Abstract
The tungsten–iron–sulfur enzyme acetylene hydratase stands out from its class because it catalyzes a nonredox reaction, the hydration of acetylene to acetaldehyde. Sequence comparisons group the protein into the dimethyl sulfoxide reductase family, and it contains a bis-molybdopterin guanine dinucleotide-ligated tungsten atom and a cubane-type [4Fe:4S] cluster. The crystal structure of acetylene hydratase at 1.26 Å now shows that the tungsten center binds a water molecule that is activated by an adjacent aspartate residue, enabling it to attack acetylene bound in a distinct, hydrophobic pocket. This mechanism requires a strong shift of pKa of the aspartate, caused by a nearby low-potential [4Fe:4S] cluster. To access this previously unrecognized W–Asp active site, the protein evolved a new substrate channel distant from where it is found in other molybdenum and tungsten enzymes.
Keywords: acetylene reduction, metalloproteins, tungsten enzymes
An estimated one-third of all proteins contain metal ions or metal-containing cofactors, and their overwhelming majority is involved in either electron transfer or the catalysis of redox reactions (1). Different metal centers typically take on specific functional roles, and although their respective substrates can vary significantly, they commonly catalyze similar kinds of chemical reactions. Molybdenum and tungsten are the only known second- and third-row transition metals to occur in biomolecules, and they are almost exclusively coordinated by the organic cofactor molybdopterin (2, 3). Mo/W proteins play important metabolic roles in all kingdoms of organisms and include prominent enzymes, such as nitrate reductase (4, 5), formate dehydrogenase (6), sulfite oxidase (7), or xanthine oxidase (8). They are involved either in oxygen atom transfer reactions or oxidative hydroxylations, whereby the metal undergoes two-electron oxidation/reduction between the states +IV and +VI (9). An exception to this rule was recently discovered for the pyrogallol:phloroglucinol hydroxyltransferase of Pelobacter acidigallici (10) that catalyzes a net nonredox reaction, but closer inspection reveals a reductive dehydroxylation and an oxidative hydroxylation as separate, consecutive events.
A true nonredox reaction has been described for the tungsten–iron–sulfur enzyme acetylene hydratase (AH) from Pelobacter acetylenicus (11), a member of the DMSO reductase family of molybdenum and tungsten enzymes (2, 9). It catalyzes the hydration of acetylene to acetaldehyde (see Eq. 1) as part of an anaerobic degradation pathway of unsaturated hydrocarbons (12).
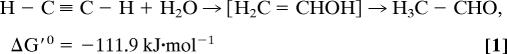 |
Chemically, the mercuric ion, Hg2+, will catalyze the addition of water to alkynes through the formation of a cyclic mercurinium ion. In a consecutive step, water attacks the most substituted carbon atom of this cyclic intermediate followed by the formation of a mercuric enol, which then rearranges to the corresponding ketone (13, 14). In the living world, AH is the only enzyme known to be able to convert acetylene other than nitrogenase (15, 16), for which, however, the alkyne is not considered a physiological substrate. Note that nitrogenase reduces acetylene by two electrons to produce ethylene. Similar to nitrogenase, AH is extremely oxygen-sensitive, and its activity is lost irreversibly upon exposure to air, concomitant with the degradation of the [4Fe:4S] cluster as documented by EPR spectroscopy (17).
Among tungstoenzymes, AH constitutes a distinct class beside the tungsten aldehyde: ferredoxin oxidoreductases (18) and the formyl methanofuran dehydrogenases (3, 19, 20). Its reaction does not involve electron transfer, and neither its W atom nor the [4Fe:4S] cluster change their oxidation states during catalysis (12, 17). However, AH activity requires activation by a strong reductant, indicating that the active form is W(IV) (17).
AH has been purified and crystallized under the strict exclusion of dioxygen in its active, reduced state (21). Here we present the crystal structure determined to a resolution of 1.26 Å by single-wavelength anomalous dispersion methods using the anomalous signal from iron and tungsten at an x-ray energy above the K-edge of iron.
Results and Discussion
Crystal Structure of AH.
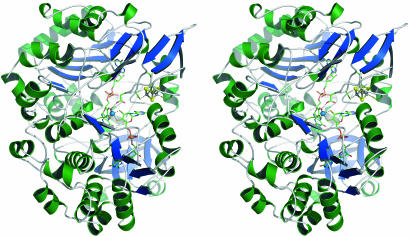
AH is a monomer of 730 aa residues (83 kDa) and contains two molybdopterin guanine dinucleotide cofactors (MGD) (designated P and Q, following the DMSO reductase nomenclature; ref. 22) in addition to a tungsten center and a cubane-type [4Fe:4S] cluster (Fig. 1). It shares the general structural features of a member of the DMSO reductase family of molybdenum and tungsten proteins: The peptide chain folds into a tertiary structure comprising four domains that are related by an internal pseudotwofold axis and that bury all cofactors deep inside the protein (Fig. 2A). Domain I comprises residues 4–60 and holds the cubane-type [4Fe:4S] cluster of the protein bound to Cys-9, Cys-12, Cys-16, and Cys-46. The subsequent domains II (residues 65–136 and 393–542) and III (residues 137–327) both display an αβα-fold with homologies to the NAD-binding fold observed in dehydrogenases (23), providing multiple hydrogen-bonding interactions to one of the MGD cofactors each, mediated by the variable loop regions at the C termini of the strands of a parallel β-sheet. The final domain IV (residues 590–730) is dominated by a seven-stranded β-barrel structure and participates in the coordination of both MGD ligands. The overall arrangement of cofactors is similar to the one observed in other members of the DMSO reductase family, with the MGD molecules in an elongated conformation and the [4Fe:4S] cluster in close proximity to QMGD.
Fig. 1.
Overall structure of acetylene hydratase from P. acetylenicus. The stereo representation shows an orientation viewing down the active site channel as seen in Fig. 4A.
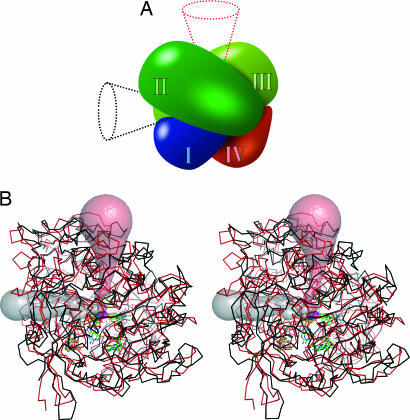
Fig. 2.
Domain structure and active site access. (A) Schematic view of the four-domain structure typically found in members of the DMSO reductase family. All members, with the exception of AH, have an active site access pathway along the red cone. AH uses the black cone, entering at the intersection of domains I, II, and III. (B) Stereoview of a superimposition of AH (black) and periplasmic nitrate reductase (red) (5), with exit pathways calculated by CAVER (24). The W atom of AH (blue) and the Mo atom of nitrate reductase (magenta) are shown as spheres.
Active Site Access.
In all of the structures of proteins of the DMSO reductase family available to date, access to the active center is provided via a funnel-like entrance whose position is conserved in DMSO and trimethylamine N-oxide reductases as well as in formate and nitrate reductases (Fig. 2A). It aligns well with the pseudotwofold axis between domains II and III that passes the central metal ion. In AH, however, the entire region connecting domains II and III (residues 327–393) is completely rearranged, resulting in a tight sealing of the substrate funnel and in a shift of the loop region ranging from residues 327 to 335 by >15 Å toward the protein surface. In formate and nitrate reductases, this loop separates the Mo/W center from the [4Fe:4S] cluster, and its displacement in AH opens up a new face of the protein surface at the intersection of domains I, II, and III, providing access to a very different portion of the metal coordination sphere than is seen in other enzymes of the DMSO reductase family. This finding is illustrated by calculating exit pathways starting from an identical position in the superimposed structures of AH and its closest structural relative, the periplasmic nitrate reductase of Desulfovibrio desulfuricans ATCC 27774 (5) with the program CAVER (Fig. 2B) (24).
Arrangement of the Active Site.
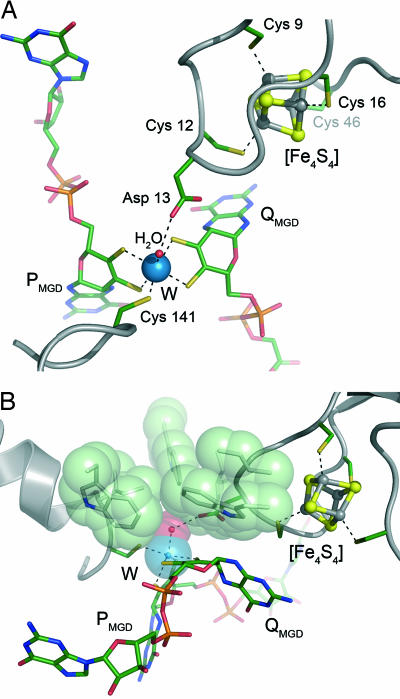
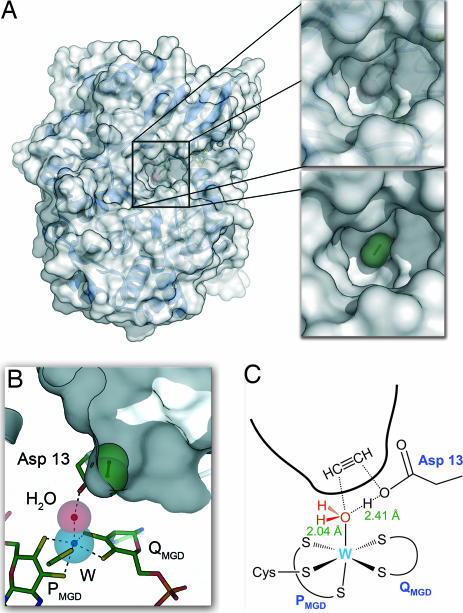
This major rearrangement within an otherwise conserved protein fold has consequences for the architecture of the active site of AH: The tungsten center, W(IV) in the active state, is canonically coordinated by the sulfur atoms of the dithiolene moieties of the PMGD and QMGD cofactors, and as in dissimilatory nitrate reductase, this coordination is completed by a cysteine residue from the protein, Cys-141. A sixth ligand to W shows a bond length of 2.04 Å, indicating a tightly coordinated water molecule (Fig. 3A). The active site geometry in this class of enzymes is commonly described either as square pyramidal or as trigonal prismatic (2). In AH, a slight rotation of the P cofactor yields a geometry that more closely resembles octahedral, or trigonal antiprismatic. Because of the displacement of the loop region 325–335 from the active site, acetylene can access the tungsten ion through a channel ≈17 Å deep and 6–8 Å wide at a position close to the N-terminal domain that harbors the [4Fe:4S] cluster. A residue from this domain, Asp-13, is then positioned in such a way as to interact closely with the water bound to the W ion, forming a short hydrogen bond of 2.4 Å (Fig. 3A). Above the tungsten atom and the coordinated water molecule, the substrate access funnel ends in a ring of hydrophobic residues, made up of Ile-14, Ile-113, Ile-142, Trp-179, Trp-293, and Trp-472 (Fig. 3B). These residues, originating from domains I, II, and III, combine to create a small binding cavity with dimensions that make it a perfect mold for binding acetylene (Fig. 4A). Although the pressurization of both reduced and oxidized crystals of AH with acetylene gas as well as soaks with acetylene or the inhibitor propargyl alcohol failed to produce a complex in the crystal, docking of an acetylene molecule at this position is possible and gives an excellent fit, positioning the two carbon atoms of the substrate exactly above the water molecule coordinated to tungsten. Through shape complementarity, the residues of the hydrophobic ring thus are a key determinant for the substrate specificity of AH (Fig. 4).
Fig. 3.
Cofactors and active site of AH. (A) The tungsten atom (blue) is coordinated by the dithiolene groups of both MGD cofactors and the side chain of Cys-141. A water molecule completes the slightly distorted octahedral geometry. This water is also hydrogen-bonded to Asp-13, a residue adjacent to the [4Fe:4S] cluster ligand Cys-12. (B) Above the bound water molecule, a ring of hydrophobic residues forms the bottom of the active site access channel (see Fig. 4).
Fig. 4.
Substrate channel and model for acetylene binding. (A) Surface representation of acetylene hydratase with the substrate access pathway (boxed). The enlarged area (upper box) shows a binding pocket that exactly fits the dimensions of an acetylene molecule (lower box). (B) The binding pocket positions an acetylene molecule directly above the water molecule and Asp-13. (C) Bond distances of 2.04 Å to W and 2.41 Å to the Oδ1 atom of Asp-13 indicate a highly activated water molecule positioned right below a binding pocket for acetylene.
Functional Modifications in Acetylene Hydratase.
Caught in its active state and defined at very high resolution, the crystal structure of AH provides the information required to understand this enzyme's mechanism. The hydration of acetylene is achieved not through an organometallic intermediate, such as the mercurinium ion (13), but rather by activation of a water ligand of the central tungsten ion through interaction with the nearby residue Asp-13. However, although such an activating residue should be a Lewis base by nature, aspartic acid is (in aqueous solution) instead the strongest acid, with a pKa of 3.83 for the COOH moiety. We propose that the solution to this problem is, on the one hand, provided by the fact that Asp-13 is deeply buried in the structure that shields it from charge stabilizing solvent effects and, on the other hand, provided by the [4Fe:4S] cluster, one of whose ligands, Cys-12, is a direct neighbor of Asp-13. Whereas in other representatives of the DMSO reductase family this cluster serves as an electron transfer center, such functionality is obviously not required in acetylene hydratase, although the lysine residue Lys-48, generally considered to be essential for electron transfer from the cluster to the QMGD, is conserved (2). In the redox-active enzymes of this family, the loop region from 325 to 335 is needed to prevent an unproductive short circuit, by separating the iron-sulfur cluster from the Mo/W ion. In AH however, the active site is found at a different side of the W ion, closer to the iron–sulfur cluster, and redox chemistry does not take place. Instead, a marked increase in pKa for Asp-13 is caused by the desolvation of this residue compared with aqueous solution and an inductive effect (+I effect) of the nearby [4Fe:4S] cluster, pushing electrons toward Asp-13 and thus increasing its proton affinity. To quantify this effect, density-functional theory-based atomic charges have been calculated and used to derive individual titration curves for all protonable residues in the protein by numerically solving the Poisson–Boltzmann equation. Most residues show clear deviations from the expected pKa values in free solution, but in almost all cases the charged form of the residues is stabilized, as is commonly observed in such calculations (25). The entire protein contains 34 Asp and 58 Glu residues, and in the calculations, only three of these show highly aberrant titration behavior: Asp-298, Glu-494, and Asp-13. Asp-298 titrates highly unusually with a pK1/2 of ≈14.2, whereas both Glu-494 and Asp-13 remained fully protonated in the pH range 0–24, indicating a substantial increase of pKa. Of these residues, only Asp-13 is close to any of the metal centers. Contrary to the situation in aqueous solution, Asp-13 will thus form a short hydrogen bond to a water molecule, leading to a positive partial charge on the oxygen atom that can then act as an electrophile.
The observed, strong increase of specific activity for AH under reducing conditions is in part explained by this finding, because the inductive effect and therefore the shift in pKa and the degree of activation of the water ligand will be stronger in a reduced cluster (that contains an additional negative charge). Partial deprotonation of Asp-13 at high pH values is indeed found in calculations carried out for the oxidized form of the enzyme. As a second effect, both W and Mo in the (+VI) state have been shown to bind oxygen preferentially in the oxo form rather than as water. In contrast, the less positively charged, reduced (+IV) state favors water, which, in AH, is required for reactivity. Determination of a structure of AH oxidized with 2 mM of K3[Fe(III)(CN)6] at a resolution of 1.6 Å showed a markedly shortened W–O distance of only 1.7 Å, indicative of an oxo ligand, whereas the rest of the structure remained essentially unchanged (data not shown). A tungsten model complex, [Et4N]2[WIVO(mnt)2], has been found to catalyze the same reaction as AH, whereas the oxidized W(VI) complex was inactive but could be reactivated by strong reductants (26).
The Mechanism of Acetylene Hydration.
A crucial question for understanding the detailed mechanism of acetylene hydration by AH concerns the nature of the water ligand bound to tungsten. Because of the proximity of the heavy scatterer tungsten, the distance derived from the crystal structure may be distorted by Fourier series termination effects. As shown for the high-resolution structure of nitrogenase, these effects can be simulated, such that their influence on the metal–ligand distance can be estimated (27). We find that at a resolution of 1.26 Å, a bond distance of 2.04 Å is observed for a true ligand distance of 2.25 Å. Mechanistically, this finding may be crucial, because the value of 2.04 Å falls between the values expected for a hydroxo ligand (1.9–2.1 Å) and a coordinated water (2.0–2.3 Å). The two possibilities lead to different mechanistic scenarios: A hydroxo ligand would constitute a strong nucleophile and would yield a vinyl anion with acetylene (14) of sufficient basicity to deprotonate Asp-13 and form the vinyl alcohol. Another water molecule could then bind to tungsten and get deprotonated by the basic Asp-13, thereby regenerating the hydroxo ligand for the next reaction cycle. Alternatively, a bound H2O molecule would gain a partially positive net charge through the proximity of the protonated Asp-13, making it an electrophile that in turn could directly attack the triple bond in a Markovnikov-type addition reaction with a vinyl cation intermediate (14). In this scheme, Asp-13 would remain protonated. A definitive distinction between both mechanisms will require further studies, but we observe that our density-functional theory calculations only converge for a water ligand in the reduced state and only for a hydroxo ligand in the oxidized state. This finding can be rationalized considering that the central metal ion is surrounded by five negative charges from its five thiolate ligands, yielding a total charge of +1 in the oxidized versus −1 in the reduced state, complementary to that of the respective ligand found to be stable in the calculations. As a consequence, and in accordance with the observed bond distances, the active W(IV) state should favor a water ligand and therefore an electrophilic addition mechanism.
Both mechanisms nevertheless require the modified architecture of the enzyme with a relocated substrate access pathway as well as the ring of hydrophobic residues to guide and orient the substrate such that the reaction can take place. A molecule of acetylene modeled into its putative binding site will be situated directly above the activated water or hydroxo ligand and will be held in place by hydrophobic interactions with the surrounding residues (Fig. 4 B and C). The initial product of acetylene hydration will be the enol, which will spontaneously tautomerize to acetaldehyde. With product thus bound in the active site, the hydrophobic constriction may present a barrier for the access of water from the side of the substrate channel, the final step required to replenish the coordination of the tungsten atom and complete the reaction cycle. However, the structure shows that water can instead be recruited from a significant reservoir of at least 16 well defined water molecules in a vestibule directly adjacent to the active site. This arrangement offers an elegant solution to the problem of a possible product inhibition by association of the enol or aldehyde to tungsten.
Far from being merely a misused redox enzyme, acetylene hydratase thus represents a highly sophisticated catalyst in which the hardware of a typical tungsten-containing reductase has been intricately modified for an entirely different function.
Materials and Methods
Data Collection and Structure Solution.
Crystals of P. acetylenicus AH were grown under strict exclusion of oxygen, as described in ref. 21, and transferred into a buffer containing the mother liquor plus 15% (vol/vol) of 2-methyl-2,5-pentanediol before flash-cooling in liquid nitrogen. Diffraction data were collected at beam lines BW6 (Max-Planck Unit for Structural Molecular Biology) and X11 (European Molecular Biology Laboratory) at Deutsches Elektronen Synchrotron, Hamburg, Germany. One data set was collected at a wavelength of 1.738 Å on the high-energy side of the K-edge of iron, and a high-resolution data set was subsequently collected from a different crystal at a wavelength of 1.05 Å (Table 1). Heavy metal sites were located with the program SHELXD (28), which produced five clear solutions corresponding to the four iron atoms of the [4Fe:4S] cluster and the tungsten atom. At the energy of the iron K-edge, the anomalous signal of tungsten corresponds to ≈6.8 e−, such that this atom appeared as the strongest peak in an anomalous difference Patterson map.
Table 1.
Data collection and refinement statistics
| Data sets | SAD | High-energy remote |
|---|---|---|
| Wavelength, Å | 1.738 | 1.050 |
| Space group | C2 | C2 |
| Unit cell dimensions | a = 120.9 Å; b = 72.1 Å; c = 106.9 Å; β = 124.3° | a = 120.8 Å; b = 72.0 Å; c = 106.8 Å; β = 124.3° |
| Resolution limits, Å | 50.0 − 1.95 (2.05 − 1.95) | 50.0 − 1.26 (1.35 − 1.26) |
| Independent reflections | 52,026 (6,985) | 194,301 (35,051) |
| Completeness, % | 93.4 (88.9) | 95.5 (92.5) |
| I/σ(I) | 29.2 (13.3) | 19.62 (1.91) |
| Rmerge | 0.053 (0.146) | 0.036 (0.333) |
| Rp.i.m.(ref. 42) | 0.021 (0.057) | 0.037 (0.332) |
| Multiplicity | 7.04 (6.58) | 1.85 (1.65) |
| Phasing power (anomalous) | 1.400 | — |
| RCullis | 0.717 | — |
| Figure of merit | 0.390 | — |
| Correlation (SOLOMON) | 0.723 | — |
| Rwork | — | 0.165 |
| Rfree | — | 0.199 |
| Cruickshank's DPI, Å (ref. 43) | — | 0.057 |
| Overall figure of merit | — | 0.836 |
| Overall correlation coefficient | — | 0.972 |
| rmsd in bond distances, Å | — | 0.010 |
| rmsd in bond angles, ° | — | 1.419 |
Numbers in parentheses represent values for the highest resolution shells. DPI, diffraction precision indicator; SAD, single-wavelength anomalous diffraction.
Model Building and Refinement.
Phase calculations were carried out with SHARP (29), and SOLOMON (30) was used for electron density modification. An initial model comprising 448 of the 730 amino acid residues of AH was built automatically by using RESOLVE (31), and all subsequent manual rebuilding steps were carried out in COOT (32). The model was refined by using REFMAC5 (33).
Density-Functional Theory and Electrostatic Calculations.
Point charges of the W-molybdopterin cofactor were calculated by using the Amsterdam Density Functional program suite (ADF 2000.02) (34). The initial structure of the tungsten ion, the molybdopterin, the cysteine thiolate, and the water or hydroxide ion were taken from the crystal structure and were subsequently energy minimized. The self-consistent field (SCF) cycles converged only for the complex of W(IV) with a water molecule coordinating the tungsten, and not with the hydroxide ion, and for the W(VI) with a hydroxide ion, not with a water. Partial charges were derived from the density-functional theory calculation results by a CHELPG-based algorithm combined with singular value decomposition. Atom radii published by Bondi (35) were used in the fitting procedure.
To calculate the pH titration of the molecule, we applied a well tested method (25). The intrinsic proton binding energies and the interaction energies between the residues were obtained by continuum electrostatic calculations. The Poisson–Boltzmann equation was solved by a finite-difference method using the MEAD program suite (36, 37). All aspartate, glutamate, lysine, arginine, cysteines, histidines, and tyrosine residues were considered as protonatable sites. Atomic partial charges for standard amino acid groups were taken from the CHARMM22 parameter set (38). The charges of the iron sulfur cluster were taken from the literature (39). The pKa values of the model compounds were taken from the literature (25). The dielectric constant of the protein was set to 4, and that of the solvent was set to 80. Continuum electrostatics calculations were performed by using the focusing technique in three steps because of the size of the system. For the protein, the electrostatic potential was calculated by focusing using three cubic grids of 1413, 1213, and 1213 grid points and grid spacings of 2.0, 1.0, and 0.25 Å, respectively. The grid with the largest spacing was centered on the protein, the other two grids on the titratable group. For the model compounds, the electrostatic potential was calculated by focusing with two grids of 1213 and with 1213 grid points and grid spacings of 1.0 Å and 0.25 Å, respectively, again with the larger grid being centered on the protein and the smaller one being centered on the titratable group. We used a Monte Carlo approach (40) to calculate the protonation probabilities of all titratable sites of AH by using our program CMCT (41).
Acknowledgments
This work was supported by the European Molecular Biology Organization Young Investigator Program (O.E.) and by the Deutsche Forschungsgemeinschaft priority program “Radicals in Enzymatic Catalysis” (B.S. and P.M.H.K.).
Abbreviations
- AH
acetylene hydratase
- MGD
molybdopterin guanine dinucleotide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The structural data reported in this paper have been deposited with the Protein Data Bank, www.pdb.org (PDB ID code 2E7Z).
References
- 1.Fráusto da Silva JJR, Williams RJP. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford, UK: Oxford Univ Press; 2001. [Google Scholar]
- 2.Dobbek H, Huber R. Molybdenum and Tungsten: Their Roles in Biological Processes. Vol 39. 2002. pp. 227–263. [PubMed] [Google Scholar]
- 3.Kisker C, Schindelin H, Baas D, Retey J, Meckenstock RU, Kroneck PMH. FEMS Microbiol Rev. 1998;22:503–521. doi: 10.1111/j.1574-6976.1998.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 4.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NCJ. Nat Struct Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 5.Dias JM, Than ME, Humm A, Huber R, Bourenkov GP, Bartunik HD, Bursakov S, Calvete J, Caldeira J, Carneiro C, et al. Structure. 1999;7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 6.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 7.Kisker C, Schindelin H, Pacheco A, Wehbi WA, Garrett RM, Rajagopalan KV, Enemark JH, Rees DC. Cell. 1997;91:973–983. doi: 10.1016/s0092-8674(00)80488-2. [DOI] [PubMed] [Google Scholar]
- 8.Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF. Proc Natl Acad Sci USA. 2000;97:10723–10728. doi: 10.1073/pnas.97.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kisker C, Schindelin H, Rees DC. Annu Rev Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- 10.Messerschmidt A, Niessen H, Abt DJ, Einsle O, Schink B, Kroneck PMH. Proc Natl Acad Sci USA. 2004;101:11571–11576. doi: 10.1073/pnas.0404378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schink B. Arch Microbiol. 1985;142:295–301. [Google Scholar]
- 12.Rosner BM, Schink B. J Bacteriol. 1995;177:5767–5772. doi: 10.1128/jb.177.20.5767-5772.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streitwieser H, Heathcock CH, Kosower EM. Introduction to Organic Chemistry. New York: Macmillan; 1992. [Google Scholar]
- 14.Yurkanis Bruice P. Organic Chemistry. Upper Saddle River, NJ: Prentice–Hall; 2004. [Google Scholar]
- 15.Burgess BK, Lowe DJ. Chem Rev. 1996;96:2983–3011. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 16.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. Philos Trans R Soc London A. 2005;363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 17.Meckenstock RU, Krieger R, Ensign S, Kroneck PMH, Schink B. Eur J Biochem. 1999;264:176–182. doi: 10.1046/j.1432-1327.1999.00600.x. [DOI] [PubMed] [Google Scholar]
- 18.Chan MK, Mukund S, Kletzin A, Adams MWW, Rees DC. Science. 1995;267:1463–1469. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- 19.Johnson MK, Rees DC, Adams MWW. Chem Rev. 1996;96:2817–2839. doi: 10.1021/cr950063d. [DOI] [PubMed] [Google Scholar]
- 20.Kletzin A, Adams MWW. FEMS Microbiol Rev. 1996;18:5–63. doi: 10.1016/0168-6445(95)00025-9. [DOI] [PubMed] [Google Scholar]
- 21.Einsle O, Niessen H, Abt DJ, Seiffert G, Schink B, Huber R, Messerschmidt A, Kroneck PMH. Acta Crystallogr F. 2005;61:299–301. doi: 10.1107/S174430910500374X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schindelin H, Kisker C, Hilton J, Rajagopalan KV, Rees DC. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 23.Lesk AM. J Mol Graphics. 1995;13:159–164. doi: 10.1016/0263-7855(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 24.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullmann GM, Knapp EW. Eur Biophys J. 1999;28:533–551. doi: 10.1007/s002490050236. [DOI] [PubMed] [Google Scholar]
- 26.Yadav J, Das SK, Sarkar S. J Am Chem Soc. 1997;119:4315–4316. [Google Scholar]
- 27.Einsle O, Tezcan FA, Andrade SL, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 28.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 29.De La Fortelle E, Irwin JJ, Bricogne G. Cryst Comp. 1997;7:1–9. [Google Scholar]
- 30.Abrahams JP, Leslie AGW. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger T. J Synchr Rad. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Guerra CF, Snijders JG, te Velde G, Baerends EJ. Theor Chem Acc. 1998;99:391–403. [Google Scholar]
- 35.Bondi A. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 36.Bashford D. Scientific Computing in Object-Oriented Parallel Environments. Vol 1343. Berlin: Springer; 1997. pp. 233–240. [Google Scholar]
- 37.Bashford D, Gerwert K. J Mol Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 38.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 39.Torres RA, Lovell T, Noodleman L, Case DA. J Am Chem Soc. 2003;125:1923–1936. doi: 10.1021/ja0211104. [DOI] [PubMed] [Google Scholar]
- 40.Beroza P, Fredkin DR, Okamura MY, Feher G. Proc Natl Acad Sci USA. 1991;88:5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bombarda E, Becker T, Ullmann GM. J Am Chem Soc. 2006;128:12129–12139. doi: 10.1021/ja0619657. [DOI] [PubMed] [Google Scholar]
- 42.Weiss MS, Hilgenfeld R. J Appl Crystallogr. 1997;30:203–205. [Google Scholar]
- 43.Cruickshank DWJ. Acta Crystallogr. 1999;D55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]