Abstract
Phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], a phosphoinositide concentrated predominantly in the plasma membrane, binds endocytic clathrin adaptors, many of their accessory factors, and a variety of actin-regulatory proteins. Here we have used fluorescent fusion proteins and total internal reflection fluorescence microscopy to investigate the effect of acute PI(4,5)P2 breakdown on the dynamics of endocytic clathrin-coated pit components and of the actin regulatory complex, Arp2/3. PI(4,5)P2 breakdown was achieved by the inducible recruitment to the plasma membrane of an inositol 5-phosphatase module through the rapamycin/FRB/FKBP system or by treatment with ionomycin. PI(4,5)P2 depletion resulted in a dramatic loss of clathrin puncta, which correlated with a massive dissociation of endocytic adaptors from the plasma membrane. Remaining clathrin spots at the cell surface had only weak fluorescence and were static over time. Dynamin and the p20 subunit of the Arp2/3 actin regulatory complex, which were concentrated at late-stage clathrin-coated pits and in lamellipodia, also dissociated from the plasma membrane, and these changes correlated with an arrest of motility at the cell edge. These findings demonstrate the critical importance of PI(4,5)P2 in clathrin coat dynamics and Arp2/3-dependent actin regulation.
Keywords: actin, dynamin, epsin, phosphoinositides, rapamycin
Phosphorylation–dephosphorylation of the inositol ring of inositol phospholipids plays a major role in cell regulation. Their phosphorylated head groups interact with a variety of cytosolic protein modules and thus mediate the recruitment and regulation of proteins at the membrane–cytosol interface (1–7). A direct interaction with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], a phosphoinositide concentrated at the plasma membrane, has been demonstrated for all of the endocytic clathrin adaptors and for several of their accessory factors, for the GTPase dynamin, and for many actin regulatory proteins (8–16). The physiological significance of these interactions has been supported by cell biological (9, 15, 17–21) and pharmacological (22) studies in living cells and by forward and reverse genetics. For example, mutations in genes encoding inositol 5-phosphatases, primarily synaptojanin family members, were shown to cause endocytic and actin defects in a variety of organisms (16, 23–28). However, genetic manipulations that result in long-lasting changes in cellular levels of PI(4,5)P2 may act indirectly or trigger compensatory mechanisms that can complicate the interpretation of results.
Conclusive evidence for the importance of PI(4,5)P2 in events that occur at the plasma membrane will come from experiments involving the acute and specific manipulation of its levels. Recently, a technique for the rapid, conditional recruitment of proteins to membranes has been used to deliver 5-phosphatase modules to the cell surface (20, 29–32). In this method, the genes encoding two proteins that can be cross-linked by the addition of rapamycin are fused in-frame to the 5-phosphatase module and to a plasma membrane-targeting sequence, respectively (31, 32). By using this approach, a massive dephosphorylation of PI(4,5)P2 in the plasma membrane upon the addition of rapamycin could be achieved (20, 29, 30). Notably, this breakdown of PI(4,5)P2, which depends on the catalytic activity of the 5-phosphatase module (30), correlated with a block of transferrin internalization, a clathrin-mediated endocytic reaction (20). We have now used this method in combination with the high spatial and temporal resolution of live total internal reflection fluorescence microscopy (TIRFM) to analyze the effect of acute PI(4,5)P2 depletion on the dynamics of clathrin coat proteins, dynamin, actin, and p20, a subunit of the Arp2/3 actin regulatory complex (33). Our findings demonstrate a requirement of PI(4,5)P2 both for the initiation and for the integrity of clathrin-coated pits (CCPs). They also prove the important role of PI(4,5)P2 in the nucleation of Arp2/3-dependent actin networks and thus in actin-dependent cell motility.
Results
Ionomycin-Induced Disruption of CCPs.
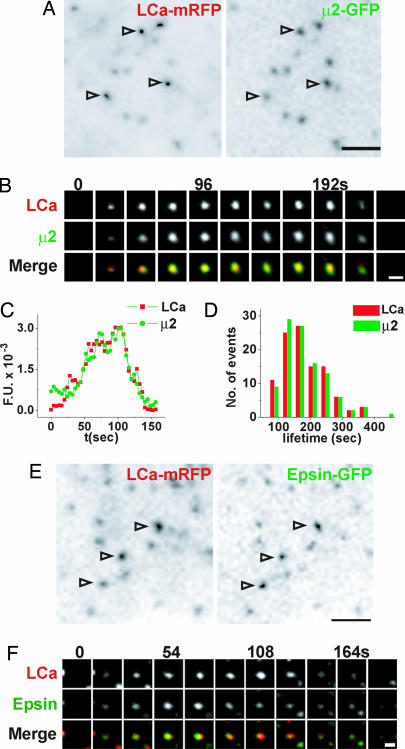
We first characterized the spatiotemporal dynamics of clathrin and clathrin adaptors in the COS-7 cells used for this work. Cells coexpressing clathrin light chain-mRFP (LCa-mRFP) and either GFP-tagged β2 or μ2 subunits of the clathrin adaptor AP2 complex (β2-GFP and μ2-GFP, respectively) were imaged by TIRFM. All three markers were found in diffraction-limited dynamic puncta with the characteristics of individual CCPs (34, 35) (Fig. 1 A and B and data not shown). They appeared together and had similar lifetimes: 174 ± 7 s for clathrin and 166 ± 7 s for μ2 (Fig. 1 B–D and data not shown). Similarly, when coexpressed, LCa-mRFP and epsin-GFP colocalized in space and time (Fig. 1 E and F), supporting the proposed role of epsin as a bona fide clathrin adaptor (36, 37).
Fig. 1.
Spatial and temporal analysis of clathrin, AP2, and epsin at CCPs. (A) TIRFM image of a COS-7 cell coexpressing LCa-mRFP (Left) and μ2-GFP (Right). Arrowheads point to individual pits. (B) Select frames from a time series of a single CCP. Merged images show colocalization of LCa-GFP and μ2-GFP throughout the vesicle lifetime. (C) Time course of LCa-mRFP (red) and μ2-GFP (green) fluorescence (fluorescence units, F.U.) at a single CCP. (D) Lifetime distribution of LCa-mRFP (red, n = 100) and μ2-GFP (green, n = 100) spots. (E) TIRFM image of a COS-7 cell coexpressing LCa-mRFP (Left) and epsin-GFP (Right). Arrowheads highlight CCPs. (F) Select frames from a time series of a single CCP. Merged images (Bottom) show colocalization of clathrin and epsin throughout the vesicle lifetime. [Scale bars, 2 μm (A and E); 1 μm (B and F).]
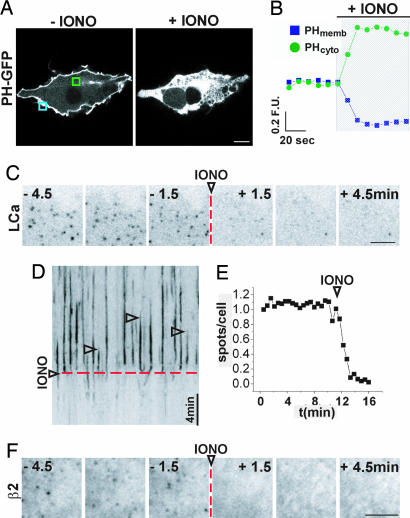
As an initial tool to manipulate membrane PI(4,5)P2 we used ionomycin, a Ca2+ ionophore that induces activation of Ca2+-dependent phospholipase C (PLC) and thus a breakdown of this phosphoinositide into diacylglycerol and inositol 3,4,5-trisphosphate (InsP3) (38). Addition of 5 μM ionomycin to COS-7 cells expressing the pleckstrin homology (PH) domain of PLCδ fused to GFP (PH-GFP), a PI(4,5)P2 reporter, produced a virtually complete redistribution of PH-GFP from the plasma membrane to the cytosol (Fig. 2 A and B; confocal microscopy) as expected (38). When the same treatment was applied to COS-7 cells expressing LCa-GFP and imaged with TIRFM, a dramatic loss of clathrin was observed [Fig. 2C and kymograph of Fig. 2D; see also supporting information (SI) Movie 1]. Automatic counting of thresholded time lapse images revealed a 90% decrease in the number of LCa-GFP spots (Fig. 2E). A similar effect was observed for β2-GFP puncta (Fig. 2F). Therefore, PI(4,5)P2 depletion by ionomycin results in a massive loss of clathrin coat components from the plasma membrane.
Fig. 2.
Disruption of CCPs by ionomycin treatment. (A) Spinning disk confocal image of a COS-7 cell expressing PH-GFP before (Left) and after (Right) addition of 5 μM ionomycin. (B) Normalized fluorescence/time plot for membrane-associated (PHmemb) and cytosolic (PHcyto) PH-GFP, measured in the regions boxed in A (blue and green, respectively) before and after (hatched region) ionomycin. (C) Frames from a detail of a COS-7 cell expressing LCa-GFP and imaged by TIRFM. The addition of ionomycin is indicated by the dashed red line. (D) Kymograph from the cell shown in C. Each vertical line (arrowheads) represents individual LCa-GFP spots. Note that they all disappear after ionomycin treatment (dashed red line). (E) Normalized number of LCa-GFP puncta over time for the cell shown in C and D automatically counted from thresholded images. (F) Frames from a detail of a COS-7 cell expressing β2-GFP and imaged by TIRFM. Dashed red line indicates the addition of ionomycin. [Scale bars, 10 μm (A); 5 μm (C and F).]
CCP Disruption by Acute PI(4,5)P2 Dephosphorylation.
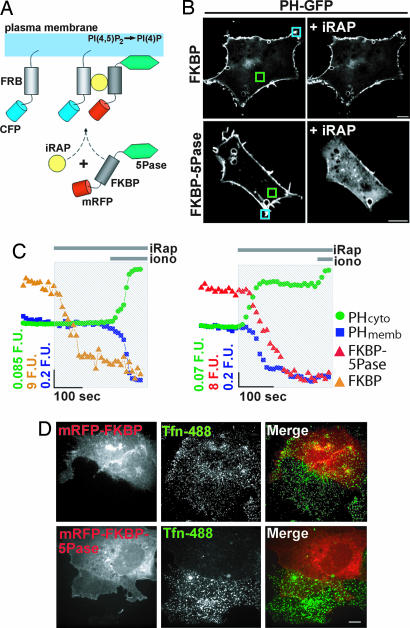
Ionomycin is a very effective tool for the depletion of PI(4,5)P2 from the plasma membrane (38). However, its action may also result from a rise in cytosolic InsP3, which could compete for PI(4,5)P2-binding sites (39), or from Ca2+-dependent effects independent of PLC activation. Thus, we used a more specific method to deplete PI(4,5)P2: the conditional recruitment of an inositol 5-phosphatase through its rapamycin-induced heterodimerization to a membrane-targeted, rapamycin-binding domain of mTOR (FRB) (Fig. 3A) (20, 29, 30, 40). COS-7 cells were triple-transfected with FRB fused to CFP (FRB-CFP), FKBP fused to mRFP and to the 5-phosphatase domain of type IV 5-phosphatase (mRFP-FKBP-5Pase), or to mRFP alone as a control (mRFP-FKBP), and PH-GFP. Addition of the indole-modified analog of rapamycin, iRAP (40), induced a rapid plasma membrane recruitment of mRFP-FKBP-5Pase and a resulting relocation of PH-GFP from the plasma membrane to the cytosol (Fig. 3B Lower; see also Fig. 4A) within 60 s as described (20), confirming PI(4,5)P2 depletion [Fig. 3C Right; PHcyto (green), PHmemb (blue)]. In contrast, PH-GFP did not detach from the plasma membrane upon the addition of iRAP in cells expressing mRFP-FKBP instead of mRFP-FKBP-5Pase (Fig. 3B Upper and 3C Left). Subsequent addition of ionomycin to cells expressing mRFP-FKBP-5Pase caused only a modest additional change in the distribution of PH-GFP (Fig. 3C Right), possibly reflecting a more complete degradation of PI(4,5)P2 or additional effects of this drug not dependent on PI(4,5)P2 hydrolysis (see Discussion). iRAP-mediated translocation of mRFP-FKBP-5Pase to the cell surface correlated with a block of transferrin-488 (Tfn-488) internalization, as reported previously (ref. 20 and Fig. 3D), consistent with an essential role of PI(4,5)P2 in the assembly and/or function of the clathrin-dependent endocytic machinery.
Fig. 3.
Plasma membrane PI(4,5)P2 depletion and block of transferrin internalization produced by iRAP-induced 5-phosphatase translocation. (A) Schematics of the experimental protocol (20, 30, 40). The addition of iRAP induces the heterodimerization of mRFP-FKBP-5Pase to plasma membrane-targeted FRB-CFP, leading to PI(4,5)P2 dephosphorylation. (B) Distribution of PH-GFP in COS-7 cells coexpressing mRFP-FKBP-5Pase (Lower) or the same construct lacking the 5Pase module, mRFP-FKBP (Upper), before and after the addition of 5 μM iRAP. (C) Normalized fluorescence/time plots of membrane and cytosolic PH-GFP from boxed regions shown in B (blue and green, respectively). Orange (Left) and red (Right) plots show a decrease in cytosolic mRFP-FKBP and mRFP-FKBP-5Pase fluorescence, respectively, upon iRAP addition. (D) COS-7 cells expressing mRFP-FKBP (Upper) or mRFP-FKBP-5Pase (Lower) were incubated sequentially with 5 μM iRAP and then with Tfn-488. The images show uptake of Tfn-488 in cells expressing mRFP-FKBP but not in those expressing mRFP-FKBP-5Pase. [Scale bars, 10 μm (B and D).]
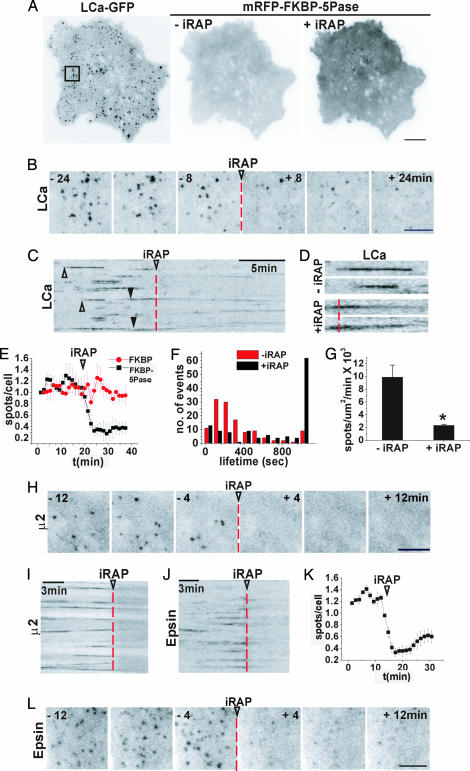
Fig. 4.
Effect of iRAP-induced acute PI(4,5)P2 dephosphorylation on CCP dynamics. (A) TIRFM image of a COS-7 cell expressing LCa-GFP (Left) before the addition of iRAP, and mRFP-FKBP-5Pase (Center and Right) before and after the addition of iRAP, respectively. (Right) mRFP-FKBP-5Pase recruitment is revealed by the increase in diffuse mRFP plasma membrane fluorescence. (B and C) Select frames and kymograph from a time series of the LCa-GFP channel from the cell shown in A before and after the addition of iRAP (arrowheads and dashed lines), illustrating the iRAP-induced loss of clathrin. (C) Note that clathrin spots appear and disappear abruptly before the addition of iRAP (open arrowheads) but dim gradually after iRAP (filled arrowheads). (D) High-power view of the LCa-GFP spots indicated by arrowheads in C. (E) Normalized and averaged number of LCa-GFP spots over time from three cells expressing mRFP-FKBP-5Pase (black) or mRFP-FKBP (red). (F) Lifetime distribution of 240 LCa-GFP puncta from three cells before (red) and after (black) iRAP delivery. (G) Number of newly formed LCa-GFP spots per μm2/min before and after iRAP. Values are averages from three cells; P < 0.017 (Student's t test). (H and I) Select frames and kymograph from a time series of a cell expressing μ2-GFP before and after iRAP. (J–L) Kymograph, number of spots, and select frames from a time series of an epsin-GFP-expressing cell before and after iRAP. [Scale bars, 10 μm (A); 5 μm (B, H, and L).]
The action of iRAP on the formation and dynamics of GFP-tagged, endocytic clathrin coats was then analyzed by TIRFM. Before iRAP, LCa-GFP had the typical dynamic punctate appearance (Fig. 4A Left). Upon iRAP-mediated recruitment of mRFP-FKBP-5Pase to the plasma membrane (Fig. 4A Right), but not upon the recruitment of mRFP-FKBP, major changes in LCa-GFP dynamics were observed. Within 5 min, ≈70% of the LCa-GFP spots disappeared (Fig. 4 B, C, and E and SI Movie 2). The remaining spots became weaker rather than undergoing abrupt disappearance as in CCP scission (Fig. 4 C and D) and were predominantly stable: 49.6% failed to turn over in a 20-min period, whereas in control cells during this time interval only 6.4% of the CCPs were static (Fig. 4F). Importantly, PI(4,5)P2 depletion decreased the rate of formation of new LCa-GFP spots, from 9.9 ± 1.9 ×10−3 pits per μm2/min to 2.3 ± 0.2 × 10−3 pits per μm2/min (Fig. 4G; P < 0.017). Moreover, the new spots never reached the fluorescence intensity of dynamic CCPs observed before the addition of iRAP. In contrast to its striking effect on CCPs at the cell surface, iRAP-mediated PI(4,5)P2 depletion did not appear to affect the distribution and dynamics of Golgi-derived clathrin structures (visible upon an increase of TIRFM depth; SI Movie 3), in agreement with selective targeting of the 5-phosphatase to the plasma membrane, but also with the proposed role of PI(4)P rather than PI(4,5)P2 in their assembly (2, 41). Some of these structures appeared as weakly fluorescent and highly mobile spots in the shallow TIRFM plane typically used (SI Movie 2).
Even more dramatic effects were observed for endocytic clathrin adaptors upon the addition of iRAP. More than 80% of all β2-GFP, μ2-GFP, and epsin-GFP spots disappeared (Fig. 4 H–L and SI Movie 4). Because the attachment of clathrin to the plasma membrane at CCPs is mediated by the adaptors, the persistence of weak, stable clathrin-positive spots may reflect, at least in part, small empty clathrin cages. In addition, Golgi-derived mobile clathrin structures at the edge of the TIRFM field may cause the action of iRAP on LCa-GFP spots at the plasma membrane to be underestimated in still images.
Dissociation of Dynamin and the Arp2/3 Complex from the Plasma Membrane.
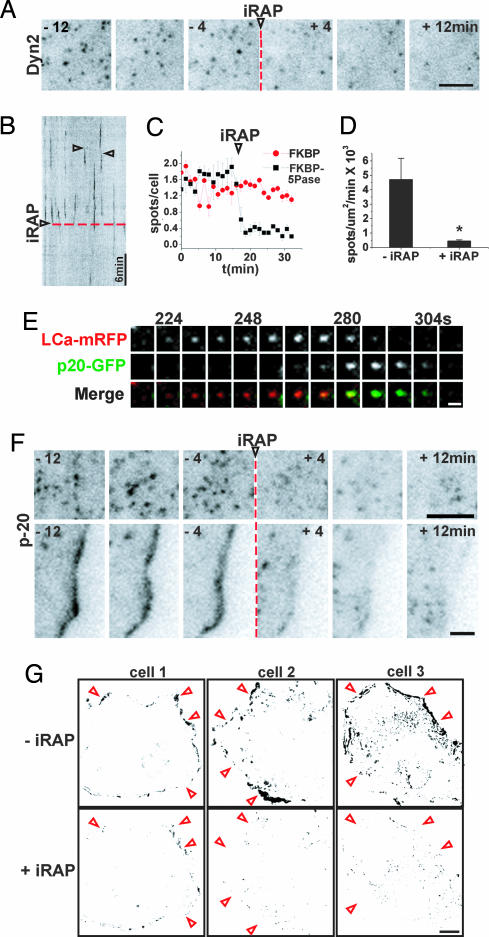
Scission of clathrin-coated vesicles requires the GTPase dynamin, which contains a PI(4,5)P2-binding PH domain (42–44). Furthermore, strong evidence indicates that the action of dynamin is tightly linked to the actin cytoskeleton, more specifically to N-WASP and thus to Arp2/3 mediated actin nucleation (45–51). N-WASP, as well as many factors that act upstream of N-WASP and Arp2/3 bind PI(4,5)P2. Some of them also bind its downstream product phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] (3, 52–54). Thus, we used TIRFM to investigate whether mRFP-FKBP-5Pase-dependent PI(4,5)P2 dephosphorylation has an effect on the membrane association of dynamin and Arp2/3-containing actin meshworks. Dynamin 2-GFP (Dyn2-GFP) had the characteristic (34, 46) punctate pattern in control cells and in cells expressing mRFP-FKBP-5Pase before the addition of iRAP (Fig. 5A). This pattern overlapped in part with the distribution of CCPs (18, 34, 46). Within 5 min after the addition of iRAP, ≈80% of total Dyn2-GFP spots disappeared from the plasma membrane (Fig. 5 A–C and SI Movie 5). Moreover, the rate of appearance of new dynamin spots dropped from 4.7 ± 1.5 × 10−3 spots per μm2/min to 0.46 ± 0.75 × 10−3 spots per μm2/min (Fig. 5D; P = 0.041).
Fig. 5.
Effect of PI(4,5)P2 depletion on dynamin and Arp2/3. (A and B) Select frames and kymograph from a TIRFM time series of a COS-7 cell expressing Dyn2-GFP and mRFP-FKBP-5Pase (channel not shown). (A–C and F) An arrowhead and a red dashed line mark iRAP delivery. (B) Examples of dynamic Dyn2-GFP spots are indicated by open arrowheads. (C) Number of Dyn2-GFP spots over time, before and after iRAP delivery, normalized and averaged from three cells expressing mRFP-FKBP (red) or mRFP-FKBP-5Pase (black). (D) Number of newly formed Dyn2-GFP spots per μm2/min before and after iRAP. Values are averages from three cells; P = 0.041 (Student's t test). (E) Select frames from a time series of a single CCP containing LCa-mRFP and p20-GFP. Recruitment of Arp2/3 (p20-GFP fluorescence) occurs at a late stage in CCP formation. (F) Select frames from a series of a p20-GFP-expressing cell showing iRAP-induced disappearance of p20-GFP fluorescence both from spots at the ventral surface of the cell (Upper) and from a lamellipodium (Lower). (G) Analysis of motility in three cells expressing actin-GFP and mRFP-FKBP-5Pase (data not shown) and imaged for 10 min before (Upper) and after (Lower) iRAP delivery. Darker pixels indicate areas of higher motility (see Methods). The profiles of the cells are roughly indicated by red arrowheads. [Scale bars, 10 μm (G); 5 μm (A and F); 1 μm (E).]
Similar observations were made with fluorescently tagged p20, a subunit of the Arp2/3 complex (p20-GFP). This protein also had a punctate pattern with 71.5% of puncta (n = 109) overlapping with CCPs and appearing during late stages of pit formation, consistent with the presence of actin at the neck of endocytic vesicles during scission (46, 47) (Fig. 5 E and F; see also ref. 55). In addition, p20-GFP was also concentrated at the cell edge in lamellipodia (Fig. 5F Lower) (56), where a pool of dynamin was also localized (SI Movie 6), in agreement with previous studies (45, 48, 57). iRAP caused a dramatic disappearance of p20-GFP puncta associated with CCPs (Fig. 5F Upper). It also caused a loss of both Dyn2-GFP (SI Movie 6) and p20-GFP from the cell edge (Fig. 5F Lower). These changes correlated with a collapse of lamellipodia and with an arrest of peripheral cell dynamics, as illustrated by a semiquantitative analysis of motility in actin-GFP-expressing cells (Fig. 5G and SI Movies 6 and 7). Collectively, these results indicate that the scission machinery of clathrin coated vesicles is disrupted by depletion of PI(4,5)P2. In addition, they provide new evidence for the “master” role of PI(4,5)P2 in Arp2/3-mediated actin nucleation and actin motility at the cell periphery.
Discussion
We have used a direct approach to test the hypothesis that PI(4,5)P2 present in the plasma membrane plays a critical role in the dynamics of CCPs and in the control of Arp2/3-containing actin meshworks. Rapid and massive PI(4,5)P2 dephosphorylation at the 5 position of the inositol ring was achieved by the iRAP-mediated recruitment to the plasma membrane of a 5-phosphatase module (20, 29, 30). As the catalytic domain of type IV phosphatase also dephosphorylates PI(3,4,5)P3 (58, 59) and because loss of PI(4,5)P2 would prevent compensatory PI(3,4,5)P3 production by phosphatidylinositol 3-kinases, a contribution by the parallel depletion of PI(3,4,5)P3 to the effects observed cannot be excluded (see also ref. 29). However, PI(3,4,5)P3 is only present at negligible levels in resting cells (60).
PI(4,5)P2 depletion led to a massive disappearance of CCPs not compensated for by the appearance of new ones, confirming the importance of PI(4,5)P2 for coat nucleation, growth, and stability. Disappearance could be explained by normal coated pit scission only at the earliest time points after the addition of iRAP because dynamin also dissociated from the plasma membrane. In addition, loss of clathrin spots seemed to result from the rapid but nonquantal loss of fluorescence from the pits, suggesting disassembly and/or fragmentation of the coats.
Ionomycin induced effects on CCPs similar to and even stronger than those generated by recruitment to the cell surface of the 5-phosphatase. Ionomycin, a Ca2+ ionophore, induces PI(4,5)P2 depletion by the Ca2+-dependent activation of PLC (38, 61), which cleaves PI(4,5)P2 but also acts on PI4P (62), thus depleting the main PI(4,5)P2 precursor. PI(4,5)P2 resynthesis after PLC cleavage requires de novo synthesis of phophatidylinositol, whereas PI(4)P generated by the action of a 5-phosphatase can be rapidly rephosphorylated to PI(4,5)P2 by PI(4)P 5-kinases. Thus, ionomycin may induce a complete depletion of PI(4,5)P2, whereas the iRAP method may allow the persistence of a very low steady-state level of this phosphoinositide. Additionally, ionomycin may affect clathrin nucleation and dynamics indirectly via the generation of InsP3 and other Ca2+-dependent effects, such as shielding of negative charges of the bilayer or changes in protein phosphorylation (63).
A rapid reversible loss of endocytic CCPs from the plasma membrane was recently observed upon addition of butanol (22), an alcohol shown to produce a depletion of PI(4,5)P2 by interfering with activation of PI(4)P 5-kinases (64, 65). However, because the action of butanol on PI(4,5)P2 levels is only indirect, effects independent of PI(4,5)P2 depletion cannot be excluded.
In summary, our data conclusively prove that plasma membrane PI(4,5)P2 has an essential role in endocytic clathrin coat assembly and budding. They also provide direct evidence for an essential role of PI(4,5)P2 in actin dynamics at the cell periphery.
Methods
Plasmids, Oligonucleotides, and Other Reagents.
The following reagents were gifts: LCa-GFP (from James Keen, Thomas Jefferson University, Philadelphia, PA), Dyn2-GFP (Mark McNiven, Mayo Clinic, Rochester, MN), β2-GFP (Pier Paolo Di Fiore, University of Milan, Italy), μ2-GFP (Alexander Sorkin, University of Colorado, Denver, CO), and p20-GFP (Jurgen Wehlands, German Research Centre for Biotechnology, Braunschweig, Germany). Epsin-GFP has been described previously (36). LCa was subcloned into a vector encoding monomeric RFP (Roger Tsien, University of California at San Diego, La Jolla, CA) to produce LCa-mRFP. Alexa 488-conjugated human transferrin was from Invitrogen (Carlsbad, CA). PM-FRB-CFP, mRFP-FKBP, and mRFP-FKBP-5Pase were described previously (20). iRAP was provided by Thomas Wandless and Gerald Crabtree (Stanford University, Palo Alto, CA).
Live Cell Imaging and Image Analysis.
TIRFM was performed as described previously (18). In some experiments, images were also acquired by using a spinning-disk confocal system (PerkinElmer, Waltham, MA), mounted onto an IX71 inverted microscope (Olympus, Center Valley, PA), equipped with a 1 kb × 1 kb National Institute on Drug Abuse EMCCD camera (Hamamatsu, Hamamatsu City, Japan). Samples were imaged by using a ×100 oil objective, yielding a spatial resolution of 0.1 μm per pixel. Excitation was achieved by using 488 argon and 568 argon/krypton lasers (Melles Griot, Carlsbad, CA). Exposure times were between 0.2 and 0.6 s. Analysis of TIRFM images was performed as described previously (ref. 18; see also SI Methods). The lifetime of fluorescently tagged proteins was measured manually from 120 fluorescent spots in three cells. A particle-counting algorithm in ImageJ (National Institutes of Health) was used to measure the number of spots over time from binarized images. A custom-written algorithm was used to determine the number of newly generated spots per μm2/min (see SI Methods). Analysis of membrane motility before and after iRAP was performed with Andor iQ (see SI Methods).
Miscellaneous Procedures.
Cell culture and transfection were performed as described previously (18). For transferrin internalization assays, COS-7 cells previously cotransfected with FRB-CFP and either mRFP-FKBP or mRFP-FKBP-5Pase were incubated with 5 μM iRAP (5 min at 37°C), exposed to Tfn-488 in imaging buffer (5 min at 37°C), then washed, fixed in 4% paraformaldehyde, and imaged by spinning disk confocal microscopy.
Supplementary Material
Acknowledgments
We thank Olympus USA for support and assistance to the Yale CINEMA laboratory. This work was supported by National Institutes of Health (NIH) Grants NS36251 and CA46128; grants from The G. Harold and Leila Y. Mathers Charitable Foundation and the Kavli Foundation, Yale/National Institute on Drug Abuse Neuroproteomic Center Grant DA018343, and Yale Diabetes and Endocrinology Research Center Grant DK45735 (to P.V.D.C.); Human Frontier Science Program Young Investigator Award RGY40/2003, a Bayer Pharmaceuticals Scholar Award, and a Ludwig Institute for Cancer Research grant (to D.T.); a Boehringer Ingelheim Fonds Ph.D. Scholarship (to R.Z.); and a JSPS Postdoctoral Fellowship for Research Abroad (to F.N.). T.B. was supported by the Intramural Research Program of the National Institute of Child Health and Human Development at NIH.
Abbreviations
- CCP
clathrin-coated pit
- Dyn2
dynamin 2
- InsP3
inositol trisphosphate
- iRAP
indole-modified analog of rapamycin
- LCa
clathrin light chain
- mRFP
monomeric red fluorescent protein
- 5Pase
5-phosphatase
- PH
pleckstrin homology
- PI(4)P
phosphatidylinositol 4-phosphate
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PI(3,4,5)P3
phosphatidylinositol 3,4,5-trisphosphate
- PLC
phospholipase C
- TIRFM
total internal reflection fluorescence microscopy.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611733104/DC1.
References
- 1.Balla T. J Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 2.De Matteis MA, Di Campli A, Godi A. Biochim Biophys Acta. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 4.Lemmon MA. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 5.Martin TF. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 6.Suh BC, Hille B. Curr Opin Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Yin HL, Janmey PA. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- 8.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 9.Gaidarov I, Keen JH. J Cell Biol. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao W, Tan Z, Prasad K, Reddy KK, Chen J, Prestwich GD, Falck JR, Shears SB, Lafer EM. J Biol Chem. 1997;272:6393–6398. doi: 10.1074/jbc.272.10.6393. [DOI] [PubMed] [Google Scholar]
- 11.Honing S, Ricotta D, Krauss M, Spate K, Spolaore B, Motley A, Robinson M, Robinson C, Haucke V, Owen DJ. Mol Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 12.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 13.Miki H, Miura K, Takenawa T. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 14.Traub LM. J Cell Biol. 2003;163:203–208. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V. J Cell Biol. 2003;162:113–124. doi: 10.1083/jcb.200301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremona O, Di Paolo G, Wenk MR, Luthi A, Kim WT, Takei K, Daniell L, Nemoto Y, Shears SB, Flavell RA, et al. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Kim H, Chang B, Ahn N, Hwang S, Di Paolo G, Chang S. FASEB J. 2006;20:2399–2401. doi: 10.1096/fj.05-4621fje. [DOI] [PubMed] [Google Scholar]
- 18.Perera RM, Zoncu R, Lucast L, De Camilli P, Toomre D. Proc Natl Acad Sci USA. 2006;103:19332–19337. doi: 10.1073/pnas.0609795104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohde G, Wenzel D, Haucke V. J Cell Biol. 2002;158:209–214. doi: 10.1083/jcb.200203103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varnai P, Thyagarajan B, Rohacs T, Balla T. J Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padron D, Wang YJ, Yamamoto M, Yin H, Roth MG. J Cell Biol. 2003;162:693–701. doi: 10.1083/jcb.200302051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucrot E, Saffarian S, Massol R, Kirchhausen T, Ehrlich M. Exp Cell Res. 2006;312:4036–4048. doi: 10.1016/j.yexcr.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuske KR, Richmond JE, Matthies DS, Davis WS, Runz S, Rube DA, van der Bliek AM, Jorgensen EM. Neuron. 2003;40:749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- 24.Singer-Kruger B, Nemoto Y, Daniell L, Ferro-Novick S, De Camilli P. J Cell Sci. 1998;111:3347–3356. doi: 10.1242/jcs.111.22.3347. [DOI] [PubMed] [Google Scholar]
- 25.Suchy SF, Nussbaum RL. Am J Hum Genet. 2002;71:1420–1427. doi: 10.1086/344517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 27.Di Paolo G, Moskowitz HS, Gipson K, Wenk MR, Voronov S, Obayashi M, Flavell R, Fitzsimonds RM, Ryan TA, De Camilli P. Nature. 2004;431:415–422. doi: 10.1038/nature02896. [DOI] [PubMed] [Google Scholar]
- 28.Dickman DK, Horne JA, Meinertzhagen IA, Schwarz TL. Cell. 2005;123:521–533. doi: 10.1016/j.cell.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 29.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suh BC, Inoue T, Meyer T, Hille B. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellano F, Montcourrier P, Guillemot JC, Gouin E, Machesky L, Cossart P, Chavrier P. Curr Biol. 1999;9:351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- 32.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 33.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 34.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Gaidarov I, Santini F, Warren RA, Keen JH. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 37.Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Dev Cell. 2006;10:329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Varnai P, Balla T. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirose K, Kadowaki S, Tanabe M, Takeshima H, Iino M. Science. 1999;284:1527–1530. doi: 10.1126/science.284.5419.1527. [DOI] [PubMed] [Google Scholar]
- 40.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Proc Natl Acad Sci USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roux A, Uyhazi K, Frost A, De Camilli P. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 43.Sever S, Damke H, Schmid SL. Traffic. 2000;1:385–392. doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 45.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Merrifield CJ, Feldman ME, Wan L, Almers W. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 47.Merrifield CJ, Perrais D, Zenisek D. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 48.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 49.Qualmann B, Kessels MM, Kelly RB. J Cell Biol. 2000;150:F111–F116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarar D, Waterman-Storer CM, Schmid SL. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salazar MA, Kwiatkowski AV, Pellegrini L, Cestra G, Butler MH, Rossman KL, Serna DM, Sondek J, Gertler FB, De Camilli P. J Biol Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- 52.Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 53.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 54.Takenawa T, Suetsugu S. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 55.Merrifield CJ, Qualmann B, Kessels MM, Almers W. Eur J Cell Biol. 2004;83:13–18. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- 56.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. J Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asano T, Mochizuki Y, Matsumoto K, Takenawa T, Endo T. Biochem Biophys Res Commun. 1999;261:188–195. doi: 10.1006/bbrc.1999.0998. [DOI] [PubMed] [Google Scholar]
- 59.Kisseleva MV, Wilson MP, Majerus PW. J Biol Chem. 2000;275:20110–20116. doi: 10.1074/jbc.M910119199. [DOI] [PubMed] [Google Scholar]
- 60.Toker A, Cantley LC. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 61.Oancea E, Teruel MN, Quest AF, Meyer T. J Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 64.Chen YG, Siddhanta A, Austin CD, Hammond SM, Sung TC, Frohman MA, Morris AJ, Shields D. J Cell Biol. 1997;138:495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris AJ, Frohman MA, Engebrecht J. Anal Biochem. 1997;252:1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.