 × 104 cells/ml in fresh media containing 5 μCi of 14C-uracil and were grown for ≈7 doublings. The cells were pelleted, resuspended in fresh media, and incubated at 35°C for 30 minutes. 3H-uracil (50 μCi) was added, and, after 30 or 90 minutes at 35°C, the cells were fixed with ethanol-toluene, were lysed, and were fractionated in 5–20% alkaline sucrose gradients (22).
RESULTS
Isolation of tah1 and tah2 Mutants with Conditional Sensitivity to DNA Topoisomerase I Poisons.
To identify mutations that affect the generation or repair of DNA lesions caused by Cpt, a genetic screen was performed to isolate mutants with enhanced sensitivity to the DNA topoisomerase I mutant, top1T722A. To avoid resistance mechanisms dependent on Cpt uptake/efflux, we focused on Top1T722Ap, which mimics the cytotoxic action of Cpt by stabilizing the cleavable complex (12). Constitutive expression of top1T722A from the TOP1 promoter is tolerated by repair-proficient cells, though increased rDNA recombination suggests Top1T722Ap-induced DNA damage (12). Mutants were selected for conditional sensitivity to the DNA damage induced by low levels of Top1T722Ap.
EKY3 (top1Δ) cells transformed with YCpSctop1T722A were mutagenized and screened for loss of viability at 35°C. Ts colonies were cured of the top1T722A plasmid, and the resulting top1-null strains were rescreened for viability at 35°C. Strains thus obtained were ts for growth when Top1T722Ap was expressed; wild-type Top1p or deletion of TOP1 had no effect. Backcrossing ensured segregation of the ts phenotype as a single gene mutation, and the mutants identified were provisionally designated tah for top1-T722A-hypersensitive. In this report, we focus on tah1 and tah2 mutants, which show specific defects in DNA replication.
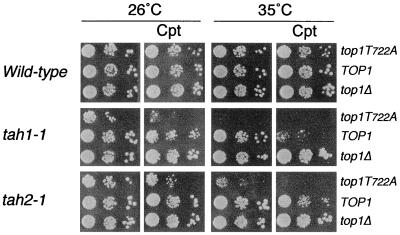
In Fig. 1, TAH+ (wild-type) cells, constitutively expressing TOP1, top1T722A, or a vector control, were viable at 26°C and 35°C (Fig. 1). The low levels of DNA topoisomerase I expressed were insufficient to render the cells sensitive to Cpt at either temperature. Although all strains in this study were top1-null, the wild-type designation refers to TAH+ (DPB11 and CDC45).
Figure 1.
top1-null strains, TAH+ (wild-type), tah1-1, or tah2-1, were transformed with ARS/CEN vectors constitutively expressing the indicated TOP1 allele. Cultures were serially diluted, were spotted onto selective media with or without 5 μg/ml Cpt, and were incubated at the permissive (26°C) or nonpermissive (35°C) temperature.
In contrast, tah1-1 and tah2-1 cells expressing top1T722A were inviable at 35°C. The top1T722A allele induced slow growth of tah1-1 cells even at 26°C, suggesting a partial loss of function at the permissive temperature. This differential was evident in the enhanced Cpt sensitivity of tah1-1 cells expressing Top1p at 35°C and Top1T722Ap at 26°C. The viability of tah2-1 cells was only slightly diminished by Cpt under these conditions. Nevertheless, the Cpt sensitivity of both mutants was augmented at higher levels of Top1p expression (data not shown), consistent with Top1T722Ap function as a Cpt mimetic. The ts phenotypes were not a consequence of alterations in enzyme function, as Top1p levels and catalytic activity were unaffected (data not shown).
tah1-1 and tah2-1 mutants exhibited increased sensitivity to other classes of DNA damaging agents at the nonpermissive temperature. UV irradiation or treatment with the DNA replication inhibitor HU induced an ≈100-fold decrease in tah1-1 and tah2-1 cell viability, relative to isogenic wild-type strains. However, only tah1-1 cells were hypersensitive to the alkylating agent MMS (data not shown). Further, the spectrum of changes in drug sensitivity appeared to be limited to DNA damaging agents, as tah1-1 and tah2-1 cell growth was unaffected by sublethal doses of cycloheximide or the mitochondrial toxin oligomycin (data not shown).
Complementation of tah1-1 by DPB11 and tah2-1 by CDC45.
TAH1 and TAH2 were cloned from the YCp-FY250 DNA library by complementation of HU sensitivity at 35°C. Two overlapping clones from chromosome X complemented tah1-1. Although common sequences contained YJL091c and DPB11, DPB11 alone complemented the ts phenotypes of tah1-1. Genetic identity of TAH1 with DPB11 was established by integration of URA3 near the DPB11 locus in a wild-type strain, mating with strain tah1-1 (dpb11-10) and observing segregation of URA3 away from the ts phenotype.
DPB11 is an essential gene, originally identified as a dosage suppressor of mutations in the POL2 and DPB2 subunits of Pol ɛ (23). Dpb11p is 48% similar to Schizosaccharomyces pombe cut5 protein, which is required for the DNA replication checkpoint (23). A conditional dpb11-1 mutant was previously described that is defective in the S. cerevisiae replication checkpoint (23).
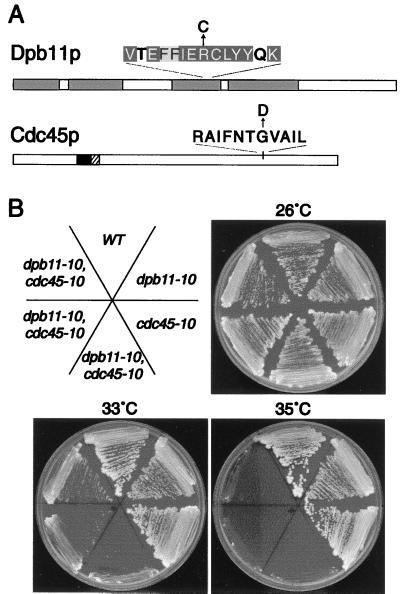
The dpb11-10 allele was recovered, and sequencing revealed a single C-to-T change, resulting in substitution of Cys for Arg-403. This substitution occurs in a region of identity between Cut5p and Dpb11p and within a BRCT motif (Fig. 2A). BRCT motifs are thought to mediate protein–protein interactions necessary for DNA repair and checkpoint functions (24).
Figure 2.
cdc45-10 and dpb11-10 mutants exhibit a synthetic lethal interaction. (A) The shaded boxes in Dpb11p indicate repeated BRCT motifs. The Arg-to-Cys substitution encoded by dbp11-10 lies within a stretch of residues conserved in the S. pombe DPB11 homologue, Cut5. Identical residues are white text on black, and conserved residues are shaded gray. The cdc45-10 mutant encodes a Gly-to-Asp substitution. The shaded areas mark a putative bipartite nuclear localization signal. (B) Wild-type (WT), dpb11-10, cdc45-10, and three independently isolated double dpb11-10, cdc45-10 mutant strains were streaked onto YPD plates and were incubated at the indicated temperatures. All strains were top1-null.
Two overlapping clones from chromosome XII complemented tah2-1. Common sequences included YLR099-YLR102 and CDC45; however CDC45 alone restored tah2-1 cell viability under restrictive conditions. The genetic identity of TAH2 as CDC45 was established as described above for DPB11. The cdc45-10 (tah2-1) allele was recovered, and sequencing defined a Gly-to-Asp change at residue 510, near the C terminus (Fig. 2A).
Cdc45p is essential for the initiation of DNA replication and assembles onto replication origins during late G1 phase of the cell cycle (25–27). Cdc45p function during replication initiation occurs after START but before the HU replication arrest checkpoint and appears to be coincident with the action of the DBF4/CDC7 kinase.
Genetic Interactions Between cdc45-10 and dpb11-10.
To determine whether Cdc45p and Dpb11p function at discrete points in a Cpt-responsive pathway, the double mutant phenotype was examined. In contrast to single mutant and wild-type strains, the dpb11-10, cdc45-10 double mutant was ts for growth in the absence of DNA damage (Fig. 2B). This synthetic lethal phenotype was irrespective of Top1p as double mutants expressing plasmid-borne wild-type TOP1 were also inviable at 35°C (data not shown).
Synthetic lethality suggests that these mutant proteins perform redundant functions in an essential cellular process, or that one mutant allele causes a lesion—such as DNA damage—that is sensed by a checkpoint function of the second gene product (for example, see ref. 28). Indeed, studies with the dpb11-10 mutant indicate Dpb11p function is required for the S-phase checkpoint as well as replication (23, 29, 30), although checkpoint function is not essential for viability.
cdc45-10 and dpb11-10 Cells Have Intact S-Phase Checkpoints yet Are Defective in DNA Replication.
Several approaches were taken to directly assess S-phase checkpoint function in dpb11-10 and cdc45-10 mutants. First, single dpb11-10 and cdc45-10 mutants and the double dpb11-10, cdc45-10 mutant arrested with short spindles and a single DNA mass in response to HU treatment (data not shown). This is in contrast to the checkpoint defect reported for the dpb11-1 mutant, which encodes a C-terminal truncation of Dpb11p (29) and exhibits elongated spindles (30). Furthermore, HU-arrested cdc45-10 and dpb11-10 strains recovered from arrest with no decrease in viability, when plated at 26°C (data not shown).
S-phase progression is also retarded in a checkpoint-dependent manner when cells are treated with a DNA damaging agent (21). dpb11-10 and wild-type strains were arrested with α factor and were released into MMS at 26 or 35°C. At 35°C, S-phase progression was slower in dpb11-10 cells in the absence of DNA damage than in untreated wild-type cells. Nevertheless, MMS treatment produced a greater defect in S-phase transit proportional to that observed with wild-type cells (data not shown). Taken together, the S-phase checkpoint appears to be intact in dpb11-10 and dpb11-10, cdc45-10 mutants.
Deletion of the RAD9 G2/M checkpoint gene in dpb11-10 and cdc45-10 strains produced a slow growth phenotype at 35°C, suggesting the production of DNA damage recognized by this checkpoint (data not shown). The phenotype of the triple rad9Δ, dpb11-10, cdc45-10 mutant resembled that of the double dpb11-10, cdc45-10 mutant. Thus, either the DNA damage produced is not recognized by the G2/M checkpoint or cell cycle progression is regulated by the S-phase checkpoint.
As functional replication checkpoints were evident in cdc45-10 and dpb11-10 mutants, the synthetic lethal phenotype of the double mutant might reflect a common essential function. Cdc45p genetically and physically interacts with components of the prereplicative complex, ORC and MCM proteins (after START), and is required for initiation of DNA replication (25–27, 31). Crosslinking studies further suggest Cdc45p and MCM proteins remain associated with the replication fork after origin firing (32). Dpb11p is critical for DNA replication and the S-phase checkpoint, genetically interacts with the Pol ɛ complex, and appears to function early during the elongation phase of DNA replication (23, 29, 30).
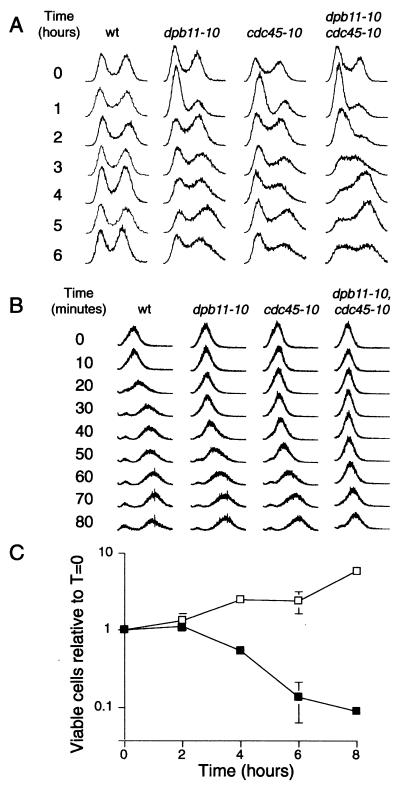
To explore dpb11-10 and cdc45-10 function in DNA replication, asynchronous cultures were shifted to 35°C and were analyzed for changes in cell cycle distribution. As shown in Fig. 3A, the cell cycle distribution of wild-type cells was unaffected at high temperature. Single dpb11-10 and cdc45-10 mutants transiently accumulate with a G1 DNA content 1 hour after shift. The majority of cells were budded, consistent with cell cycle arrest in early S-phase. By 2 hours at 35°C, the populations returned to a relatively normal distribution, although the data indicated a slower rate of S-phase progression. The double dpb11-10, cdc45-10 mutant exhibited a more persistent arrest in early S-phase, with little effect on cell viability at 2 hours (Fig. 3 A and C). After 3 hours, the cells proceeded through S-phase, coincident with a rapid drop in viability (Fig. 3C).
Figure 3.
cdc45-10 and dpb11-10 mutants affect S-phase progression. (A) Asynchronous cultures of EKY4 (wt), RRY71 (dpb11-10), RRY72 (cdc45-10), and MSY32 (dpb11-10, cdc45-10), grown at 26°C, were split, and half of the sample was shifted to 35°C. Each hour, aliquots were fixed, stained with propidium iodide, and sorted on a Coulter Profile II flow cytometer. (B) α factor-arrested cultures were shifted to 35°C for 2 hours and were released into the cell cycle. At 10-minute intervals, aliquots were collected and examined for cell cycle distribution. (C) At t = 0, an asynchronous culture of dpb11-10, cdc45-10 cells was split; half remained at 26°C, and half was shifted to 35°C. At the times indicated, aliquots were serially diluted and plated. The number of viable cells forming colonies at 26°C were counted and plotted relative to t = 0. All strains were top1-null.
To further explore S-phase progression, cells were synchronized at START with α factor, were shifted to 35°C, and were released into the cell cycle. As shown in Fig. 3B, wild-type cells entered S-phase after 10 minutes, with the majority of the cells achieving a 2N DNA content by 30 minutes. In contrast, the single mutants, though viable, experienced a delay in early S-phase progression and took upwards of 60 minutes to acquire a 2N DNA content. The dpb11-10, cdcd45-10 double mutant exhibited a persistent, terminal defect in S-phase transit, and with <2N DNA content at 80 minutes.
Similar defects in the cell cycle transit of cdc45-10 and dpb11-10 cells are consistent with a common essential function in replication. Although Cdc45p plays an essential role at replication origins during initiation (25–27), recent evidence suggests Cdc45p also associates with advancing replication forks, coincident with Pol ɛ and presumably Dpb11p (32). Such an expanded role for Cdc45p might involve interactions with distinct DNA polymerases: Pol α for replication priming and Pol ɛ or δ for elongation.
Cdc45p and Dpb11p Are Required for Okazaki Fragment Maturation.
Processive replication by Pol ɛ/δ requires loading of proliferating cell nuclear antigen (PCNA) onto a DNA template that has been primed by Pol α/primase (reviewed in refs. 14 and 15). Replication factor C is required to open the stable PCNA (Pol30p) trimer and close it around the template in an ATP-dependent process. pol30 mutants, with diminished loading onto DNA, accumulate immature Okazaki fragments on the lagging DNA strand similar to DNA ligase mutants (22). To examine the role of Dpb11p and Cdc45p in this stage of replication, the maturation of Okazaki fragments was assessed.
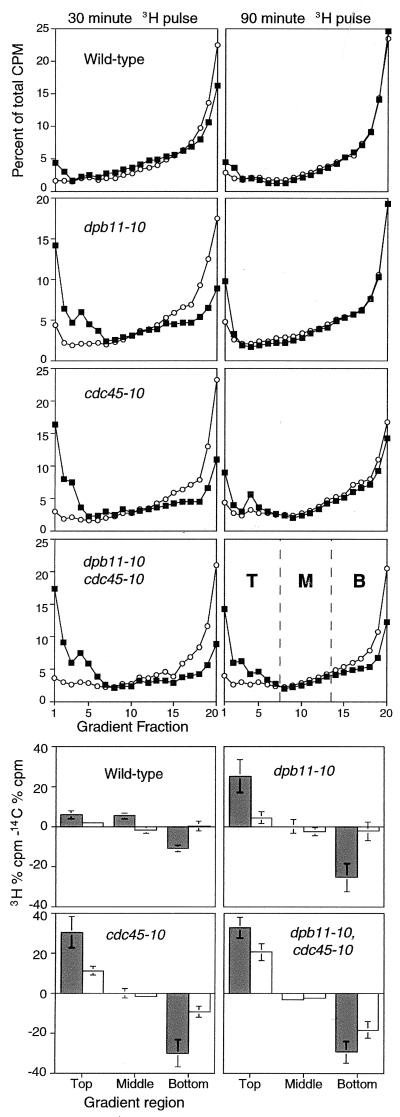
Wild-type and mutant strains were grown in the presence of 14C-uracil to uniformly label DNA at the permissive temperature. The cultures were shifted to 35°C, and newly replicated DNA was labeled with 3H-uracil for 30 or 90 minutes. Total cellular DNA was fractionated in alkaline sucrose gradients (Fig. 4). The relative accumulation of Okazaki-sized DNA fragments, and the proportional decrease in large DNA molecules at 35°C is displayed in Fig. 4 (lower panels).
Figure 4.
cdc45-10 and dpb11-10 mutants accumulate Okazaki-sized DNA fragments. (A) Exponentially growing rho° cultures, continuously labeled with 14C-uracil, were shifted to 35°C, and 3H-uracil was added for 30 minutes (left panels) or 90 minutes (right panels). The DNA was fractionated in alkaline sucrose gradients. Sedimentation is from left to right (fraction 1 is the top). φX174 and λ DNA migrated to fractions 6 and 11, respectively. Acid precipitable 3H (■) and 14C (○) cpm are plotted as the percent total cpm. Each graph is the average of at least three experiments. (B) The relative accumulation of Okazaki-sized DNA fragments (fractions 1–7) or full length DNA molecules (fractions 14–20) at 35°C was assessed by summing the percentage of 3H cpm in the top, middle, and bottom portions of each gradient and subtracting the percentage of 14C cpm obtained with continuous labeling at 26°C. Results from the 30-minute 3H gradients are gray, and those from 90-minute 3H gradients are white. Values >0 indicate an accumulation of 3H label at 35°C whereas values <0 indicate a deficit.
Newly synthesized DNA in wild-type cells was rapidly assembled into large molecules. In Fig. 4, the 3H and 14C traces for the 30 and 90 gradients were practically superimposable. cdc45-10 and dpb11-10 mutants exhibited an initial defect in Okazaki fragment maturation, as indicated by the accumulation of 3H-label at the top of the gradients after the 30′ pulse. However, consistent with the transient accumulation of cdc45-10 and dpb11-10 cells in S-phase 1 hour after temperature shift (Fig. 3A), this deficiency was mostly resolved by 90 minutes (Fig. 4). The double dpb11-10, cdc45-10 mutant showed a similar defect in Okazaki fragment maturation at 30 minutes that persisted at 90 minutes and mirrored the sustained lag in S-phase progression seen in Fig. 3.
These data suggest Cdc45p and Dpb11p function are required for processive DNA replication, possibly in promoting the switch from Pol α/primase to the processive polymerases (δ or ɛ). Along these lines, wild-type CDC45 was unable to dosage suppress the top1T722A-lethality in dpb11-10 cells, but DPB11 could partially dosage suppress top1T722A-induced lethality in cdc45-10 cells (data not shown). Because Cdc45p presumably acts at a point in replication before Dpb11p, this result suggests that the downstream events controlled by Dpb11p can partially compensate for the earlier deficiency caused by mutant Cdc45p.
DISCUSSION
A genetic screen was designed to identify yeast genes that normally function to suppress cell sensitivity to the antitumor drug Cpt. Using Top1T722Ap as a Cpt mimetic, ts mutants hypersensitive to this form of DNA damage were selected. Two genes defined by this approach encode proteins that play a key role in DNA replication, Cdc45p and Dpb11p. At 35°C, single amino acid substitutions in Dpb11p or Cdc45p impaired the ability of yeast cells to survive the DNA lesions induced by alterations in DNA topoisomerase I function, yet cell viability was unaffected in the absence of DNA damage. However, the conditional lethality of the dpb11-10 and cdc45-10 mutants was not restricted to top1T722A, as the strains were also hypersensitive to other DNA damaging agents.
DPB11 suppresses mutations in Pol ɛ subunits and likely associates with the Pol ɛ complex (23). Dpb11p plays a critical role in DNA replication and is a component of the S-phase checkpoint (23, 29, 30). Cdc45p function is essential for the initiation of DNA replication (25–27, 31) and is recruited to replication origins in an S-phase cyclin-Cdk-dependent manner (25). Genetic and physical interactions of Cdc45p with several ORC and MCM proteins have been defined (15, 26, 31).
Here, we show that cdc45-10 and dpb11-10 were synthetically lethal in the absence of DNA damage. Further, cdc45-10 and dpb11-10 mutants exhibit a delay in early S-phase resulting from a transient defect in Okazaki fragment maturation. The persistence of this defect in the double mutant proves lethal as the cells transit S-phase, suggesting a deficiency in switching from priming to processive polymerization.
Several recent reports support our observations. First, chromatin immunoprecipitation experiments indicate a redistribution of Cdc45p during S-phase, coincident with Pol ɛ in advancing replication forks (32). Second, Kamimura et al. (29) identified mutant alleles of CDC45 and SLD2 in a synthetic lethal screen with dpb11-1. Sld2p physically associates with Dpb11p, and the authors argue that these gene products function early in S-phase, but after replication initiation (29). This is consistent with the arrest in early S-phase observed with cdc45-10 and dpb11-10 single mutants after temperature shift. Finally, Xenopus Cdc45p physically associates with Pol α and is essential for loading the polymerase onto chromatin in a cell free system (33). Taken together, these results support a model in which Cdc45p is important for assembling Pol α onto chromatin and for removing the polymerase after its function is complete. Our data further indicate Dpb11p acts downstream of Cdc45p and is required for processive replication, possibly through the loading of processive polymerases.
Replication factor C and PCNA also function in the displacement of Pol α and the recruitment of Pol δ and probably Pol ɛ (15). Although Pol δ and ɛ are both required for DNA replication, the specific function of Pol ɛ remains unclear. Previous studies suggest Pol δ and Pol ɛ are associated with different strands (15); however, the direct assignment of either polymerase with leading or lagging strand synthesis has not been made.
PCNA further stimulates Rad27p, the FEN1 nuclease implicated in the nucleolytic processing of RNA primers (15). PCNA function in the cdc45-10 and dpb11-10 mutants has not been addressed. However, a direct effect on Rad27p is unlikely because deletion of RAD27 had little effect on the Cpt sensitivity of yeast cells expressing wild-type TOP1 or the viability of cells expressing top1T722A (R.J.D.R., M.S., and M.-A.B., unpublished data).
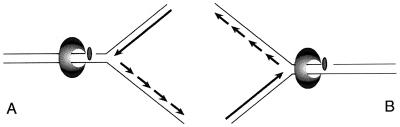
The question remains of how a defect in polymerase processivity, and the resultant accumulation of Okazaki fragments, enhances cell sensitivity to Cpt. Collision of the advancing replication fork with Cpt-stabilized Top1p-DNA complexes leads to irreparable lesions, cell cycle arrest, and death (reviewed in refs. 3 and 6). Further studies imply the importance of replication fork polarity (6, 10), where formation of the ternary Cpt-Top1p-DNA complex on the leading strand template (Fig. 5A) is more likely to produce irreparable lesions than Cpt-induced breaks of the lagging strand template (Fig. 5B). Our studies indicate the accumulation of Okazaki fragments on the lagging strand exacerbates the cytotoxicity of DNA topoisomerase I covalent complexes. Whether replication fork collision with these complexes produces double strand DNA breaks or stalled replication forks, homologous recombination coupled with replication is required for the repair of either (34, 35). In cdc45-10 or dpb11-10 cells, fragmentation of the newly replicated lagging strand, i.e., accumulation of Okazaki fragments, might preclude the effective repair of Top1p-induced DNA damage. Here, invasion of the nicked leading template strand into homologous sequences of the sister chromatid would displace the unligated Okazaki fragments, such that they could no longer serve as a substrate for either replication or homologous recombination. An alternative model is that the interaction is indirect, that increased DNA lesions induced by Cpt, coupled with the accumulation of Okazaki fragments exceeds the repair capacity of the cell.
Figure 5.
Model for the enhanced sensitivity of cdc45-10 and dpb11-10 mutants to Cpt.
Nevertheless, the enhanced Cpt sensitivity of rad52Δ cells implicates homologous recombination in the repair of DNA (2, 3). Rad52p is also required for the formation of Holiday junctions during DNA replication (36) and for the viability of pol30 mutants defective in promoting processive DNA replication (22), consistent with the repair of collapsed replication forks. Experiments are currently underway to determine the specific DNA lesions induced by Cpt in these mutant strains.
Acknowledgments
We thank H. Askari, J. Fertala, and other members of the lab for their assistance. This work was supported in part by Fellowship 203.04.15 from the Consiglio Nazionale delle Ricerche (Italy) (to P.F.), National Institutes of Health Grants CA58755 and CA70406 (to M.-A.B.), CA21675 Cancer Center Grant, and the American Lebanese Syrian Associated Charities.
ABBREVIATIONS
- Cpt
camptothecin
- HU
hydroxyurea
- MMS
methyl methanesulfonate
- PCNA
proliferating cell nuclear antigen
- Pol α
DNA polymerase α
- Pol δ
DNA polymerase δ
- Pol ɛ
DNA polymerase ɛ
- ts
temperature-sensitive
- Top1p
DNA topoisomerase I
- YPD
yeast extract/peptone/dextrose
References
- 1.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Nitiss J. Biochim Biophys Acta. 1998;1400:63–82. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 3.Reid R J D, Benedetti P, Bjornsti M-A. Biochim Biophys Acta. 1998;1400:289–300. doi: 10.1016/s0167-4781(98)00142-0. [DOI] [PubMed] [Google Scholar]
- 4.Stewart L, Redinbo M R, Qiu X, Hol W G J, Champoux J J. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y, Pourquier P, Fan Y, Strumberg D. Biochim Biophys Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 6.Chen A, Liu L F. Annu Rev Pharmacol Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 7.Nitiss J, Wang J. Mol Pharmacol. 1996;50:1095–1102. [PubMed] [Google Scholar]
- 8.Hsiang Y, Lihou M G, Liu L F. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 9.Holm C, Covey J M, Kerrigan D, Pommier Y. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 10.Tsao Y P, Russo A, Nyamuswa G, Silber R, Liu L F. Cancer Res. 1993;53:5908–5914. [PubMed] [Google Scholar]
- 11.Shao R-G, Cao C-X, Zhang H, Kohn K W, Wold M S, Pommier Y. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megonigal M D, Fertala J, Bjornsti M-A. J Biol Chem. 1997;272:12801–12808. doi: 10.1074/jbc.272.19.12801. [DOI] [PubMed] [Google Scholar]
- 13.Snapka R M, Permana P A. BioEssays. 1993;15:121–127. doi: 10.1002/bies.950150208. [DOI] [PubMed] [Google Scholar]
- 14.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 15.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 16.Reid R J D, Kauh E A, Bjornsti M-A. J Biol Chem. 1997;272:12091–12099. doi: 10.1074/jbc.272.18.12091. [DOI] [PubMed] [Google Scholar]
- 17.Kauh E A, Bjornsti M-A. Proc Natl Acad Sci USA. 1995;92:6299–6303. doi: 10.1073/pnas.92.14.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sikorski R, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strathern J N, Higgins D R. Methods Enzymol. 1991;194:319–329. doi: 10.1016/0076-6879(91)94024-7. [DOI] [PubMed] [Google Scholar]
- 20.Haase S B, Lew D J. Methods Enzymol. 1997;283:322–341. doi: 10.1016/s0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- 21.Paulovich A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 22.Merril B J, Holm C. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araki H, Leem S-H, Phongdara A, Sugino A. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind L, Walker D R, Koonin E V. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 26.Zou L, Mitchell J, Stillman B. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens J C, Detweiler C S, Li J J. Proc Natl Acad Sci USA. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 29.Kamimura Y, Masumoto H, Sugino A, Araki H. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Elledge S J. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton S, Hopwood B. Mol Cell Biol. 1997;17:5867–5875. doi: 10.1128/mcb.17.10.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 33.Mimura S, Takisawa H. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes A M, Haber J E. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 35.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 36.Zou H, Rothstein R. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 37.Fiorani, P., Amatruda, J. F., Silvestri, A., Butler, R. H., Bjornsti, M.-A. & Benedetti, P. (1999) Mol. Pharmacol., in press. [DOI] [PubMed]