Abstract
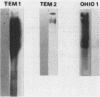
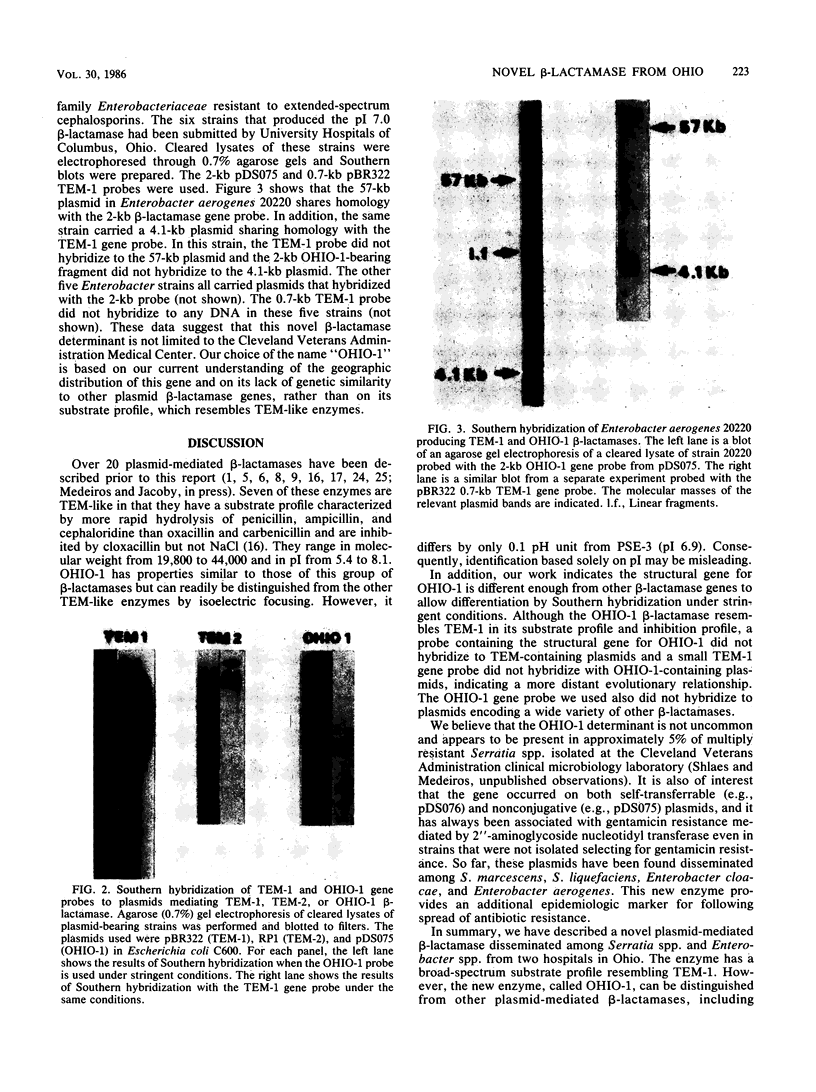
Epidemiologic studies of plasmid-mediated resistance at the Cleveland Veterans Administration Medical Center revealed that related plasmids had disseminated among members of the family Enterobacteriaceae. We studied the beta-lactamases encoded by these plasmids in Escherichia coli C600 transformants or transconjugants. Substrate and inhibition profiles of the enzymes determined by two of these plasmids suggested an activity resembling TEM-1; however, isoelectric focusing revealed a pI of 7.0. These two plasmids were originally found in a Serratia marcescens (pDS076) and an Enterobacter cloacae (pDS075) strain isolated from the same sink in the medical intensive care unit and later, in an Enterobacter cloacae (pDS142 identical to pDS076) isolate colonizing a patient in the same unit. The plasmids also carried the aminoglycoside resistance determinant, 2"-aminoglycoside nucleotidyl transferase. A 2-kilobase AvaI restriction endonuclease digestion fragment of pSD075 known to carry the beta-lactamase determinant was used as a molecular probe. This probe did not recognize sequences of any plasmid-mediated beta-lactamase tested including the recently described determinants ROB-1, TLE-1, and OXA-4-7. A TEM-1 probe derived from the 0.7-kilobase PstI-EcoRI fragment of pBR322 failed to recognize the new beta-lactamase gene. Four additional Enterobacter cloacae and two Enterobacter aerogenes strains isolated in Columbus, Ohio, have been shown to produce a pI 7.0 beta-lactamase and to carry plasmids recognized by the 2-kilobase probe. These data suggest dissemination of a novel plasmid-mediated beta-lactamase among members of the family Enterobacteriaceae in Ohio and demonstrate the development and utility of a molecular probe for the new determinant. We suggest that the novel beta-lactamase be named OHIO-1.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bobrowski M. M., Matthew M., Barth P. T., Datta N., Grinter N. J., Jacob A. E., Kontomichalou P., Dale J. W., Smith J. T. Plasmid-determined beta-lactamase indistinguishable from the chromosomal beta-lactamase of Escherichia coli. J Bacteriol. 1976 Jan;125(1):149–157. doi: 10.1128/jb.125.1.149-157.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey R. C., Clark N. C., Thornsberry C. A gene probe for TEM type beta-lactamases. Antimicrob Agents Chemother. 1985 Jul;28(1):154–156. doi: 10.1128/aac.28.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson I., Kamme C. Characterization of the plasmid-mediated beta-lactamase in Branhamella catarrhalis, with special reference to substrate affinity. J Antimicrob Chemother. 1985 Feb;15(2):139–149. doi: 10.1093/jac/15.2.139. [DOI] [PubMed] [Google Scholar]
- Furth A. J. Purification and properties of a constitutive beta-lactamase from Pseudomonas aeruginosa strain Dalgleish. Biochim Biophys Acta. 1975 Feb 19;377(2):431–443. doi: 10.1016/0005-2744(75)90323-x. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1974 Jan;117(1):56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Medeiros A. A., Cohenford M., Jacoby G. A. Genetic and biochemical properties of AER-1, a novel carbenicillin-hydrolyzing beta-lactamase from Aeromonas hydrophila. Antimicrob Agents Chemother. 1985 Apr;27(4):479–484. doi: 10.1128/aac.27.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Matthew M. The distribution of beta-lactamase genes on plasmids found in Pseudomonas. Plasmid. 1979 Jan;2(1):41–47. doi: 10.1016/0147-619x(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Labia R., Andrillon J., Le Goffic F. Computerized microacidimetric determination of beta lactamase Michaelis-Menten constants. FEBS Lett. 1973 Jun 15;33(1):42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- Mathew A., Harris A. M., Marshall M. J., Ross G. W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975 May;88(1):169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- Matthew M., Hedges R. W., Smith J. T. Types of beta-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979 Jun;138(3):657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M., Sykes R. B. Properties of the beta-lactamase specified by the Pseudomonas plasmid RPL11. J Bacteriol. 1977 Oct;132(1):341–345. doi: 10.1128/jb.132.1.341-345.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros A. A., Cohenford M., Jacoby G. A. Five novel plasmid-determined beta-lactamases. Antimicrob Agents Chemother. 1985 May;27(5):715–719. doi: 10.1128/aac.27.5.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin L. G., Medeiros A. A., Yolken R. H., Moxon E. R. Ampicillin treatment failure of apparently beta-lactamase-negative Haemophilus influenzae type b meningitis due to novel beta-lactamase. Lancet. 1981 Nov 7;2(8254):1008–1010. doi: 10.1016/s0140-6736(81)91214-9. [DOI] [PubMed] [Google Scholar]
- Shlaes D. M., Currie C. A. Endemic gentamicin resistance R factors on a spinal cord injury unit. J Clin Microbiol. 1983 Aug;18(2):236–241. doi: 10.1128/jcm.18.2.236-241.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Vartian C., Currie C. A. Variability in DNA sequence of closely related nosocomial gentamicin-resistant plasmids. J Infect Dis. 1983 Dec;148(6):1013–1018. doi: 10.1093/infdis/148.6.1013. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I., Tsukamoto K., Harada M., Sawai T. Carbenicillin-hydrolyzing penicillinases of Proteus mirabilis and the PSE-type penicillinases of Pseudomonas aeruginosa. Microbiol Immunol. 1983;27(12):995–1004. doi: 10.1111/j.1348-0421.1983.tb02934.x. [DOI] [PubMed] [Google Scholar]
- Vecoli C., Prevost F. E., Ververis J. J., Medeiros A. A., O'Leary G. P., Jr Comparison of polyacrylamide and agarose gel thin-layer isoelectric focusing for the characterization of beta-lactamases. Antimicrob Agents Chemother. 1983 Aug;24(2):186–189. doi: 10.1128/aac.24.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]