Abstract
Analysis of lung cancer response to chemotherapeutic agents showed the accumulation of a Taxol-induced protein that reacted with an anti-phospho-MEK1/2 antibody. Mass spectroscopy identified the protein as nucleophosmin/B23 (NPM), a multifunctional protein with diverse roles: ribosome biosynthesis, p53 regulation, nuclear-cytoplasmic shuttling, and centrosome duplication. Our work demonstrates that following cellular exposure to mitosis-arresting agents NPM is phosphorylated and its chromatographic property is altered, suggesting changes in function during mitosis. To determine the functional relevance of NPM, its expression in tumor cells was reduced by siRNA. Cells with reduced NPM were treated with Taxol followed by microarray profiling accompanied by gene/protein pathway analyses. These studies demonstrate several expected and unexpected consequences of NPM depletion. The predominant downstream effectors of NPM are genes involved in cell proliferation, cancer, and the cell cycle. In congruence with its role in cancer, NPM is over-expressed in primary malignant lung cancer tissues. We also demonstrate a role for NPM in the expression of genes encoding SET (TAF1β) and the histone methylase SET8. Additionally, we show that NPM is required for a previously unobserved G2/M upregulation of TAF1A, which encodes the rDNA transcription factor TAFI48. These results demonstrate multi-faceted functions of NPM that can affect cancer cells.
Keywords: Nucleophosmin, G2/M, rDNA, TAF1A, transcriptional profiling
INTRODUCTION
During our analysis of Taxol-induced protein and gene changes, we observed the G2/M induction of a protein that cross reacts with an antibody produced against phosphorylated MEK1/2 (pMEK). This protein is smaller than pMEK and migrates as a 37kDa moiety. After extensive biochemical analysis, this band was identified as nucleophosmin (numatrin, B23). This revelation largely coincided with two published report showing the same [1, 2]. The induction of NPM by Taxol is intriguing, thus this report pursues the functional relevance of NPM in cancer and its global effects on gene profiles.
The relevance of NPM is underscored by the attention it has received from researchers concerned with very different aspects of cell biology. Initial research on the protein centered on its nucleolar distribution and role in ribosome synthesis though alternate unrelated roles for NPM have also been uncovered [3-5]. Among these are its capacity to act as a chaperone for nuclear-cytoplasmic shuttling and its participation in the ARF-p53 tumor suppressor pathway [6-10]. A recent report has also implicated nucleophosmin in the control of transcription by chromatin remodeling [11]. These are exciting aspects of NPM research, but the protein is chiefly implicated in the regulation of cell proliferation and transformation. A more thorough examination of its involvement in these latter two processes is critical to understanding the significance of NPM to the cell.
NPM has long been implicated in cellular transformation, particularly in leukemias and lymphomas. These types of cancer are typically characterized by chromosomal abnormalities such as translocations, which often result in fusion proteins combining the N-terminus of NPM with one of several genes that disrupt its localization and function, including the retinoic acid receptorα (RARα), myelogenous leukemia factor 1 (MLF-1), and the anaplastic lymphoma kinase (ALK) [12]. Recent studies have also identified mutations in the C-terminus of NPM that are associated with acute myelogenous leukemia even in the absence of chromosome rearrangement [13]. Moreover, NPM is abundant in certain hepatomas, and overexpression has been shown to promote the transformation of NIH3T3 cells [14, 15]. The dysregulation of NPM in cancer cells highlights its importance in regulating cell proliferation.
Proliferation depends on ribosome synthesis, and evidence for NPM’s role in this process has been accumulating for over two decades [16]. Several lines of evidence have pointed to a role for NPM in the maturation of pre-ribosomal RNA, including its interaction with pre-rRNA particles in cell lines, its in vitro ribonuclease activity, and its localization to the granular region of the nucleolus, where pre-RNA processing takes place. Intriguingly, NPM is also found in the dense fibrillar area of the nucleolus, where transcription of rDNA occurs, suggesting that NPM may play a role not only in the maturation of pre-rRNAs but in their transcription as well [4, 5, 17-19].
NPM is also important to the segregation of chromosomes and the physical division of daughter cells. During mitosis, the microtubule organizing center duplicates in an NPM-dependent fashion to form the centrosomes, poles from which the mitotic spindle extends [20]. Knockout studies have demonstrated that the absence of NPM results in abnormal accumulation of centrosomes in mouse embryonic fibroblasts [21].
Since we observed Taxol-induced NPM and likely post-translational modification of NPM during mitosis, we focused our attention on its differential functions at that stage of the cell cycle in comparison to interphase. To do so we employed the chemotherapeutic agent Taxol. The primary chemotherapeutic value of Taxol is its ability to bind the β-subunit of tubulin and suppress microtubule dynamics. This prevents dissolution of the mitotic spindle, arresting proliferating cells in mitosis and eventually resulting in their death [22]. While its pro-apoptotic effect is of great clinical importance, Taxol is often used in the laboratory as a mitotic-arresting reagent. Shorter periods of Taxol exposure were used to halt progression of the cell cycle while minimizing cytotoxicity. This allowed us to study the mitosis-specific role of NPM without generating confounding effects such as apoptosis. To understand the role of NPM in transformed cells we performed our experiments in the H157 cell line, which is derived from advanced non-small cell lung carcinoma (NSCLC).
We reasoned that depletion of nucleophosmin would have consequences on several known and/or novel functions, and that these consequences would be reflected at the level of gene transcription. It is also possible that modification prevents activity of nucleophosmin. We studied NPM function during the cell cycle by depleting it with NPM-using specific small interference siRNA (siRNA) oligonucleotides, then treating cells with Taxol to arrest them in mitosis. After examining the effect of NPM-knockdown on cell cycle progression and apoptosis, we performed large scale microarray analysis to investigate global changes in gene expression. Based on previous studies we expected NPM to be crucial to cell cycle progression. We found novel roles for NPM in regulating the expression of genes associated with cell proliferation and the progression of cancer. We also show that NPM is upregulated in cancerous tissue, suggesting that it can be used as a marker for NSCLC. Additionally, we present evidence that NPM modulates the transcription of pre-rRNAs by enhancing the transcription of TAF1A, a component of the RNA Polymerase I machinery, supporting the supposition that not only is NPM important the maturation of pre-rRNAs but in their transcription as well.
MATERIALS AND METHODS
Cell Lines and Reagents -
The H157 human lung carcinoma line was obtained from the American Type Culture Collection (ATCC), and cultured in RPMI 1640 media (Gibco) with 8% FBS, 10 units/ml penicillin, and 100μg/ml streptomycin. Cells were maintained at 37°C with 5%CO2. Paclitaxel (Sigma) and PMA (Sigma) were maintained in a stock solution in dimethyl sulfoxide (DMSO) (Sigma). Anti-pMEK (#9121), anti-total MEK 1/2 (#9122), anti-pNPM (#3541), and anti-pH3 (#9706) antibodies were purchased from Cell Signaling, NPM antibodies were purchased from Santa Cruz Biotechnology (H-106) and Zymed (32-5200), anti-GAPDH (MAB374) was purchased from Chemicon, and anti-tubulin antibody was a gift from the laboratory of Lishan Su. Fluorescent secondary antibodies were purchased from Molecular Probes. Lambda phosphatase was purchased from Calbiochem.
Propidium Iodide Staining, Immunoblot Analysis, and Immunoprecipitation -
PI staining and immunoblots were performed as described previously [23]. Immunoprecipitation was performed as described previously [10].
Thymidine Block -
Cells were incubated in serum-free RPMI 1640 with 2mM thymidine (Sigma) for 16 hours. Media was then replaced with RPMI 1640 containing 2% serum. Lysates and cell pellets for cell cycle analysis were collected at the indicated time points following release.
Intracellular Staining for Flow Cytometry -
Following treatment, cells were collected by scraping into PBS. Scraped cells were fixed in 2% paraformaldehyde (Electron Microscopy Sciences) at room temperature in the dark for 15 minutes. Cells were pelleted and resuspended in ice-cold MeOH, kept in the dark at 4°C overnight, then moved to -20°C and stored for 2 hours or longer. After rinsing, cells were incubated with primary antibody for 1.5 hours at room temperature, rinsed, incubated with fluorescent conjugated secondary antibody for 45 minutes in the dark, and rinsed again. Cells were analyzed using a FACScan (Becton Dickinson).
Phosphatase Assay -
Parallel lysates were generated as for western blots, though phosphatase inhibitors were excluded from the samples subjected to phosphatase treatment. In addition, phosphatase buffer was added to a final concentration of 50mM Tris-HCl pH 7.5, 0.1mM EDTA, 5mM DTT, and 2mM MnCl2. Lysates were then boiled for 1 minute with frequent vortexing. 800 units of λ phosphatase were added to 80 μl of lysate. Samples were incubated at 30°C for 30 minutes.
Gel Filtration -
Lysates were prepared by sonication in PBS supplemented with EDTA-free protease cocktail (Roche). Lysates were injected over a Superdex 200 or Superose 6 packed column equilibrated with lysis buffer. Fractions were collected at one minute intervals and analyzed by immunoblot. To determine molecular weights, each column was calibrated with commercially available gel filtration standards.
Coomassie Blue staining -
The gel was fixed for one hour in a solution of 25% isopropanol, 10% acetic acid, and 65% ddH2O. It was then stained overnight with shaking in Coomassie Blue solution (BioRad). Acetic acid (10%) was used to destain.
Proteomics -
Tandem mass spectroscopy for protein identification was performed at the University of North Carolina at Chapel Hill Proteomics Facility. siRNA - Double-stranded NPM siRNA oligonucleotides (Ambion) were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Sense sequences are: 5′-GGAGGAUGUGAAACUCUU-3′ and 5′-GGAAGUCUCUUUAAGAAAA-3′.
Microarray Analysis -
Cells were transfected with siRNA oligonucleotides or a commercially available control that activates the RISC complex (Dharmacon). After 24 hours, cells were treated with either 100nM Taxol or an equivalent volume of DMSO. Cells were harvested 16 hours later. RNA was collected with Trizol and purified using the QIAgen RNAeasy kit according to manufacturer’s instructions. RNA isolations were performed on three independent sets of biological replicates. Microarray analysis was performed using the Operon 3.0 human transcript chip, which includes 35,000 transcripts, at the Duke University Microarray Facility. Analysis was performed using GeneSpring (Agilent), Cluster (http://rana.lbl.gov/EisenSoftware.htm), and Java Treeview (http://jtreeview.sourceforge.net). Data was derived from the ratio of transcript measured in an experimental sample to that found in a commercially-available universal reference set of RNA (Stratagene). This was done to reduce experimental variability and simplify comparisons between samples. Only normalized data with a standard error within 0.5 between replicates was considered. Individual spots were flagged present or absent following the initial scan of the microarray. If any of the 18 spots per transcript were flagged as absent or marginal the transcript was excluded from further consideration. In performing comparisons, raw value cutoffs were set at 50 for those transcripts considered to be higher in expression. The limit on fold change in expression was set to 1.5. To graphically represent normalized values as up- or downregulated relative to our si control sample we used the formula: log2 [(experimental/reference)/(si control/reference)] Genes were clustered for presentation by non-centered averaging. Normalized values are included in the supplemental data tables.
Network Generation -
Those genes which passed the filtering scheme were uploaded into the Ingenuity Pathways analysis application, which overlaid them onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Each network was algorithmically generated based on the connectivity of genes. P-values, which indicate the likelihood that the same number of genes taken from a random set would appear in the network, were calculated using Fischer’s exact test. The maximum number of genes in any network is limited to 35.
Functional Analysis of a Network -
Functional analysis of the merged network was undertaken to identify biological functions and/or diseases most significant to the genes in the network. Only those network genes in the Ingenuity Pathways Knowledge Base were considered for analysis. P-values, indicating the probability that each biological function and/or disease assigned to that network is due to chance alone, were calculated using Fischer’s exact test. The three most significant functions are listed in Figure 5. As each of these is a higher-level function comprised of multiple processes, significance is represented as a range of p-values indicating association to the most and least significant component process.
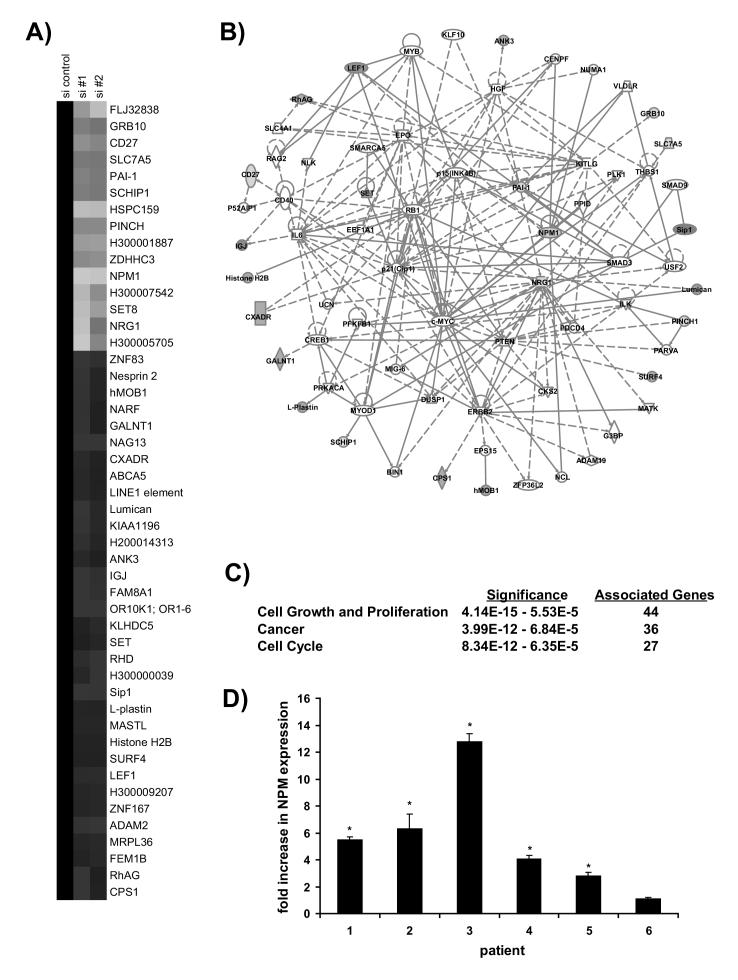
Figure 5.
NPM depletion alters gene expression patterns. A) Incubation with siNPM oligonucleotides changes the expression of 45 genes. Genes with altered expression in NPM-depleted cells were identified as indicated in the Materials and Methods and clustered for graphical representation. Downregulation is indicated by green and upregulation by red. B) The change in gene expression following NPM depletion corresponds to a network of genes implicated in cell proliferation and cancer. Ingenuity Systems pathway analysis software was used to determine two potential interaction pathways for the products of these transcripts. These pathways were merged to form a single network. Direct interactions are indicated by solid lines, indirect interactions by dashed lines. C) The merged network is associated with cancer and cell cycle progression. Ingenuity Systems pathway analysis software was used to identify functional associations for the network. The three most significant associations are listed. D) Nucleophosmin message is overexpressed in cancerous lung tissue. NPM expression was measured by real-time PCR analysis using RNA isolated from biopsies of non-small cell lung carcinoma transformed NSCLC tissue and adjacent normal tissue. Data is presented as relative to the expression of the housekeeping gene cyclophilin A1. Error bars represent standard deviation. Asterisks indicate p<0.01.
Real Time PCR -
RNA was isolated as above. Reverse transcription was performed on 1μg of each RNA sample to generate cDNA as described previously [24]. Samples were normalized by dividing copies of the indicated gene by copies of a gene with unchanged expression - either cyclophilin A1 (Figure 5D) or 18s rRNA (Figure 6D). Gene expression was quantified via quantitative real-time RT-PCR using an ABI PRISM 7900 (Perkin-Elmer Applied Biosystems). Primers are listed in Supplementary Table 3.
Figure 6.
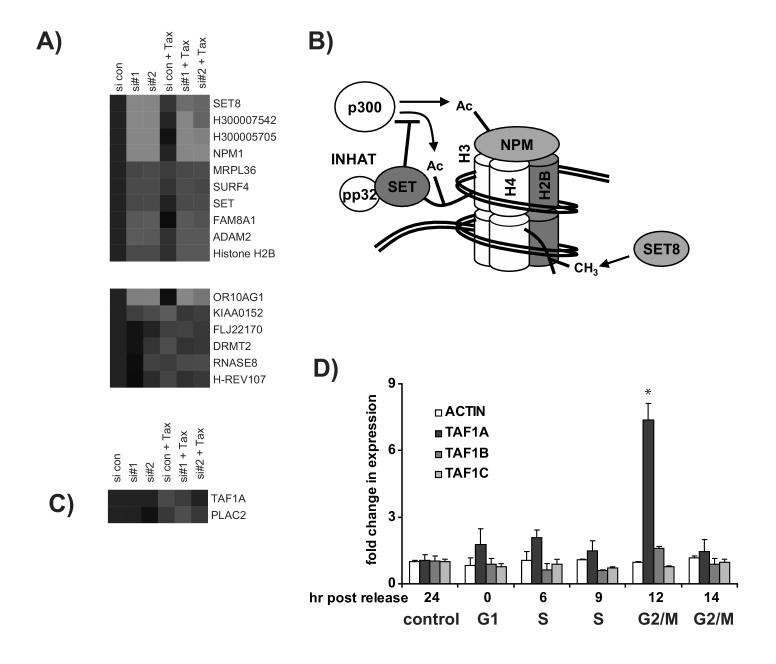
NPM depletion alters gene expression patterns following mitotic-arrest by Taxol. (A) The expression patterns of genes that are altered in NPM siRNA-containing cells regardless of the presence or absence of Taxol. Four of these genes demonstrate close interaction with DNA. These include several genes encoding proteins that are involved in DNA architecture. (B) A model is drawn based on the existing literature regarding roles of SET, H2B, SET, and SET8 in chromatin structure. Transcripts for these proteins were altered in the absence of NPM regardless of exposure to Taxol. Proteins representing upregulated transcripts are represented in red, and those with downregulated transcripts are green. As part of the INHAT complex, the SET protein can inhibit the activity of the acetylase (-Ac) p300, which can act on histone H3 and nucleophosmin [11, 39]. SET8 is a histone methylase (-CH3) that acts on histone H4 [36]. Nucleophosmin itself can interact with core histones [11]. C) NPM siRNA changes the expression of several genes following Taxol treatment that are not changed in control cells. Only one transcript demonstrated strong nucleophosmin-dependent expression following Taxol-treatment. TAF1A is upregulated in control siRNA control cells exposed to Taxol, but unchanged in the siNPM knockdown cells following Taxol-treatment. No transcripts with an NPM-dependent downregulation in Taxol-treated cells were identified. D) TAF1A upregulation is a normal feature of G2/M. Real time PCR was performed following release of cells from thymidine synchronization as shown in Fig. 1B. TAF1A was upregulated at a time point corresponding to G2/M, though TAF1B, TAF1C, and actin remained stable. Error bars represent standard deviation. The asterisk indicates p<0.03.
RESULTS
Taxol induces a form of nucleophosmin recognized by an antibody to phospho-MEK1/2
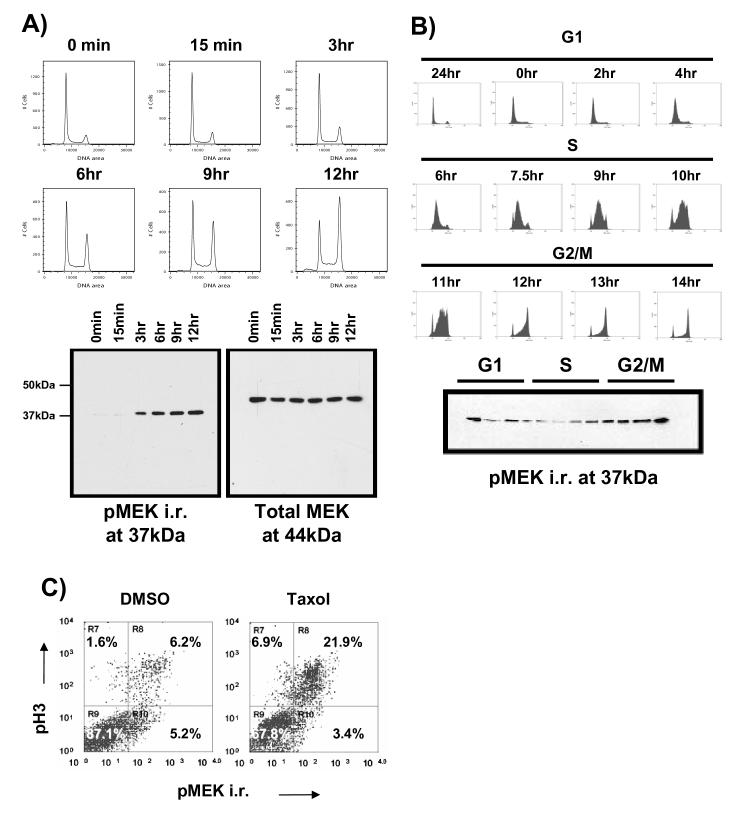
When treated with Taxol, the H157 lung cancer cell line arrests in the G2/M phase of the cell cycle. This arrest correlates with increased intensity of a 37kDa band recognized by anti-phospho-MEK1/2 antibody (Figure 1A). However an immunoblot of total MEK1/2 revealed a band that migrated at the expected size of 44kDa. The intensity of this band does not fluctuate greatly in Taxol-treated cells. This result initially suggested the possibility of a smaller MEK1/2 isoform specific to mitosis [25]. However, Taxol is a powerful chemotherapeutic agent with potential effects secondary to its ability to induce G2/M arrest. We used thymidine synchronization, an alternate method for isolating a mitotic cell population, to rule out any other effects of Taxol that might cause an increase in pMEK immunoreactivity (hereafter referred to as pMEK i.r.). Briefly, cells were synchronized by thymidine block then collected at various stages of the cell cycle. Similar to Taxol treated cells, those cells with a 4N (G2/M) DNA content demonstrated higher levels of pMEK i.r. (Figure 1B). Intracellular staining with the pMEK antibody and an antibody against the mitotic marker phospho-histone 3 (pH3) showed that pMEK i.r. and pH3 staining tend to co-segregate into the same population. This supports the conclusion that expression of the pMEK immunoreactive protein is indeed mitotic (Figure 1C).
Figure 1.
An antibody directed against phospho-MEK1/2 recognizes a protein induced by Taxol in G2/M at mitosis. A) Phospho-MEK immunoreactivity (pMEK i.r.) at 37kDa increases following exposure to Taxol. H157 cells were treated with 100nM Taxol for the indicated periods of time, then collected, fixed, and stained with propidium iodide to follow DNA content. Lysates were at the same time points then immunoblotted as indicated. The increase in pMEK i.r. corresponded to an increase in G2/M. B) The 37kDa pMEK i.r. is enriched during G2/M. Cells were synchronized by thymidine block then released to progress through the cell cycle as a population. Cells were collected at the indicated time points following release for both DNA content analysis and immunoblotting. The pMEK i.r. diminished during G1 progression, but was enriched during G2/M progression. C) pMEK i.r. is highest at G2/M. Cells were collected following exposure to either DMSO or 100nM Taxol for 12 hours, then double stained for pMEK i.r. and the G2/M marker phospho-histone 3 (pH3). Fluorescence was measured with flow cytometry. Cells that reacted with anti-pH3 were also pMEK i.r. positive.
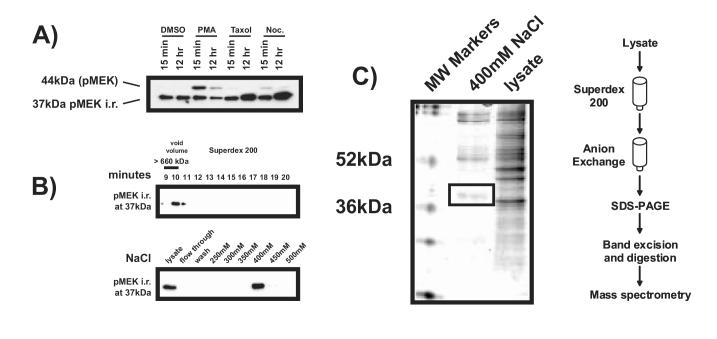
To determine if the putative smaller MEK isoform was induced by the known MEK stimulant PMA (Phorbol Myristyl Acetate), or to nocodazole, an alternate microtubule inhibitor, additional cell treatments were performed. Treatment with Taxol and nocodazole, both mitotic arresting agents, enhanced the intensity of the smaller 37kDa band. However, the commercially available pMEK antibody recognized an upper band at the known MEK molecular weight of 44kDa was induced only following treatment with PMA (Figure 2A). This suggests that the lower band represents either a smaller isoform of MEK lacking PMA-responsive elements found in the full-length protein or a different protein altogether. We used peptide sequencing to definitively identify the lower band. Lysates made from Taxol-treated cells were fractionated according to both size and charge (Figure 2B), then separated on an SDS-PAGE gel. The appropriate sized band (Figure 2C) was excised and subjected to MALDI-TOF analysis. The protein was identified as nucleophosmin (NPM). Coincident with our own study, cross-reactivity of the pMEK1/2 antibody with NPM was observed by two other groups [1, 2].
Figure 2.
The 37kDa band recognized by anti-phospho MEK antibody is nucleophosmin. A) Intensity of the 37kDa band does not increase following exposure to the known MEK stimulant PMA, but is increased by Taxol and nocodazole. Cells were treated with DMSO control, 100nM PMA, 100nM Taxol, or 100nM nocodazole (noc.) for the indicated periods of time. Lysates were collected and immunoblotted for pMEK i.r. PMA induced the 44kDa phospho-MEK band in congruagreement with the literature [46]. However Taxol and Nocadazole both induced the 37kDa band. B) Fractionation of the pMEK i.r. band for biochemical identification. Lysates were made from H157 cells treated with 100nM Taxol for 12 hours, then fractionated by molecular weight with a Superdex 200 column (top panel). The fraction in which pMEK i.r. was strongest was then refractionated according to charge using an anion exchange column (bottom panel). C) The charge fraction containing the strongest pMEK i.r. was denatured and separated by weight on an SDS-PAGE gel, which was then stained for total protein with Coomassie Blue. The band appearing at 37kDa was excised. Tandem mess spectroscopy performed on the gel slice identified the protein as nucleophosmin.
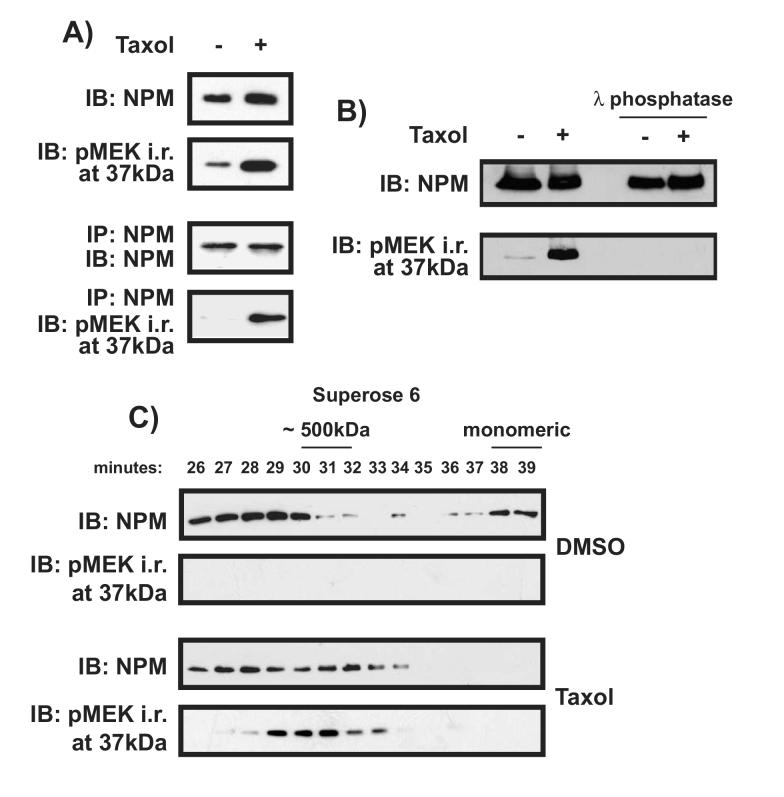
The mitotic nucleophosmin species recognized by an antibody to phospho-MEK1/2 is phosphorylated and appears in a complex of distinct size
Immunoblot analysis was performed with an antibody raised against total NPM. The results revealed that NPM protein expression was slightly increased following Taxol treatment, but not to the same extent as pMEK i.r. (Figure 3A). This result suggests that the pMEK1/2 antibody might be detecting a post-translationally modified NPM that is preferentially induced during mitosis. To address this issue further, NPM from control and Taxol-treated cells was immunoprecipitated with an anti-NPM antibody, then immunoblotted with either an antibody raised against total NPM or the pMEK1/2 antibody. As measured with the total NPM antibody, there was no difference in NPM expression between control and Taxol-treated cells, indicating that the total level of NPM is not changed by Taxol. In contrast, NPM recognized by pMEK1/2 crossreactivity was greatly enhanced in Taxol-arrested cells. Coupled with the cellular redistribution of nucleophosmin observed in Taxol-treated cells (Figure 1), this suggests that the NPM protein is modified during mitosis. Nucleophosmin is a known target of several kinases [26-28]. Treatment with the broad-specificity λ phosphatase abolished the intensity of pMEK i.r, confirming that the observed modification is phosphorylation (Figure 3B). We pursued these observations by using gel filtration to examine the chromatographic properties of NPM at different points of the cell cycle. In cells incubated with DMSO, NPM was detected in both a large complex and in a monomeric state, while the pMEK i.r. is negligible or undetectable. In Taxol treated cells, monomeric NPM was not found. NPM still appeared in complexes of higher molecular weight, though the apparent weight of these complexes extends over a different range than that of control treated cells; some NPM appears in smaller complexes than those identified in control cells. Taxol treatment caused the appearance of the pMEK i.r., and its elution pattern coincides with the smaller NPM complexes unique to Taxol-treated cells (Figure 3C). These results suggest that upon Taxol treatment, a large fraction of cellular NPM is represented by a modified form of NPM identified by pMEK i.r.. This form is found in a protein complex caused by Taxol treatment, and is negligible or absent in control cells.
Figure 3.
The nucleophosmin recognized by pMEK1/2 antibody is phosphorylated and appears in a complex distinct in size from the interphase NPM complex. A) The intensity of Taxol-induced mitotic NPM revealed by an antibody to pMEK1/2 is greater than that revealed by an antibody to total NPM. In the top two panels, cells were treated with either DMSO or 100nM Taxol for 12 hours. Lysates were then collected and immunoblotted with antibodies to either total NPM or pMEK1/2. In the two lower panels, lysates were initially precipitated with anti-NPM, and then immunoblotted to visualize the pMEK i.r. Only pMEK i.r. and not total NPM was substantially enhanced by Taxol. B) Phosphatase treatment abolishes NPM immunoreactivity with the pMEK1/2 antibody. Lysates were generated as above then subjected to λ phosphatase treatment as outlined in Materials and Methods. C) Lysates from cells treated with either DMSO or 100nM Taxol for 16 hours were fractionated according to size using a Superose 6 column and immunoblotted to reveal total NPM and pMEK i.r. as indicated. Standards eluted at the following times: Thyroglobulin (670 kDa) 29 minutes, Ferratin (440 kDa) 32 minutes, Catalase (232 kDa) 35 minutes, LDH (140 kDa) 36 minutes, BSA (60 kDa) 36-37 minutes.
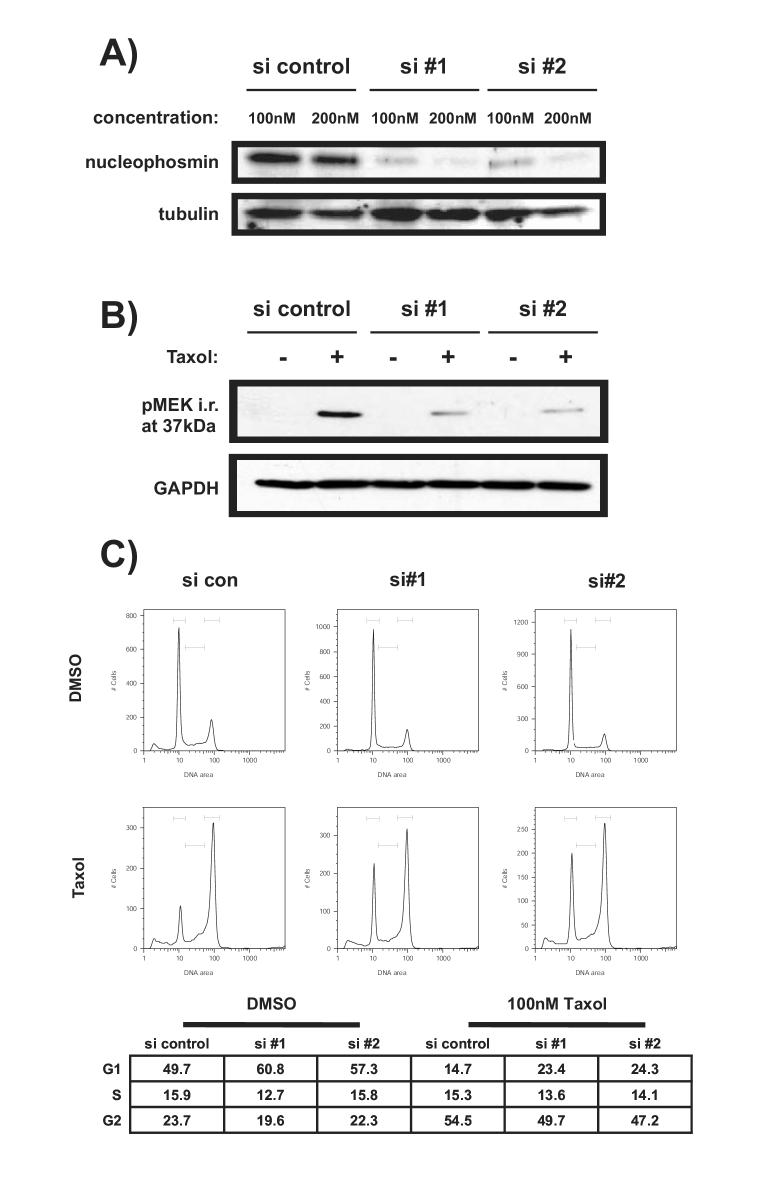
siRNA mediates knockdown of NPM but allows for mitotic arrest
Two different small interfering (si) RNA oligonucleotides were used to promote the degradation of NPM mRNA. Protein expression was severely diminished in the presence of siRNA oligonucleotides specific for NPM but not control siRNA 42 hours after transfection (Figure 4A). At a higher oligonucleotide concentration, expression was nearly abolished. However, the amount of transfection reagent alone required for this concentration of oligonucleotide proved cytotoxic after several hours of additional incubation (data not shown). Therefore, the lower concentration was used in all subsequent studies. This concentration of siRNA effectively diminished expression of the phosphorylated NPM recognized by anti-pMEK antibody following exposure to Taxol (Figure 4B). Cell cycle analysis was performed to determine the effect of NPM knockdown on H157 cells. Knockdown of NPM expression affected progression through the cell cycle by promoting accumulation in G1, though this effect was modest. Delayed progression did not prevent mitotic arrest by Taxol (Figure 4B).
Figure 4.
SiNPM-mediated knockdown of decreases NPM expression but allows for mitotic arrest by Taxol. A) NPM-specific siRNA oligonucleotides effectively diminish NPM protein expression. H157 cells were transfected with the indicated concentrations of control or specific oligonucleotide (NPM si #1 or si#2) or left untransfected. Two NPM specific siRNAs were used to assure specificity. Lysates were collected after 42 hours. Immunoblotting was performed with antibodies against NPM or tubulin. B) NPM specific siRNA oligonucleotides diminish expression of the modified NPM recognized by anti-pMEK antibody. H157 cells were transfected with 100nM siRNA. After 36 hours 100nM Taxol was added to the indicated samples. Lysates were collected after 48 hours and immunoblotted with either anti-pMEK1/2 or anti-GAPDH antibody. C) Knockdown of NPM does not prevent mitotic arrest. H157 cells were transfected as in A then treated with 100nM Taxol for 16 hours prior to DNA content analysis.
Nucleophosmin depletion alters the expression of genes associated with cell proliferation and cancer
To more globally address the function of NPM, microarray analysis was performed on RNA isolated from control (DMSO) or Taxol-treated cells in the presence or absence of siNPM oligonucleotides. The limit for change in expression was set to 1.5 fold. Complete lists of transcripts altered in NPM depleted cells in the absence or presence of Taxol, along with their associated expression patterns, are presented in Supplementary Tables 1 and 2 respectively. In some instances, multiple transcripts were identified for a single gene. To simplify presentation, only one representative transcript for each gene is shown. Extensive transcriptional profiling of cells treated with Taxol has been published elsewhere and will not be expounded on here [29].
The expression of 45 unique transcripts was altered in NPM-depleted cells (Figure 5A). Among these, the NPM transcript is reduced in NPM siRNA-containing cell lines as expected. Subsequently the Ingenuity Systems Pathway Analysis software was used to determine interaction networks in which the products of these transcripts participate. Two such networks were identified and merged to form a larger network (Supplementary Figure 1 and Figure 5B).
Certain genes within the pathway act as hubs, forming many connections. Among those hubs with altered mRNA expression in the presence of NPM siRNA are neuregulin, plasminogen-activator inhibitor (PAI-1), and NPM itself. Neuregulin, which is downregulated at the transcript level, is an EGF-like ligand for human epidermal growth factor receptors. Heavily impicated in cancer, it triggers pro-survival and pro-differentiation signaling in epithelia and is implicated in cytoskeletal rearrangement [30, 31]. It has also been shown to promote S phase entry from G1 [32]. This process may involve the product of another transcript altered by NPM siRNA, the transcription factor LEF-1 [33]. PAI-1 inhibits uPa, a protease that cleaves the zymogen plasminogen to form plasmin. Active plasmin is a serine protease that degrades the extracellular matrix (ECM) to allow for cell spreading and mobility. Dysregulation of the plasminogen activator system has been widely implicated in cancer metastasis [34].
To determine the functional importance of the merged network, the Ingenuity Systems software was employed to perform a literature-based functional analysis, revealing significant association with cell proliferation, cell cycle regulation, and cancer (Figure 5C). These results reinforce the proposition that NPM expression is linked to cancer progression and suggest that NPM may be a marker for NSCLC and potential candidate for therapeutic targeting. To address this possibility, we used real-time RT-PCR to examine the expression of NPM message in malignant NSCLC tumor tissue. For each sample, histologically nontransformed tissue adjacent to the primary tumor was isolated to provide a healthy matched control. Compared with adjacent matched control normal tissue, NPM is overexpressed in 5 of the 6 NSCLC cases analyzed, by an average of 5.4-fold (Figure 5D).
NPM depletion alters the expression of genes associated with DNA architecture and rDNA transcription following exposure to Taxol
To determine the consequence of NPM-depletion during mitosis, we examined those transcripts that exhibited altered expression in siNPM cells following Taxol-induced mitotic arrest. Ten genes demonstrated strong changes in expression following siNPM transfection in either the presence or absence of Taxol (Figure 6A - Top). Notable among them is NPM itself. Another of these transcripts, H30005705, is listed in the Operon database as a paralog of NPM located on chromosome 16. This gene has over 98.5% identity with NPM and has been previously identified as one of several NPM-related pseudogenes [35]. Because of the high degree of sequence identity, it is likely that we propose that siRNA to NPM mRNA also reduced any transcripts that arise from this pseudogene, and what appears to be a downregulation of this product is actually further confirmation of the targeted degradation of NPM mRNA. Strikingly, three of the four remaining genes in this list for which functional data is available play a role in regulating the architecture of DNA (Figure 6B - Top). These include the genes encoding the histone methylase SET8 and SET/TAF1β, a protein with several known functions. These findings may relate to an emerging role for nucleophosmin in regulating transcription [11].
According to our criteria, six additional transcripts demonstrated altered transcription in NPM-depleted cells only in the presence of Taxol but showed the same trend in unarrested cells. Five of these transcripts, including that for the growth suppressive gene H-REV107, were upregulated relative to treatment with Taxol alone. Three were downregulated (Figure 6A - Bottom).
Two additional transcripts showed expression patterns uniquely altered following Taxol treatment (Figure 6B). One of them is TAF1A, which encodes the ribosomal DNA transcription factor TAFI48. Based on our criteria, this gene was also identified as upregulated in the presence of Taxol alone. It is thus the only transcript identified which demonstrated an NPM-dependent induction following exposure to Taxol. As mitotic induction of TAF1A has not been previously observed, Taxol-induced NPM-dependent expression was subsequently verified by real time PCR (data not shown). Since exposure to Taxol may have cytological consequences outside of mitotic arrest, we used thymidine synchronization to determine the expression of TAF1A mRNA during the cell cycle (Figure 6D). Real time PCR analysis demonstrated that the expression of TAF1A, but not the functionally related transcripts TAF1B or TAF1C, increases during early G2/M. This is consistent with Taxol-mediated arrest, which occurs at the metaphase to anaphase transition. This identifies TAF1A as a potentially new biomarker for G2/M.
DISCUSSION
Nucleophosmin is a multifunctional protein with roles in a variety of cellular processes, perhaps most importantly in the regulation of cell cycle progression. NPM has long been studied for its participation in ribosome biosynthesis, a necessary step in cell proliferation. Additional evidence points to further roles for NPM in the cell cycle. NPM fusion proteins are common to acute myelogenous leukemia, a disease characterized by chromosome rearrangement, and NPM is also a known regulator of centrosome duplication, which is a crucial step in mitotic progression [20, 21].
We and others have shown that a cross-reacting antibody raised against phospho-MEK1/2 recognizes a modified form of NPM specific to mitosis [1, 2]. The phosphorylation of NPM, along with a change in its apparent molecular weight, suggests altered function for this protein during mitosis. To explore the significance of NPM during the cell cycle, we developed siRNA oligonucleotides to target NPM mRNA for degradation. Analysis of NPM in tumor cells is important because we also demonstrated enhanced NPM in primary lung tumors. We used microarray technology to explore the nature of delayed cell cycle progression in NPM-depleted cells. An unbiased knowledge-based approach was used to organize the data into a relevant framework, providing new insight into the regulatory consequences of NPM-depletion. Pathway analysis software identified an interaction network comprised of 60 gene products.
Based on an unbiased software-based analysis, the interaction network demonstrates a strong association with cancer. To further examine this connection, we measured the expression of NPM in transformed tissue. In this study, we present the first evidence that NPM is overexpressed in primary NSCLC. Our findings strengthen the links between NPM function and cell proliferation and highlight its importance to G1 progression. Therefore NPM might be considered as a potential anti-cancer target.
We next examined NPM knockdown cells that were incubated in the presence or absence of Taxol, a microtubule stabilizing agent that arrests cells in mitosis. NPM is implicated in the progression of mitosis, particularly with regard to regulating the division of centrosomes, and the consequences of NPM depletion on mitosis were expected to be severe. To explore these consequences, we first examined those transcripts that demonstrated altered expression in siNPM knockdown cells incubated with or without Taxol. As expected, NPM itself appears in this list. Intriguingly, three of the other genes in this list for which functional data is available, SET8, SET, and histone H2B, interact with DNA or chromatin. The transcriptional regulator silencer SET8, or PR-Set7, is a histone methyltransferase that specifically targets lysine 20 of histone H4. The activity of SET8 is primarily restricted to late S phase, G2, and mitosis, and it has been demonstrated to physically associate with mitotic chromosomes [36, 37]. Moreover, its silencing activity is a requirement for mitosis [38]. Its reduction by NPM siRNA and the reduction of NPM siRNA-containing cells in S/G2/M is consistent with this observation. The SETet/TafIβ protein was recently discovered to participate with pp32 in a complex that mediates transcriptional repression by inhibiting the activity of the histone-acetyltransferases [39]. SET is upregulated in NPM siRNA-containing cells, suggesting that NPM normally reduces the expression of SET to alleviate transcriptional repression. The Yin-Yang pattern of NPM-dependent SET and SET8 expression suggests that control of these two proteins by NPM may be coordinated, but in opposing directions. NPM depletion also affects chromatin at an even more fundamental level. Core histones, which comprise the nucleosomes around which DNA is wrapped, have recently been reported to bind NPM [11]. Included among the core histones is histone H2B, which is upregulated in siNPM cells. This raises the possibility that a feedback mechanism exists whereby the lack of NPM binding to histones promotes induction of H2B.
We also examined those genes that were altered by Taxol and also exhibited a NPM-dependent change. Only TAF1A exhibited this pattern. NPM siRNA most significantly reduced the Taxol-induction of this gene, indicating that mitotic TAF1A induction relies on intact NPM expression. The TAF1A gene encodes TATA-binding protein (TBP) Associated Factor 1A, also called TAFI48. TAFI48 is a component of the Selectivity Factor 1 (SL1) transcription initiation complex, and is required for the recruitment of the Upstream Binding Factor (UBF) to ribosomal DNA. Together, these proteins are responsible for initiating the transcription of ribosomal RNA by RNA Polymerase I in the nucleolus [40-42]. This finding is the first evidence that NPM, long considered an activator of ribosome maturation, might be involved in the transcription of rRNAs, and presents a remarkable and fundamental new function for NPM. It is also the first demonstration that TAF1A is upregulated by Taxol-induced arrest in mitosis. The latter finding is especially interesting given that transcription of rDNA is inhibited during this phase of the cell cycle by inactivating phosphorylation of UBF and one of the other two TAF components of SL1, TAFI110 [43-45]. Notably, neither TAF1B nor TAF1C, which encode the other two TAF components of SL1, were upregulated during mitosis. The transience of TAF1A upregulation suggests that TAFI48 may have an unrecognized function, perhaps related to the transition from early into late mitosis.
Together, these findings implicate NPM in a number of new and unexpected roles in transformed cells. Analysis of NPM depleted cells revealed that its absence affects proteins involved in chromosome architecture, including two remodeling proteins and the core histone H2B. NPM depletion also prevents mitotic transcription of the transcription factor TAFI48, which is required for the transcription of ribosomal DNA. NPM has been implicated in ribosomal maturation almost since its identification, but this is the first evidence linking it to the genesis of pre-rRNA by transcription. Altered regulation of proteins involved in cancer and cell cycle progression was observed in NPM-knockdown cells, and we have also shown overexpression of NPM in primary transformed tissue. Our findings not only identify NPM as a biomarker for NSCLC but raise the possibility that NPM is a candidate target for anticancer therapy.
Supplementary Material
Acknowledgements
The authors thank Evangeline Reynolds at the Lineberger Comprehensive Cancer Center Tumor Procurement Facility for providing primary lung carcinoma tissues. This work was supported by an NCI Breast Cancer SPORE program grant.
Footnotes
This work was supported by NIH Grant CA-58223.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cha H, Hancock C, Dangi S, Maiguel D, Carrier F, Shapiro P. Phosphorylation regulates nucleophosmin targeting to the centrosome during mitosis as detected by cross-reactive phosphorylation-specific MKK1/MKK2 antibodies. Biochem J. 2004;378:857–65. doi: 10.1042/BJ20031173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayne C, Xiang X, Luo Z. MEK inhibition and phosphorylation of serine 4 on B23 are two coincident events in mitosis. Biochem Biophys Res Commun. 2004;321:675–80. doi: 10.1016/j.bbrc.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Olson MO, Guetzow KA. Nucleolar phosphoprotein phosphatase from Novikoff hepatoma and rat liver: characterization and partial purification. Biochim Biophys Acta. 1978;526:174–85. doi: 10.1016/0005-2744(78)90302-9. [DOI] [PubMed] [Google Scholar]
- 4.Herrera JE, Savkur R, Olson MO. The ribonuclease activity of nucleolar protein B23. Nucleic Acids Res. 1995;23:3974–9. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savkur RS, Olson MO. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–15. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–90. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 7.Szebeni A, Olson MO. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–12. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529–33. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 9.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–96. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady SN, Yu Y, Maggi LB, Jr., Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–38. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swaminathan V, Kishore AH, Febitha KK, Kundu TK. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol Cell Biol. 2005;25:7534–45. doi: 10.1128/MCB.25.17.7534-7545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–53. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 13.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci PG, Martelli MF. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 14.Chan WY, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, Chan PK. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989;28:1033–9. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- 15.Kondo T, Minamino N, Nagamura-Inoue T, Matsumoto M, Taniguchi T, Tanaka N. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene. 1997;15:1275–81. doi: 10.1038/sj.onc.1201286. [DOI] [PubMed] [Google Scholar]
- 16.Kistler J, Duncombe Y, Laemmli UK. Mapping nucleolar proteins with monoclonal antibodies. J Cell Biol. 1984;99:1981–8. doi: 10.1083/jcb.99.6.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biggiogera M, Fakan S, Kaufmann SH, Black A, Shaper JH, Busch H. Simultaneous immunoelectron microscopic visualization of protein B23 and C23 distribution in the HeLa cell nucleolus. J Histochem Cytochem. 1989;37:1371–4. doi: 10.1177/37.9.2768807. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzacher HG, Wachtler F. The functional significance of nucleolar structures. Ann Genet. 1991;34:151–60. [PubMed] [Google Scholar]
- 19.Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol Biol Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda M. The role of nucleophosmin in centrosome duplication. Oncogene. 2002;21:6170–4. doi: 10.1038/sj.onc.1205708. [DOI] [PubMed] [Google Scholar]
- 21.Grisendi S, Pandolfi PP. NPM mutations in acute myelogenous leukemia. N Engl J Med. 2005;352:291–2. doi: 10.1056/NEJMe048337. [DOI] [PubMed] [Google Scholar]
- 22.Bergstralh DT, Ting JP. Microtubule stabilizing agents: Their molecular signaling consequences and the potential for enhancement by drug combination. Cancer Treat Rev. 2006 doi: 10.1016/j.ctrv.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Bergstralh DT, Taxman DJ, Chou TC, Danishefsky SJ, Ting JP. A comparison of signaling activities induced by Taxol and desoxyepothilone B. J Chemother. 2004;16:563–76. doi: 10.1179/joc.2004.16.6.563. [DOI] [PubMed] [Google Scholar]
- 24.Wong AW, Ghosh N, McKinnon KP, Reed W, Piskurich JF, Wright KL, Ting JP. Regulation and specificity of MHC2TA promoter usage in human primary T lymphocytes and cell line. J Immunol. 2002;169:3112–9. doi: 10.4049/jimmunol.169.6.3112. [DOI] [PubMed] [Google Scholar]
- 25.Harding A, Giles N, Burgess A, Hancock JF, Gabrielli BG. Mechanism of mitosis-specific activation of MEK1. J Biol Chem. 2003;278:16747–54. doi: 10.1074/jbc.M301015200. [DOI] [PubMed] [Google Scholar]
- 26.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–40. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 27.Szebeni A, Hingorani K, Negi S, Olson MO. Role of protein kinase CK2 phosphorylation in the molecular chaperone activity of nucleolar protein b23. J Biol Chem. 2003;278:9107–15. doi: 10.1074/jbc.M204411200. [DOI] [PubMed] [Google Scholar]
- 28.Jiang PS, Chang JH, Yung BY. Different kinases phosphorylate nucleophosmin/B23 at different sites during G(2) and M phases of the cell cycle. Cancer Lett. 2000;153:151–60. doi: 10.1016/s0304-3835(00)00362-1. [DOI] [PubMed] [Google Scholar]
- 29.Taxman DJ, MacKeigan JP, Clements C, Bergstralh DT, Ting JP. Transcriptional profiling of targets for combination therapy of lung carcinoma with paclitaxel and mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor. Cancer Res. 2003;63:5095–104. [PubMed] [Google Scholar]
- 30.Chausovsky A, Waterman H, Elbaum M, Yarden Y, Geiger B, Bershadsky AD. Molecular requirements for the effect of neuregulin on cell spreading, motility and colony organization. Oncogene. 2000;19:878–88. doi: 10.1038/sj.onc.1203410. [DOI] [PubMed] [Google Scholar]
- 31.Stove C, Bracke M. Roles for neuregulins in human cancer. Clin Exp Metastasis. 2004;21:665–84. doi: 10.1007/s10585-004-6917-6. [DOI] [PubMed] [Google Scholar]
- 32.Daly JM, Olayioye MA, Wong AM, Neve R, Lane HA, Maurer FG, Hynes NE. NDF/heregulin-induced cell cycle changes and apoptosis in breast tumour cells: role of PI3 kinase and p38 MAP kinase pathways. Oncogene. 1999;18:3440–51. doi: 10.1038/sj.onc.1202700. [DOI] [PubMed] [Google Scholar]
- 33.Graham NA, Asthagiri AR. Epidermal growth factor-mediated T-cell factor/lymphoid enhancer factor transcriptional activity is essential but not sufficient for cell cycle progression in nontransformed mammary epithelial cells. J Biol Chem. 2004;279:23517–24. doi: 10.1074/jbc.M314055200. [DOI] [PubMed] [Google Scholar]
- 34.Sheng S. The urokinase-type plasminogen activator system in prostate cancer metastasis. Cancer Metastasis Rev. 2001;20:287–96. doi: 10.1023/a:1015539612576. [DOI] [PubMed] [Google Scholar]
- 35.Liu QR, Chan PK. Characterization of seven processed pseudogenes of nucleophosmin/B23 in the human genome. DNA Cell Biol. 1993;12:149–56. doi: 10.1089/dna.1993.12.149. [DOI] [PubMed] [Google Scholar]
- 36.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–13. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 37.Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–30. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–5. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 40.Beckmann H, Chen JL, O’Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–9. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 41.Hannan KM, Hannan RD, Rothblum LI. Transcription by RNA polymerase I. Front Biosci. 1998;3:d376–98. doi: 10.2741/a282. [DOI] [PubMed] [Google Scholar]
- 42.Dynes JL, Xu S, Bothner S, Lahti JM, Hori RT. The carboxyl-terminus directs TAF(I)48 to the nucleus and nucleolus and associates with multiple nuclear import receptors. J Biochem (Tokyo) 2004;135:429–38. doi: 10.1093/jb/mvh051. [DOI] [PubMed] [Google Scholar]
- 43.Klein J, Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci U S A. 1999;96:6096–101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhn A, Vente A, Doree M, Grummt I. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J Mol Biol. 1998;284:1–5. doi: 10.1006/jmbi.1998.2164. [DOI] [PubMed] [Google Scholar]
- 45.Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. Embo J. 1998;17:7373–81. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Troppmair J, Bruder JT, Munoz H, Lloyd PA, Kyriakis J, Banerjee P, Avruch J, Rapp UR. Mitogen-activated protein kinase/extracellular signal-regulated protein kinase activation by oncogenes, serum, and 12-O-tetradecanoylphorbol-13-acetate requires Raf and is necessary for transformation. J Biol Chem. 1994;269:7030–5. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.