Abstract
Gastrointestinal stromal tumor (GIST) has emerged as a clinically distinct type of sarcoma with frequent overexpression and mutation of the c-Kit oncogene and a favorable response to imatinib mesylate [also known as STI571 (Gleevec)] therapy. However, a significant diagnostic challenge remains in the differentiation of GIST from leiomyosarcomas (LMSs). To improve on the diagnostic evaluation and to complement the immunohistochemical evaluation of these tumors, we performed a whole-genome gene expression study on 68 well characterized tumor samples. Using bioinformatic approaches, we devised a two-gene relative expression classifier that distinguishes between GIST and LMS with an accuracy of 99.3% on the microarray samples and an estimated accuracy of 97.8% on future cases. We validated this classifier by using RT-PCR on 20 samples in the microarray study and on an additional 19 independent samples, with 100% accuracy. Thus, our two-gene relative expression classifier is a highly accurate diagnostic method to distinguish between GIST and LMS and has the potential to be rapidly implemented in a clinical setting. The success of this classifier is likely due to two general traits, namely that the classifier is independent of data normalization and that it uses as simple an approach as possible to achieve this independence to avoid overfitting. We expect that the use of simple marker pairs that exhibit these traits will be of significant clinical use in a variety of contexts.
Keywords: cancer, classification, diagnostic, machine learning, molecular signature
Gastrointestinal stromal tumors (GISTs) and leiomyosarcomas (LMSs) are common mesenchymal tumors with remarkably similar phenotypic features (1, 2). Until recently, the differentiation between these two entities had not been thought to be clinically relevant. Chemotherapeutic agents, such as doxorubicin and ifosfamide used in the treatment of soft-tissue sarcomas have resulted in response rates of 0–10% in patients with advanced GIST (3–5). However, the use of the selective tyrosine kinase inhibitor imatinib mesylate [also known as STI571 (Gleevec; Novartis Pharmaceuticals Corp., East Hanover, NJ)] has resulted in response rates of >50% for patients with GIST (6, 7, **). Conversely, patients with advanced LMS expect response rates of 27–53% when treated with doxorubicin or newer regimens combining gemcitabine with docetaxel (8, 9) but do not benefit from imatinib therapy (10, 11, ††). Thus, there is clear clinical importance in distinguishing between these two entities to guide the most effective therapy. Currently, the best marker to differentiate GIST from LMS is Kit immunostaining, which is subjective and variable due to cellular heterogeneity that may result in false-negative diagnoses. Kit-negative GISTs and Kit-expressing LMS have been reported on the basis of tumor cell morphology and other markers such as CD34, desmin, and smooth muscle actin (‡‡). The occurrence of Kit-negative GIST in the literature is ≈4–10% (2, 12).
We used whole human genome microarray data of 68 well characterized GIST and LMS samples to identify a simple gene expression classifier that would differentiate these tumor types with high accuracy. We chose to use a supervised top scoring pair (TSP) analysis (13, 14), which finds pairs where the relative expression of a gene pair is reversed between the two cancers. This method is advantageous because it provides the simplest possible classifier that is independent of data normalization, helps to avoid overfitting, and results in a very simple experimental test that is easy to implement in the clinic. We identified a single gene set (OBSCN and C9orf65) that accurately classified GIST from LMS with an estimated accuracy by using leave-one-out cross-validation of 97.8% on future cases on the basis of the microarray data and of 19 of 19 additional cases diagnosed correctly using RT-PCR. We conclude that this two-gene set provides a rapid, PCR-based assay that reliably distinguishes GIST from LMS and has the potential to aid in diagnosis and in the selection of appropriate therapies. The use of marker pairs based on relative expression reversals that are independent of data normalization holds tremendous promise as a method for the development of clinically relevant biomarkers.
Results
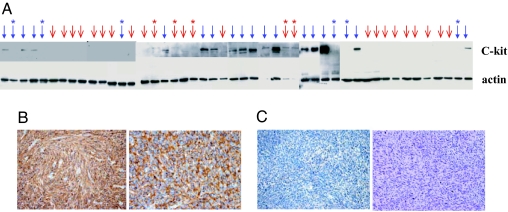
We selected 22 cases of GIST and 25 cases of LMS and isolated total proteins from the tumor tissues and measured Kit protein expression with a Western blotting assay. Only 16 of 22 GIST cases had detectable levels of Kit protein (Fig. 1A). In contrast, 5 of 25 LMS cases had (weak) Kit expression. An immunohistochemistry staining assay showed that, in low Kit-expressing tissues, staining of c-Kit protein was weak and heterogeneous (Fig. 1B). For these cases, immunostaining for other markers such as CD34, desmin, and smooth muscle actin had to be performed for pathological determination, significantly increasing the time and effort required for accurate diagnosis. Thus, Kit expression as a marker for GIST is useful, but not always adequate.
Fig. 1.
Kit protein expression in GIST and LMS. (A) The expression of Kit detected by Western blotting for 47 tumor samples (22 GIST and 25 LMS). Blue arrows indicate samples diagnosed as GIST, and red arrows indicate LMS. Each GIST sample for which no Kit protein was observed and each LMS sample that (weakly) expressed Kit are marked with a ∗. (B) Immunohistochemistry staining of two Kit-positive GIST samples. (C) Immunohistochemistry staining of two Kit-negative GIST samples.
Identification of a Two-Gene Classifier to Distinguish GIST from LMS.
The goal of this study was to identify accurate and simple diagnostic markers that differentiate GIST from LMS through genomic profiling and comparison. This study was composed of two major steps. The first step was to use microarray data from 68 tumor samples to find a potentially simple gene expression classifier to distinguish GIST from LMS with a high degree of accuracy. The second major step was to test this classifier by using RT-PCR on a new set of correctly diagnosed tumor samples to verify a simple yet accurate gene expression-based test for diagnosis.
Microarray data (68 tissue samples and 43,931 transcripts) were used to find a classifier to distinguish GIST from LMS with the primary goal of identifying a simple pattern that has a high degree of likelihood of performing well on future cases. The method we used for supervised classification is the TSP approach (13, 14). This approach has been shown to provide comparable accuracies to support vector machines and other sophisticated methods, as well as to provide a very robust gene-expression marker for prostate cancer across multiple array platforms (15).
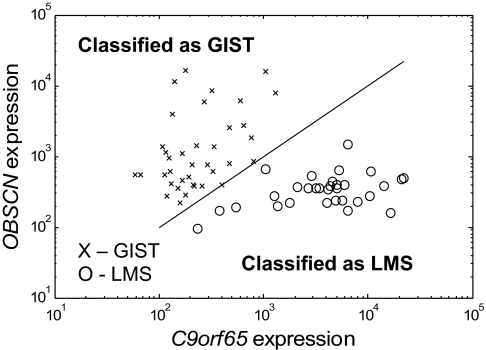
Training a classifier on our 68 samples led to the discovery of the following TSP classification rule: If OBSCN expression is greater than C9orf65 expression, then classify as GIST; all else classify as LMS. This TSP classifier was correct on all of the samples except for one for which the measured expressions of OBSCN and C9orf65 were the same on the microarray (Fig. 2). This indeterminate case was scored as a random guess (50%), resulting in a 99.3% (67.5 of 68 samples) accuracy of the data set.
Fig. 2.
Expression values of the two genes involved in the TSP classifier on the Agilent microarrays after quantile normalization. (Note: The classification is independent of normalization, because the decision is based only on which gene is higher, but the magnitude of the expression shown does vary somewhat with normalization technique.) The separating line (slope = 1) represents the cutoff for which gene is more highly expressed. It is not a fit to the data.
In addition to finding the two-gene classifier, we also used leave-one-out cross-validation to assess the expected error of this classifier on future data. Thus, we left out each of the 68 samples to evaluate how a classifier trained on the remaining 67 samples would perform on the left-out sample and then averaged the results. The simple two-gene tests picked within each cross-validation loop classified each test sample correctly, except for one case for which the outcome was indeterminate (the same sample as discussed above) and one case for which the selected pair misdiagnosed the held-out sample. This misdiagnosis occurred because, in one of the 68 cross-validation loops, a different pair was selected than OBSCN/C9orf65 and then subsequently misdiagnosed the held-out sample. Thus, although the OBSCN/C9orf65 classifier did not miss on any of the samples, the method we applied to the data set (TSP) has a slightly higher than expected error on future cases. Thus, the accuracy of the TSP approach as applied to this data set was estimated at 97.8% (66.5 of 68 samples) for future cases. With these promising results, we proceeded to the RT-PCR validation step.
Validation of the Classifier by Using RT-PCR and Independent Samples.
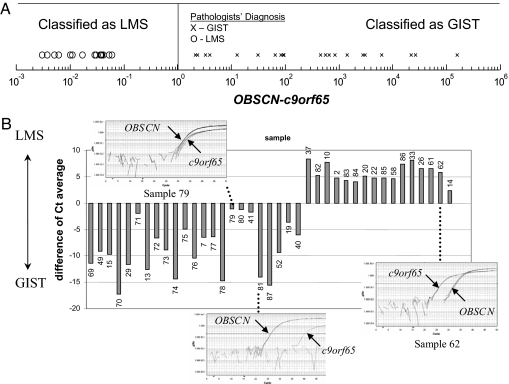
We next performed RT-PCR on (i) a subset of the samples used in the microarray experiment, including the sample with identical OBSCN and C9orf65 expression on the microarray, and (ii) an independent set of 19 additional samples that were not included in the microarray experiment. The RT-PCR results showed that the relative expression of OBSCN and C9orf65 (i.e., which gene had higher expression) was the same as what was shown on the microarray in all cases tested (Fig. 3). When the sample that was indeterminate based on the microarray study was tested using RT-PCR, the expression of OBSCN was found to be slightly higher than the expression of C9orf65, resulting in the classifier correctly matching the clinicopathologic diagnosis of GIST. Thus, the accuracy of the classifier based on RT-PCR data confirmed the expectations from the microarray expression data and showed that, with a more precise measurement, it was also correct on the sample previously classified as indeterminate.
Fig. 3.
Relative expression (OBSCN/C9orf65) as measured using RT-PCR. (A) Results are shown from 20 samples also used in the microarray experiments and an additional independent set of 17 samples. Classifier prediction as LMS or GIST is determined by which gene (OBSCN or C9orf65) is more highly expressed. Clinical diagnosis is shown as “X” for GIST and “O” for LMS. (B) The raw data in terms of the distance of Ct average from the RT-PCR experiments (OBSCN Ct − C9orf65 Ct).
The independent set of 19 additional samples provided the most important test of our classifier, with the clinicopathologic diagnosis of GIST or LMS being made before running the RT-PCR of the classifier genes. On the independent set of 19 samples, consisting of 14 GIST samples and 5 LMS samples, the classifier agreed with the clinicopathologic diagnosis in every case. Moreover, two of the 14 GIST samples were from fine needle aspirates, indicating that this test can be used with small tissue samples. (The clinicopathogical diagnosis and classifier performance for each sample used in this study can be found at www3.mdanderson.org/∼genomics/SupplementalTableSampleClassifications.pdf.) Thus, the total accuracy of the classifier on the data set combining the independent samples with those used in the microarray study was 100%. (One of the samples was initially diagnosed by histopathology as a LMS but was later determined to be most consistent with a GIST in view of the finding that it arose from the ileum, metastasized to the liver and peritoneum, did not stain for desmin by immunohistochemistry, and was resistant to LMS chemotherapy.) Thus, the two-gene classifier chosen has yet to fail on any sample we have tested. Of course, this result does not mean that we can expect the classifier to perform perfectly on all future cases, but based on the evidence accumulated to date, there is a strong expectation of high accuracy.
Classifier Performance Relative to Kit Expression.
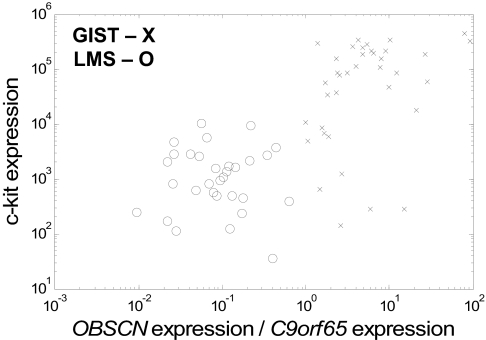
We also compared the performance of the two-gene relative expression classifier with c-Kit expression from the microarray experiments. The data show greatly increased effectiveness of separation of GIST and LMS by using the TSP classifier over c-Kit expression (Fig. 4). We noted that the expression of c-Kit had an accuracy of 87.3% (cutoff determined by using 1D linear discriminate analysis) compared with 97.8% of the TSP classification procedure in leave-one-out cross-validation. Examples of GIST samples with low Kit expression and LMS samples with high Kit expression are shown in Table 1. Therefore, the TSP gene expression classifier was more accurate than c-Kit at both the protein and the RNA levels.
Fig. 4.
Comparison of TSP classifier with Kit gene expression for separating samples of GIST (X) and LMS (O). The values shown are the expression values from the Agilent micorarrays after quantile normalization. (Note: The OBSCN/C9orf65 classifier is independent of the normalization chosen, but any classification based on c-Kit expression alone would not be.)
Table 1.
Abnormal Kit expression in GIST and LSM
| Sample ID nos. | Kit exp, arbitrary units | OBSCN exp/C9orf65 exp (>1 = >GIST) | Kit exp/median Kit exp in all samples, % |
|---|---|---|---|
| GIST with low c-Kit exp | |||
| 21 | 140 | 2.6 | 3 |
| 34 | 634 | 1.5 | 13 |
| 50 | 1,243 | 2.7 | 25 |
| 67 | 280 | 6.0 | 6 |
| 68 | 283 | 15.2 | 6 |
| LMS with high c-Kit exp | |||
| 10 | 10,156 | 0.06 | 207 |
| 41 | 5,535 | 0.07 | 113 |
| 43 | 9,127 | 0.22 | 186 |
exp, expression.
Discussion
GIST was previously thought to be best grouped with spindle cell and other soft-tissue sarcomas, including LMS, but in recent years it has emerged as a distinct entity. Moreover, GISTs continue to be misclassified as LMS or other soft-tissue sarcomas. GISTs have mutated and activated c-Kit or PDGFR oncogene and are exquisitely sensitive to therapy with imatinib mesylate or sunitinib but resistant to cytotoxic chemotherapy. Conversely, LMSs are most effectively treated with cytotoxic chemotherapy and are resistant to tyrosine kinase inhibitors. Thus, the appropriate diagnosis of these histopathologically similar entities is at times a life-and-death decision. Because mutated Kit is frequently expressed at a relatively elevated level in GIST cells, Kit expression has become a key diagnostic marker supporting the diagnosis of GIST. However, use of Kit as a diagnostic marker poses a number of problems, as previously discussed. Thus, we drew on the wealth of information encoded in the transcriptomes of relevant human tissues to develop a global search approach to identify an accurate, robust, simple-to-use method to accurately distinguish GIST from LMS. We applied a recently developed analytical method to 68 well characterized GIST and LMS tumors and identified a single pair of genes whose relative expression patterns accurately differentiated GIST from LMS.
The genes in the TSP classifier, OBSCN and C9orf65, are not well characterized. The OBSCN (obscurin) gene is located on chromosome 1. It is a relatively large gene spanning >150 kb and containing >80 exons. The protein product is ≈720 kDa, with 68 Ig domains, 2 fibronectin domains, 1 calcium/calmodulin-binding domain, 1 RhoGEF domain with an associated PH domain, and 2 serine–threonine kinase domains. OBSCN belongs to the family of giant sarcomeric signaling proteins that includes titin and nebulin and appears to mediate interactions between the sarcoplasmic reticulum and myofibrils. OBSCN binds to the sarcoplasmic reticulum by interaction with small ankyrin-1 and by the contractile myofibril via titin and sarcomeric myosin (16, 17).
However, the C9orf65 gene is yet to be characterized and is only named after its location on chromosome 9 (ORF 65). Unpublished information provided in the GeneCard (www.genecards.org/cgi-bin/carddisp.pl?gene=C9orf65&search=C9orf65) suggests that C9orf65 interacts with reticulocalbin 3, a member of the Cab45/reticulocalbin/ERC45/calumenin (CREC) family of multiple EF hand Ca2+-binding proteins localized to the secretory pathway (18). In a recent study, Meza-Zepeda et al. (19) reported chromosomal region copy number differences between 7 GIST and 12 LMS cases. Interestingly, one of the regions reported, 9q21.11–9q34.3, includes one of the two gene pairs, C9orf65 (located at 9q21.2).
The biological reason why OBSCN is essentially always expressed at a higher level than C9orf65 in GISTs and with the inverse occurring in LMS remains unknown. It is interesting to speculate that perhaps OBSCN has functional importance for the GIST cell. GIST cells are thought to arise from the interstitial cell of Cajal, the intestinal pacemaker cell, or a closely related cell in this lineage. Therefore, if GISTs do arise from a neuromuscular pacemaker cell, than it would not be surprising to find the altered expression of certain genes found in muscle cells, such as OBSCN. However, because OBSCN expression seems to play a role in normal muscle processes, an alternative possibility is that, in the process of tumorigenesis, the LMS cells may have phenotypically diverged from normal smooth muscle such that these cells no longer express the proteins a normal muscle cell requires for its function. For instance, leiomyoma typically retain expression of smooth muscle markers such as α-actin, myosin heavy chain, and total myosin that are frequently lost in LMS (20). Thus, it would not be surprising that LMS would have a relatively low expression level of the muscle protein OBSCN. Although not yet proven, it is conceivable that the two genes with reversed expression patterns represent two key nodes in the gene regulatory network such that their relative expression has a major impact on the network state and the resulting cellular phenotype. In this sense, the relative expression approach is poised to help identify key genes that drive important cellular processes.
The use of genomics-based molecular approaches in determining the diagnosis, prognosis, and appropriate therapeutic approach is already impacting clinical care for breast cancer patients. Paik et al. (21) reported the discovery of a 21-gene biomarker set that could be used to predict the risk for recurrence of breast cancer after adjuvant hormonal therapy. Thus, some clinicians are now adding chemotherapy to a patient's therapeutic regimen if the 21-gene classifier predicts that the patient has a high chance of recurrence if treated with hormonal therapy alone. Moreover, patients with a low risk of recurrence can be spared the toxicity of chemotherapy. Other investigators have developed prognostic tests for breast cancer that rely on overall patterns of gene expression in microarrays using very large gene sets (22). In our approach, we used paired gene set discriminators that allow the use of fewer genes to effectively separate populations.
The accuracy of this two-gene classifier method is near 100% in both the training and independent validation groups. The assay is superior to Kit-based diagnosis and accurately diagnoses Kit-negative GISTs and those GISTs with weak or heterogeneous Kit expression. Additionally, the patchy pattern of Kit expression in some cases renders current diagnostic methods used on biopsy samples unreliable, whereas the two-gene relative expression classifier we identified was highly accurate in the diagnosis of biopsy samples. The probable reason for the predictive power of the classifier in cases for which it was not trained is due to two inherent characteristics. First, the classifier is very simple, so it is not prone to overfitting the data, which can commonly occur when using more complex classifiers. The second major advantage of this approach to classification is the use of pairs of genes to eliminate normalization issues rather than relying on absolute gene expression levels. In particular, the use of relative expression makes unnecessary the establishment of a population-wide threshold, as is needed for a single marker, or for parameter weightings, as is needed for more complex multiple-parameter classifiers.
In addition to the success of differentiating GIST and LMS detailed herein, the k-top scoring pair (k-TSP) method was shown to perform comparably with the best multigene classification methods using a number of published cancer transcriptome data sets (13). Further, the method had remarkable ability to correctly predict results across different mRNA measurement platforms (15). It is possible that in the future this approach may be used not only to differentiate ambiguous histologies and to assess prognosis but also to determine who will benefit from chemotherapy, which type of chemotherapy to use, and which patients are at risk for local or distant relapse. Similarly, the relative expression reversal approach should be applicable to the development of robust protein-based markers from complex proteomic measurements from tumor tissues or bodily fluid such as blood. Because quantification of any single protein is subject to uncertainties caused by measurement variability, normal fluctuations, and individual related variation in baseline expression, identification of pairs of markers that may be under coordinated, systematic regulation should prove to be more robust for individualized diagnosis and prognosis.
In summary, we have developed an approach to discriminate GIST from LMS that may lead to a better understanding of the biologic differences of these two histologically similar entities. Moreover, utilization of this technology may aid clinicians and pathologists in diagnosis and treatment of patients who have tumors that cannot be clearly classified as either GIST or LMS. We believe that this approach will be widely applicable to molecular marker identifications from genomics and proteomics studies and will accelerate the translation of results from high-throughput exploratory studies to the clinic.
Materials and Methods
Patients and Samples.
All of the samples were obtained from surgical specimens at the M. D. Anderson Cancer Center and stored at the Institutional Tumor Tissue Repository with patient consent and an Institutional Review Board-approved protocol. The tissues were snap-frozen within 20 min of surgical resection.
Pathology Evaluation.
Hematoxylin- and eosin-stained slides of formalin-fixed paraffin-embedded tissue blocks of all cases were reviewed by one of the authors (A.K.E.-N.). Previous diagnosis and the immunostaining results, if any, between 4 and 36 (mean 14) slides per case were evaluated. Immunohistochemical staining for smooth muscle actin (SMA) was performed on 45 cases.
Because intra-abdominal spindle cell malignant neoplasms comprise a wide spectrum of morphologic and biological entities, including GIST and LMS, multidisciplinary attempts were made to segregate individual tumors on the basis of cellular features by light microscopy; immunostaining for Kit, CD34, and SMA; and clinical observations, including the site of the primary tumor, the pattern of metastatic spread, and the efficacy of systemic therapy. Leiomyosarcoma was diagnosed when a tumor manifested intersecting fascicles of elongated spindle cells with cigar-shaped, elongated nuclei with amphophilic cytoplasm with at least 5 of 10 high power field mitotic figures and positive SMA and after negative CD34 immunostaining in patients with the appropriate clinical setting. GIST diagnosis was made when a patient had the clinical presentation consistent with GIST, and the tumor was composed of spindled, epithelioid, or mixed cell proliferations with positive Kit, positive cytoplasmic CD34, and negative SMA. The few tumors that were negative for Kit, SMA, and CD34 were classified on the basis of the light microscopic features and clinical pattern of disease.
Western Blot Analysis.
Standard procedure was used for protein isolation and Western blot analysis. The membranes were probed with primary antibodies [anti-Kit (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-β-actin to control for protein loading] followed with horseradish peroxidase-conjugated secondary antibodies (at a dilution of 1:2,000). Membranes were washed and incubated in enhanced chemiluminescence solution (Amersham Life Science, Piscataway, NJ), and subjected to autoradiography.
RNA Isolation and Quantitative RT-PCR Assays.
Total RNA was isolated and quantified as described in ref. 23. Assays-on-Demand from ABI (Applied Biosystems, Foster City, CA) were used to quantify RNA levels of OBSCN (Hs00405789_m1) and c9orf65 (Hs00373436_m1) on an ABI 7900 HT with a 96-well block. PPIA, also known as cyclophilin A endogenous control assay (4326316E), was used to verify the integrity of each sample. We assayed each sample in triplicate with 25 ng of input RNA per well in a volume of 25 μl reaction containing 1× TaqMan One-Step RT-PCR Master Mix (Applied Biosystems) and 1× gene expression assay. The following cycling conditions were used: 48°C for 30 min for reverse transcriptase reaction then PCR, 10 min at 95°C, then 50 cycles of 95°C for 15 seconds and 60°C for 1 min. Cycle threshold values (Ct) generated from Sequence Detection System 2.2 (Applied Biosystems) default parameters were exported to determine relative mRNA abundances between the two genes in the classifier. The lower the Ct value, the more abundant the RNA was because fewer PCR cycles were required to amplify the RNA.
Microarray Experiments.
Microarray experiments were carried out using whole human genome oligo arrays with 44k 60-mer probes (Agilent Technologies, Palo Alto, CA) with 500 ng of total RNA starting material according to the manufacturer's protocol. Hybridized arrays were scanned with Agilent's dual laser-based scanner. Then, Feature Extraction software version 8.0 (Agilent Technologies) was used to link a feature to a design file and to determine the relative fluorescence intensity between the two samples. The microarray data are publicly available at www3.mdanderson.org/∼genomics/sarcoma_data_matrix_for_supplemental.zip.
Classification Algorithm.
The method used herein for supervised classification is TSP (13, 14). The basic idea of the TSP approach is to select markers in pairs that exhibit relative expression reversal between the classes being compared. In its simplest form, the marker is thus dependent on only the following question: is the expression of gene A higher than the expression of gene B in the sample? If so, the diagnosis is class 1 (i.e., GIST). If the expression of gene B is higher than for gene A, then the diagnosis is for class 2 (i.e., LMS). If additional pairs had been needed to obtain better classification, they would have been combined and each pair rule would have been assigned a vote, with a majority vote for a given class determining the diagnosis; this is called the k-TSP approach (13). For our data set, TSP outperformed k-TSP. The computation of the TSP classifier and the error estimation were done using the k-TSP program downloaded from https://jshare.johnshopkins.edu/atan6/public_html/KTSP (13). The estimation of the classification error on future cases was performed using leave-one-out cross-validation. This estimate was then verified using an independent test on an additional set of patient samples. All other numerical analyses presented herein were performed using Matlab (Mathworks, Natick, MA).
Genes Included in Selecting the Classifier.
A subset of genes was removed before final analysis because, although they were potentially among the best predictive genes from a computational standpoint, they were not amenable to RT-PCR amplification for the validation stage (a robust multiexon assay was not available in Assays-on-Demand from ABI). Thus, a subset of 20 genes was removed. The classifier was then trained on only the remaining set of genes. Given the near perfect behavior of our classifier, however, we could hardly have improved on the effectiveness of the gene pair found, even if robust RT-PCR amplification was available for all of the genes on the microarray.
Acknowledgments
We thank Dr. Aik Choon Tan for helpful assistance with code for the k-TSP algorithm. This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) Grant R01 CA098570-01 (to W.Z.), a grant from the Commonwealth Foundation for Cancer Research (to W.Z. and J.T.), and NIH/National Institute of General Medical Sciences Grant P50 GM076547 (to L.H.). N.D.P. was supported by a postdoctoral fellowship from the American Cancer Society (no. PF-06-062-01-MGO). J.T. was supported by an Institutional Physician-Scientist Award and by NIH/NCI Grant 1K23CA109060-01. The Cancer Genomics Core Facility is supported by the Tobacco Settlement Fund to the M. D. Anderson Cancer Center, as appropriated by the Texas Legislature, by grants from the Michael and Betty Kadoorie Foundation and from the Goodwin Fund, and by NCI Cancer Center Support Grant CA-16672.
Abbreviations
- Ct
cycle threshold value
- GIST
gastrointestinal stromal tumor
- LMS
leiomyosarcoma
- SMA
smooth muscle actin
- TSP
top scoring pair.
Footnotes
The authors declare no conflict of interest.
Rankin, C., von Mehren, M., Blanke, C. D., Benjamin, R., Fletcher, C. D., Bramwell, V. H., Crowley, J., Borden, E., Demetri, G. (2004) Am. Soc. Clin. Oncol. 23:815 (abstr. 9005).
Chugh, R., Thomas, D., Wathen, K., Thall, P., Benjamin, R., Maki, R., Samuels, B., Keohan, M., Priebat, D., Baker, L. (2004) 2004 ASCO Annual Meeting Proceedings 22:818s (abstr. 9001).
Blackstein, M. E., Rankin, C., Fletcher, C., Heinrich, M., Benjamin, R., von Mehren, M., Blanke, C., Fletcher, J. A., Borden, E., Demetri, G. (2005) Proceedings of the American Society of Clinical Oncology 23:9010 (abstr. 9010).
References
- 1.Clary BM, DeMatteo RP, Lewis JJ, Leung D, Brennan MF. Ann Surg Oncol. 2001;8:290–299. doi: 10.1007/s10434-001-0290-3. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP, et al. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 3.Patel S, Vadhan-Raj S, Burgess M, Papadopoulos N, Plager C, Jenkins J, Benjamin R. Am J Clin Oncol. 1998;21:317–321. doi: 10.1097/00000421-199806000-00025. [DOI] [PubMed] [Google Scholar]
- 4.Patel SR, Gandhi V, Jenkins J, Papadopolous N, Burgess MA, Plager C, Plunkett W, Benjamin RS. J Clin Oncol. 2001;19:3483–3489. doi: 10.1200/JCO.2001.19.15.3483. [DOI] [PubMed] [Google Scholar]
- 5.Trent JC, Beach J, Burgess MA, Papadopolous N, Chen LL, Benjamin RS, Patel SR. Cancer. 2003;98:2693–2699. doi: 10.1002/cncr.11875. [DOI] [PubMed] [Google Scholar]
- 6.Dematteo RP, Heinrich MC, El-Rifai WM, Demetri G. Hum Pathol. 2002;33:466–477. doi: 10.1053/hupa.2002.124122. [DOI] [PubMed] [Google Scholar]
- 7.Verweij J, Casali PG, Zalcberg J, LeCesne A, Reichardt P, Blay JY, Issels R, van Oosterom A, Hogendoorn PC, Van Glabbeke M, et al. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 8.Gottleib J, Baker L, O'Bryan R, Sinkovics J, Hoogstraten B, Quagliana J, Rivkin S, Bodey G, Rodriquez B, Blumenschein G, et al. Cancer Chemother Rep. 1974;6:271–282. [Google Scholar]
- 9.Hensley ML, Maki R, Venkatraman E, Geller G, Lovegren M, Aghajanian C, Sabbatini P, Tong W, Barakat R, Spriggs DR. J Clin Oncol. 2002;20:2824–2831. doi: 10.1200/JCO.2002.11.050. [DOI] [PubMed] [Google Scholar]
- 10.Serrano C, Mackintosh C, Herrero D, Martins AS, de Alava E, Hernandez T, Perez-Fontan J, Abad M, Perez A, Serrano E, et al. Clin Cancer Res. 2005;11:4977–4979. doi: 10.1158/1078-0432.CCR-04-2461. author reply 4979–4980. [DOI] [PubMed] [Google Scholar]
- 11.Silvestris N, Parra HS, Angelini F, Di Cosimo S, D'Aprile M, Santoro A. Tumori. 2005;91:103. doi: 10.1177/030089160509100123. [DOI] [PubMed] [Google Scholar]
- 12.Medeiros F, Corless CL, Duensing A, Hornick JL, Oliveira AM, Heinrich MC, Fletcher JA, Fletcher CD. Am J Surg Pathol. 2004;28:889–894. doi: 10.1097/00000478-200407000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Bioinformatics. 2005;21:3896–3904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geman D, d'Avignon C, Naiman DQ, Winslow RL. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1071. Article19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Tan AC, Naiman DQ, Geman D, Winslow RL. Bioinformatics. 2005;21:3905–3911. doi: 10.1093/bioinformatics/bti647. [DOI] [PubMed] [Google Scholar]
- 16.Armani A, Galli S, Giacomello E, Bagnato P, Barone V, Rossi D, Sorrentino V. Exp Cell Res. 2006;312:3546–3558. doi: 10.1016/j.yexcr.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 17.Kontrogianni-Konstantopoulos A, Bloch RJ. J Muscle Res Cell Motil. 2005;26:419–426. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- 18.Tsuji A, Kikuchi Y, Sato Y, Koide S, Yuasa K, Nagahama M, Matsuda Y. Biochem J. 2006;396:51–59. doi: 10.1042/BJ20051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meza-Zepeda LA, Kresse SH, Barragan-Polania AH, Bjerkehagen B, Ohnstad HO, Namlos HM, Wang J, Kristiansen BE, Myklebost O. Cancer Res. 2006;66:8984–8993. doi: 10.1158/0008-5472.CAN-06-1972. [DOI] [PubMed] [Google Scholar]
- 20.Valenti MT, Azzarello G, Vinante O, Manconi R, Balducci E, Guidolin D, Chiavegato A, Sartore S. J Cancer Res Clin Oncol. 1998;124:93–105. doi: 10.1007/s004320050140. [DOI] [PubMed] [Google Scholar]
- 21.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 22.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 23.Shmulevich I, Hunt K, El-Naggar A, Taylor E, Ramdas L, Laborde P, Hess KR, Pollock R, Zhang W. Cancer. 2002;94:2069–2075. doi: 10.1002/cncr.10425. [DOI] [PubMed] [Google Scholar]