Abstract
The assembly of proteins that display complementary activities into macromolecular complexes is critical to cellular function. One such enzyme complex, of environmental significance, is the plant cell wall degrading apparatus of anaerobic bacteria, termed the cellulosome. The complex assembles through the interaction of enzyme-derived “type I dockerin” modules with the multiple “cohesin” modules of the scaffolding protein. Clostridium thermocellum type I dockerin modules contain a duplicated 22-residue sequence that comprises helix-1 and helix-3, respectively. The crystal structure of a C. thermocellum type I cohesin-dockerin complex showed that cohesin recognition was predominantly through helix-3 of the dockerin. The sequence duplication is reflected in near-perfect 2-fold structural symmetry, suggesting that both repeats could interact with cohesins by a common mechanism in wild-type (WT) proteins. Here, a helix-3 disrupted mutant dockerin is used to visualize the reverse binding in which the dockerin mutant is indeed rotated 180° relative to the WT dockerin such that helix-1 now dominates recognition of its protein partner. The dual binding mode is predicted to impart significant plasticity into the orientation of the catalytic subunits within this supramolecular assembly, which reflects the challenges presented by the degradation of a heterogeneous, recalcitrant, insoluble substrate by a tethered macromolecular complex.
Keywords: cellulosome structure, cellulosome assembly, Clostridium thermocellum, cohesin-dockerin
The microbial degradation of the plant cell wall is a fundamental biological process that is of considerable, and increasing, industrial importance. This process is critical to the cycling of carbon between microbes, herbivores and plants and the enzymes that catalyze this process are now used in several biotechnology-based industries (1–3). The major and evolving potential application of these biocatalysts, however, is the conversion of plant biomass into bio-ethanol and other forms of energy (4, 5). A common feature of all plant cell wall degrading organisms is that they harness a consortium of enzymes, acting in synergy, to degrade the otherwise recalcitrant substrate (1). The plant cell wall degrading apparatus of aerobic and anaerobic microorganisms, however, differ considerably in their macromolecular organization. The plant cell wall hydrolases synthesized by anaerobes frequently assemble into a large (molecular mass >3 MDa) multienzyme complex termed the “cellulosome” (6, 7).
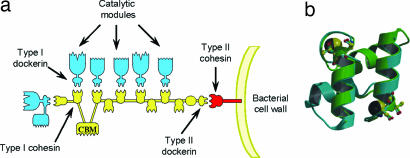
The cellulosome of Clostridium thermocellum is the paradigm for such enzyme complexes (8–12). The grafting of the catalytic entities, primarily glycoside hydrolases but also carbohydrate esterases and polysaccharide lyases, onto the macromolecular scaffold CipA, contributes to enzyme-substrate targeting and enhances the synergistic interactions between the hydrolases. CipA is a noncatalytic protein composed of nine modules known as type I cohesins (13) which display high affinity for the type I dockerins present in the cellulosomally destined plant cell wall degrading enzymes (14). CipA also contains a type II dockerin at its C terminus, which maintains the cellulosome on the bacterial cell surface by its binding to the type II cohesin modules located in proteins anchored to the bacterial proteoglycan layer (15). Significantly, there is no cross-specificity between type I and type II cohesin-dockerin partners (15). Cellulosome assembly is therefore mediated by the interaction of the type I dockerins of the enzymes each with one of the complementary type I cohesin modules of CipA (8–10, 14). In CipA the nine type I cohesins exhibit a high level of sequence identity and the type I C. thermocellum dockerins seem to display little discrimination between their receptors in the protein scaffold of the cellulosome (16, 17) (Fig. 1a). Indeed, the composition of C. thermocellum cellulosomes is dictated by the expression of the ≈70 type I dockerin-containing proteins produced by the bacterium in response to different plant cell wall derived inducers.
Fig. 1.
The cellulosome. (a) Schematic of the cellulosome. The type I dockerins, appended to the catalytic subunits, interact with the cohesin modules on the scaffoldin (CipA) leading to the formation of the supramolecular cellulosome complex. The type II dockerin on CipA, by binding to a type II cohesin on the bacterial membrane, tethers the cellulosome to the surface of C. thermocellum. (b) Internal symmetry of the WT dockerin in complex with cohesin. Not only do residues 1–22 overlap with 35–56, but the reverse is also true, because the dockerin shows internal 2-fold symmetry (panel b adapted from ref. 18).
The crystal structures of C. thermocellum type I (18) and type II (19) cohesin-dockerin complexes have been solved providing insight into the mechanism of cellulosome assembly and cell surface attachment, respectively. In both complexes, cohesin-dockerin recognition is dominated by hydrophobic interactions, augmented through an extensive hydrogen-bonding network. Within the 60-residue C. thermocellum dockerins there is a tandem duplication of a 22-residue sequence, each contributing an α-helix with a calmodulin-like fold. The structure of the complex showed that the type I dockerin binds to its cognate cohesin primarily through its C-terminal α-helix. In the type I complex, the dockerin residues that dominate electrostatic contact with the cohesin are Ser-45 and Thr-46 in the C-terminal helix, whereas the corresponding Ser-Thr pair in the first duplicated sequence (within the N-terminal helix), located at positions 11 and 12, does not contribute to protein-protein interactions in the crystal structure.
One of the most interesting features of the type I cohesin-dockerin complex is that, in addition to sequence homology, the duplicated dockerin regions also display significant structural conservation, with an rmsd for the internally repeated segments of just 0.36 Å for all main-chain atoms. Significantly this structural conservation includes the EF hand motifs and the two Ser-Thr pairs. This structural homology is further manifested in an internal near-perfect two-fold symmetry within the dockerin molecule (Fig. 1b); the structure of the first duplicated segment, which contains the N-terminal helix (helix-1), can be superimposed precisely over the structure of the second segment, containing the C-terminal helix (helix-3), and vice versa. Based on these data we predicted that a 180° rotation of the dockerin would lead to cohesin recognition by the N-terminal helix, in which Ser-11 and Thr-12 would play an equivalent role, in ligand binding, to Ser-45 and Thr-46. To demonstrate and visualize the alternative binding mode the crystal structure of a dockerin mutant, in which the helix-3 Ser/Thr pair (Ser-45-Thr-46) had been substituted with alanine, in complex with its cognate cohesin was determined. The resultant structure at 2 Å resolution indeed shows that the dockerin module interacts with its cognate cohesin through a dual binding mode. The implications of the plasticity in dockerin-cohesin interactions are discussed in light of the functional synergy between the catalytic components of the cellulosome, one of the most efficient plant cell wall degrading systems known.
Results
To produce the cohesin–dockerin S45A–T46A mutant complex (designated Coh-DocS45A-T46A) the genes encoding the WT cohesin and the S45A-T46A dockerin mutant were co-expressed in Escherichia coli, allowing for dockerin stabilization in vivo. The final 2-Å structure of the Coh-DocS45A-T46A complex model comprises 2 × 149-aa residues belonging to two cohesin modules (termed molecules A and C), 2 × 62 residues of the dockerin modules (modules B and D), two phosphate molecules, 2 × 2 calcium atoms, coordinated by residues of the dockerin modules, and 562 water molecules. The two complexes in the asymmetric unit are related by a 2-fold noncrystallographic axis and overlay with an rmsd of 0.3 Å, for 140 aligned residues of the cohesin modules, and 0.2 Å for 62 aligned residues of the dockerin modules [crystallographic statistics are in supporting information (SI) Table 2].
Architecture of the Type I Coh-DocS45A-T46A Complex: A Dual Mode of Binding.
Superimposition of the WT cohesin-dockerin complex (designated Coh-DocWT) and Coh-DocS45A-T46A reveals that the structure of the cohesin (rmsd of 0.5 Å for 140 Cα residues) and dockerin (rmsd of 0.5 Å, for 55 aligned residues) modules are very similar in the two crystal structures (Fig. 2). In the WT and mutant complexes, the type I cohesin contains nine β-strands, which form two β-sheets aligned in an elongated β-barrel that displays a classical jelly roll fold. The first β-sheet comprises β-strands 5, 6, 3 and 8 and defines the interacting surface with the dockerin, whereas β-strands 4, 7, 2, 1 and 9 define the second β-sheet. Comparison of both type I or type II cohesin structures of C. thermocellum, solved either as discrete entities (10, 20–25), or in complex with the corresponding dockerin modules (18, 19), show that the conformation of both cohesins do not change upon binding its protein ligand. Coh-DocS45A-T46A contains three α-helices (1–3) and two calcium ions coordinated by residues at the C- and N-terminal ends of the dockerin that display typical EF-hand Ca2+-binding motifs. Calcium coordination in Coh-DocS45A-T46A is essentially identical to the Coh-DocWT complex reported previously (18), and details of this coordination are provided in supplementary information (SI Table 3).
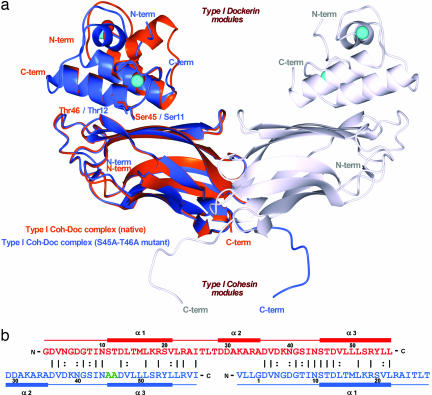
Fig. 2.
The dual binding mode of the Xyn10B dockerin. (a) Ribbon representation of the superposition of the type I Coh-DocWT complex (in orange) with its S45A-T46A mutant complex (in blue). In the mutant complex, helix-1 (containing Ser-11 and Thr-12) dominates binding whereas, in the WT complex, helix-3 (containing Ser-45 and Thr-46) plays a key role in ligand recognition. Ser-11, Thr-12, Ser-45, and Thr-46, which interact with the cohesin module, are depicted as stick models and colored accordingly. The second molecule of the mutant complex, generated by the 2-fold NCS, is represented in light-gray ribbon. The Ca2+ ions are depicted as spheres and colored orange, in the case of the WT complex, and light blue, in the case of the mutant. The N- and C-terminal ends are labeled and colored accordingly. (b) The structure-based sequence alignment of the WT (in red) and S45A-T46A mutant (in blue) type I dockerins. Mutated residues, Ala-45 and Ala-46, are shown in green. Because of internal 2-fold symmetry of each dockerin module, the two structures overlap almost perfectly in their α1/α3 regions. The N- and C-terminal ends of each module are indicated, as well as the α-helix regions. Numbering is indicated for every 10th residue.
The dockerin in the Coh-DocS45A-T46A structure presents the symmetry observed for the WT module in Coh-DocWT (18), with α-helices 1 and 3 rotated 180° with respect to each other and overlapping almost perfectly, Fig. 2a. Although interpreting electron density maps of a protein that displays dyad symmetry can be problematic, the following differences in the duplicated sequence of the dockerin in Coh-DocWT and Coh-DocS45A-T46A, respectively, enabled the orientation of the protein to be unambiguously assigned: Gly-5/Lys-39, Ser-11/Ala-45, Thr-12/Ala-46, Thr-15/Leu-49, Met-16/Leu-50, Lys-18/Ser-52 and Ser-20/Tyr-54. The orientation of the dockerin in the two protein complexes indicate that α-helices 1 and 3 indeed play equivalent roles in cohesin recognition. Consistent with this view is the observation that in Coh-Doc S45A-T46A the major region of the dockerin that interacts with its cohesin partner is not the C-terminal helix-3, as occurs in the WT complex, but helix-1, Fig. 2a. As expected, the two alanine residues (Ala45B and Ala46B) created by the mutation, are now very distant from, and thus do not interact with, the cohesin. Indeed, Ser-11B and Thr-12B in the mutant protein, fulfil the same role as Ser-45B and Thr-46B in the WT dockerin (see Fig. 2b) making extensive hydrogen bonds with its cohesin partner (see below).
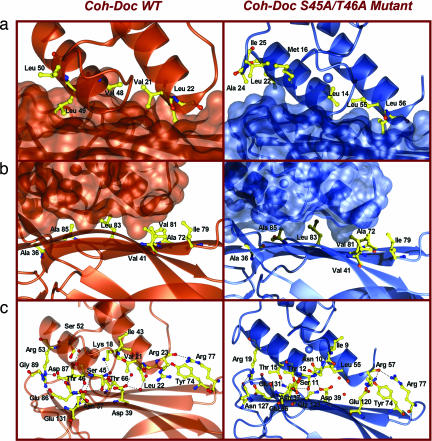
The equivalent hydrophobic character of the interactions between the protein partners is also evident in both complexes (Fig. 3a and SI Table 4). The cohesin hydrophobic residues participating in complex formation (Ala-36A, Val-41A, Ala-72A, Ile-79A, Val-81A, Leu-83A, Ala-85A and Leu-129A from β-strands 3, 5, and 6) remain unchanged in Coh-DocWT and Coh-DocS45A-T46A (Fig. 3). In both the WT and mutant dockerin the equivalent residues from α-helix 1 and 3 make hydrophobic contacts with the cohesin. Thus, in the dockerin mutant S45A-T46A, Leu-55B and Leu-56B from helix-3 and the equivalent residues in helix-1 of the WT protein (Val-21 and Leu-22, respectively) make analogous hydrophobic interactions with the cohesin. Similarly, Leu-14B, Thr-15B and Met-16B from helix-1 of the dockerin mutant, and the corresponding amino acids in helix-3 of the WT protein (Val-48, Leu-40 and Leu-50, respectively) make apolar interactions with aliphatic residues in the cohesin (Fig. 3a and SI Table 4).
Fig. 3.
The Coh-Doc interface of the native (in orange) and S45A-T46A mutant (in blue) type I complexes. (a) Stick representation of the hydrophobic residues on the surface of the cohesin modules (in ribbon representation). The dockerin modules are represented by their molecular surfaces. (b) Stick representation of the hydrophobic residues on the surface of the dockerin modules (in ribbon representation). The cohesin modules are represented by their molecular surfaces. (c) Stick representation of the hydrogen-bond network in the interface of the Coh-DocS45A-T46A complex (in ribbon representation). Carbon atoms are shown in yellow, oxygens are shown in red, and nitrogens are shown in blue. All pictures were produced with the CCP4 mg program (42).
The hydrogen-bonding network between the dockerin and cohesin in the two complexes is also very similar; the equivalent residues in helix 3 and helix 1 of the WT versus the mutant dockerins, respectively, make near-identical hydrogen bonds with cohesin residues (Fig. 3c and SI Table 5). The only differences in the hydrogen bonding network in the mutant and WT cohesin-dockerin complexes are as follows: In the S45A-T46A dockerin mutant Asn-10B (Nδ2) interacts with Glu-131A (Οε2), Thr-15B (Oγ1) hydrogen bonds to Asn-37A (Nδ2), Lys-18B (Nζ) interacts with the carbonyls of Ala-85A and Asp-87A, whereas Arg-57Β Nη2 makes a hydrogen bond with the carboxylate of Glu120A. The equivalent residues in the WT dockerin; Asn-44B, Thr-49B, Ser-52B and Arg-23B (Nη2), respectively, do not make direct interactions with the cohesin (SI Table 5). Mutagenesis studies support the importance of the polar interactions between the cohesin residues Ala-36A, Asn-37A, Asp-39A, Tyr-74A, Glu-86A, Gly-89A and Glu-131A (26, 27) and type I dockerins, although the functional significance of Arg-77A, which seems to make important hydrogen bonds with Arg-23B and Arg-57B at the C-terminal end of the two dockerin binding sites, has not previously been explored.
In addition to the direct polar interactions there are also several solvent-mediated hydrogen bonds, which are generally conserved in the two complexes (Fig. 3). Whereas the role of indirect hydrogen bonds in protein ligand recognition is controversial (28, 29), mutating the cohesin residue Asp-39A, which makes solvent-mediated hydrogen bonds with the backbone carbonyl and amines of the dockerin residues Ile-43B, Ile-19B, Val-21B and Leu-55B, reduces affinity 1000-fold (26), suggesting that these interactions may be important in cohesin-dockerin binding. Furthermore, the cohesin residue Thr66A, which makes a solvent-mediated hydrogen bond with Lys18B, when the C-terminal binding site of the dockerin interacts with its protein ligand, is not invariant in the CipA cohesins. In cohesins 3, 4, 5, 6 and 8 this residue is replaced by an Asp. It remains to be established whether this amino acid replacement influences the affinity of these CipA cohesins for the C-terminal binding site of type I dockerins.
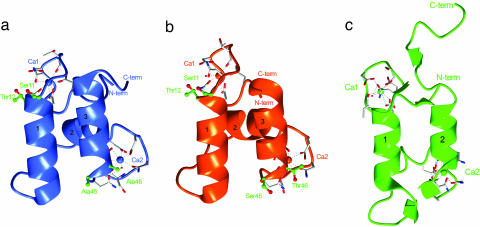
The recent resolution of the type II cohesin-dockerin complex, which recruits the C. thermocellum cellulosome onto the surface of the bacterium, reveals structural similarities with the corresponding type I proteins (19). Comparison of the type I and type II dockerins, bound to their cognate cohesin partners, reveal differences in the relative position of the two helices, which results in marked differences at the complex interface, Fig. 4. Upon complex formation the two helices of the type II dockerin contact the cohesin by forming a parallel interacting hydrophobic platform. The multiple contacts made with the cohesin module by both helices and, decisively, the lack of symmetry of type II dockerin amino acids at the interface (19) indicates that the module is unlikely to display the dual binding mode exhibited by the corresponding type I module.
Fig. 4.
The type I and type II dockerin modules. (a) The S45A-T46A mutant type I dockerin module. (b) The WT type I dockerin from the type I Coh-DocWT complex (PDB code 1ohz). (c) The type II dockerin from the type II Coh-Doc complex (PDB code 2b59). The α-helices in each module are numbered. Residues in positions 11, 12, 45, and 46 are shown as ball-and-stick models (carbon atoms in green and oxygen atoms in red) and labeled in green. The calcium ions in each module are colored according to structure and labeled as Ca1 and Ca2. Residues coordinating each calcium ion are depicted as stick models (carbon atoms are shown in white, oxygens are shown in red, and nitrogens are shown in blue). The N- and C-terminal ends are labeled and colored accordingly.
Relative Affinity of the Two Binding Sites of the Dockerin for Its Cohesin Ligand.
The structural data presented above supports the hypothesis that type I dockerins contain two, highly conserved, cohesin-binding surfaces, consistent with mutagenesis data showing that cohesin recognition is only disrupted when both Ser-Thr pairs are replaced with bulky amino acids or when multiple mutations are introduced into both of the duplicated segments of the dockerin (refs. 27 and 30 and Table 1). The observation that there is no significant change in affinity when either of the Ser-Thr pairs is mutated on their own (Table 1 and refs. 27 and 30) indicates that both binding sites display similar affinity for the cohesin partner. It is interesting to note that the apparently more polar nature of ligand binding via helix 1 is entropically driven, whereas the more hydrophobic dockerin helix-3 cohesin interaction is associated with a gain in enthalpy (27). Whereas it is currently unclear why the thermodynamic forces driving ligand binding are not reflected in the nature of the amino acids that mediate cohesin-dockerin recognition, the thermodynamic parameters are likely to be influenced by changes in solvation, which cannot easily be explained by static crystal structures.
Table 1.
Binding of WT and mutants of the Xyn10B dockerin to cohesins
| Dockerin | Cohesin | Ka, M−1 | Stoichiometry |
|---|---|---|---|
| WT | CipA Cohesin-2 | 8 × 107 | 1.0 |
| S11A T12A | CipA Cohesin-2 | 7 × 107 | 1.1 |
| S45A T46A | CipA Cohesin-2 | 7 × 107 | 1.1 |
| S11A T12A S45A T46A | CipA Cohesin-2 | 8 × 107 | 0.9 |
| S11Q S45Q | CipA Cohesin-2 | 2 × 106 | 1.1 |
| S11L T12LS45Q | CipA Cohesin-2 | 5 × 105 | 1.0 |
| WT | EAM46162 | 8 × 108 | 1.2 |
| WT | OlpA | 3 × 107 | 1.0 |
T = 65°C.
Inspection of the draft genome sequence of C. thermocellum revealed the presence of 71 polypeptides containing type I dockerins (31). Indeed, the residues in the two cohesin-binding sites in the Xyn10B dockerin are highly conserved in most of the identified dockerins. The one exception to this high degree of conservation is the residue corresponding to Arg-19B (N-terminal helix) or Arg-53B (C-terminal helix), which is often substituted for a lysine, although this amino acid change is unlikely to influence the functionality of either the N- or C-terminal ligand-binding site. More dramatic variation from this high degree of conservation is observed in the dockerins of Cel9D-Cel44A and two cellulosomal proteins of unknown function (accession numbers BAA12070, EAM47344 and EAM44539, respectively), where the first Ser-Thr pair is replaced by Ala/Asp-Val/Ile/Glu, whereas the other residues participating in cohesin recognition at both binding sites are conserved. In a fourth cellulosomal polypeptide (accession number EAM46149) the second Ser-Thr pair is replaced by the residues Asp-Ile. The introduction of these mutations into helix-1 (Ala/Asp-Val/Ile/Glu) and helix-3 (Asp-Ile) destroys cohesin recognition by the N- and C-terminal binding faces, respectively. These type I C. thermocellum dockerins will therefore interact with their cohesin partners via only a single surface, whereas the remaining 67 modules will contain two sites that display very similar affinities for the protein ligand.
The crystal structure reported here sheds light on the biochemical properties of cohesin and dockerin mutants. The crystal structure of the mutant also clarifies the role of Arg-57B in cohesin recognition. Mutagenesis studies suggest that the equivalent residue to Arg-57B plays a key role in cohesin recognition (30), but in the WT cohesin-dockerin complex this amino acid is disordered. Consistent with the proposal of Carvalho et al. (18) the data reported here show that Arg-57B plays an equivalent role to Arg-23B when cohesin recognition is mediated by the N-terminal binding site. The most surprising feature of the mutagenesis studies reported here (Table 1) and elsewhere (30) is that substitution of both Ser-Thr pairs with small residues does not cause a dramatic reduction in affinity, even though these hydroxy amino acids make a significant contribution to the hydrogen-bonding network between the dockerin and cohesin partner. Crystals of cohesin bound to the N-terminal dockerin binding site were only obtained when the S45A-T46A double mutation was introduced into the dockerin (18) suggesting that the orientation of binding is exquisitely poised such that although mutation weakens binding only slightly, the population is modified sufficiently to reverse the cohesin recognition trapped crystallographically. Only by replacing both of these Ser/Thr pairs with bulky amino acids could ligand binding be significantly reduced (Table 1 and ref. 27).
Type I Dockerins Bind to Cohesins with a Stoichiometry of 1.
To determine the stoichiometry of the cohesin-dockerin complex, the binding of the Xyn10B dockerin, fused to the enzyme's CBM22, to the cohesin was determined by using isothermal titration calorimetry (ITC). The data revealed a stoichiometry of 1 indicating that both ligand-binding sites on the dockerin cannot be occupied simultaneously. Overlaying Coh-DocWT and Coh-DocS45A-T46A indicates that two cohesin molecules would indeed be unable to bind to a single dockerin molecule simultaneously because residues in the cohesin loop extending from Pro-63 to Ser-68 will make steric clashes. Inspection of the nine type I cohesins in CipA show that this sequence is completely conserved. It would seem, therefore, that all of the CipA cohesin modules bind to the dockerins of the catalytic subunits with a stoichiometry of one. In addition to CipA, three membrane-bound C. thermocellum proteins also contain type I cohesins in which the loop predicted to cause the steric clash, discussed above, is truncated, and thus these modules may be able to interact with the two dockerin ligand binding sites simultaneously. To test this hypothesis, two of these proteins were produced in Escherichia coli and their binding to the Xyn10B dockerin determined by ITC. The data reveal that the three cohesins bind to the Xyn10B dockerin with a stoichiometry of one (Table 1) indicating that all of the type I C. thermocellum cohesins cannot occupy the two dockerin ligand-binding sites simultaneously.
Discussion
The construction of multiprotein complexes is one of the key emerging fields in modern chemistry. The clostridial cellulosome is a stunning example of a naturally occurring multienzyme complex in which key catalytic elements may be grafted onto a framework scaffold. The cohesin-dockerin interaction is central to the cellulosomal architecture; both in integrating the enzyme components into the complex, and in its anchoring to the cell surface. Within a given cellulosome-producing species such as C. thermocellum, the conservation of the amino acid sequence between the two repeated units of the dockerin is remarkable. Intriguingly, crystallographic data pointed to asymmetric binding of type I dockerins to its protein ligand, even though these modules displayed structural symmetry consistent with the observed sequence duplication. Here we have demonstrated that the internal symmetry of the dockerin is not merely structural but also functional. By changing the Ser-Thr dyad that was previously shown to interact with the cohesin (18), the equilibrium between the two binding modes was altered sufficiently to allow the mutated dockerin to interact with the cohesin via the alternative Ser-Thr pair in the N-terminal helix.
The extremely tight sequence conservation within the dockerins of C. thermocellum, which results in >90% identity in the residues involved in calcium coordination and cohesin interaction, demonstrates that there is a strong selective advantage for the duplication of the two dockerin segments leading to an internal structural symmetry and a dual mode of cohesin binding. The functional significance of this plasticity of cohesin-dockerin recognition in cellulosome assembly is thus intriguing, not only in light of cellulose assembly and function but as a template for nonnatural engineered protein complexes elsewhere. Although a dual binding mode points to a single dockerin interacting with two cohesin molecules simultaneously, biochemical and structural studies presented in this report indicate that steric clashes between the cohesin molecules would prevent the formation of such a tri-molecular complex.
The dual binding mode displayed by type I dockerins will confer flexibility in cellulosome assembly. Although there is no selectivity between specific cohesin-dockerin partners (16, 17, 32), the steric constraints imposed by the appended catalytic (and/or ancillary) modules will restrict the combination of enzymes that can be assembled into a single cellulosome complex. The dual binding mode displayed by dockerins is predicted to overcome these steric constraints and thus increase the range of enzymes that can be integrated into discrete cellulosome molecules. This flexibility in the arrangement of catalytic subunits within cellulosomes is important as different (and potentially temporally evolving) enzyme combinations are required to degrade the myriad of composite structures displayed by plant cell walls (1). Indeed, the incorporation of complementary enzyme activities into single cellulosomes potentiates the synergistic interactions between these biocatalysts and is a key element for the optimization of plant cell wall degradation (33–35). In addition, the switching of the mode of binding from one site to another can introduce quaternary flexibility into the multienzyme complex and enhance substrate targeting and hydrolysis. For example, the optimal juxtaposition of the catalytic subunits could vary during the degradative process, and the flexibility introduced by binding face switching can contribute to the optimization of the quaternary structure of the enzyme complex for the specific composite substrate presented to the bacterium. For example, the established synergistic interaction between endo- and exo-acting cellulases demands that these enzymes attack the same cellulose microfibril. The dual binding mode displayed by the dockerin modules facilitates the optimization of the orientation of these enzymes such that they can attack a common microfibril structure. In that sense the cellulosome is an unusual protein complex in that it consists of a tethered enzymatic microarray acting upon a solid substrate. The dual orientation facilitated by the twin faces of the dockerin should allow the flexibility in position that is always harnessed by free enzyme systems.
One of the key issues raised by the demonstration that the vast majority of C. thermocellum type I dockerins display structural and functional dyad symmetry is the generic significance of these findings. Inspection of type I dockerins appended to the catalytic subunits of cellulosomes from a variety of organisms reveals a similar sequence duplication as observed in the C. thermocellum modules (8, 10). Therefore, the dual binding mode displayed by the C. thermocellum type I dockerins seems to be replicated in other bacterial dockerins and thus we propose that the plasticity in dockerin-cohesin recognition is fundamental to the mechanism by which these complexes catalyze plant cell wall degradation.
Methods
Cloning and Expression.
Previously we obtained the WT Coh-Doc complex from the coexpression of the genes encoding the xylanase 10B dockerin (residues 733–791) and CipA cohesin-2 (residues 182–328) organized in tandem, in plasmid pCF1 (18). Recombinant cohesin contained a C-terminal His6 tag. The Coh-Doc S45A-T46A complex was generated from pCF2, which is an engineered version of pCF1 produced by using the PCR-based QuikChange site-directed mutagenesis kit (Stratagene). The encoded dockerin contains the three N-terminal dockerin residues, Val-730-Leu-731-Leu-732, and the mutations S45A and T46A. To study cohesin-dockerin binding, the dockerin was also expressed independent of its cohesin partner, fused to a His-tagged family 22 carbohydrate binding module (CBM22) (18), whereas the His-tagged CipA cohesin-2 was also expressed from a plasmid lacking the dockerin sequence (20). DNA encoding additional type I cohesins, derived from the C. thermocellum membrane protein (OlpA) and an ORF identified from the draft C. thermocellum genome (EAM46162) were amplified by PCR and cloned into the BamHI and EcoRI cloning sites of the T7-based E. coli expression vector pRSET, which supplies an N-terminal His-tag to the recombinant protein. The various cohesin-dockerin constructs were expressed in E. coli BL21 at 37°C and cell pellets were prepared by using standard methodology (18).
Protein Purification.
The Coh-Doc S45A-T46A complex was purified by metal-ion affinity chromatography (18), buffer exchanged into 20 mM Tris·HCl, pH 8.0, containing 2 mM CaCl2, and then further purified by anionic exchange chromatography by using a Source 30Q column and a gradient elution of 0–1 M NaCl (Amersham Biosciences), to separate the complex from unbound cohesin. Fractions containing the complex were buffer exchanged and then concentrated in 2 mM CaCl2 to a final concentration of 5 mg/ml. The discrete His-tagged cohesin and dockerin-CBM22 were purified by metal-ion affinity chromatography (18, 20).
ITC.
ITC was carried out essentially as in (18). The WT and mutants of the Xyn10B dockerin fused to CBM22 (20 μΜ) were retained in the reaction cell and was titrated with 25 × 10-μl aliquots of various cohesin proteins (350 μΜ) in the syringe, at 65°C in 50 mM NaHepes (pH 7.5) containing 2 mM CaCl2. Integrated heat effects, after correction for heats of dilution, were analyzed by nonlinear regression by using a single-site binding model (Microcal ORIGIN Ver. 5.0, Microcal Software, Northampton, MA). The fitted data yield the association constant (KA) and the enthalpy of binding (ΔH). Other thermodynamic parameters were calculated by using the standard thermodynamic equation: −RTlnKA = ΔG = ΔH − TΔS. The c values (product of the molar concentration of binding × the association constant) were ≈100.
Complex Crystallization, Data Collection and Structure Determination.
The S45A-T46A Coh-Doc complex was crystallized by using vapor diffusion techniques and the hanging drop method. With the protein at a concentration of 5 g/liter, crystals suitable for subsequent analysis were obtained from 20% (wt/vol) PEG 8000 in the presence of 0.05 M of potassium dihydrogen phosphate. Crystals were flash-cooled in a nitrogen stream at 110 K, incorporating 30% (vol/vol) glycerol as cryoprotectant, and data were collected to 2Å resolution on European Synchrotron Radiation Facility (ESRF; Grenoble, France) beamline ID14-3. Data (SI Table 2), were processed by using programs MOSFLM (36) and SCALA (37) from the CCP4 suite (38) and 10% of the observations flagged for cross-validation. Crystals are in space group P21 with cell dimensions a = 48.6 Å, b = 92.6 Å, and c = 49.9 Å, with a β = 94.0 °. The structure was solved by molecular replacement, by using the program MOLREP (CCP4 suite; ref. 39) with the type I cohesin module alone (PDB accession code 1ohz; ref. 18) which yielded a solution with two cohesins in the asymmetric unit. Manual inspection of the electron density map, clearly revealed the presence of two S45A-T46A mutated dockerin molecules, one bound to each of the two cohesin molecules. The dockerin modules were built manually by using the COOT program (40) and the two complexes refined by using REFMAC5 (41). Water molecules were added and final refinement included translation, libration and screw-rotation of the four independent groups (molecules A, B, C, and D).
Supplementary Material
Acknowledgments
We thank the staff of the European Synchrotron Radiation Facility (Grenoble, France). This work was funded by Fundação para a Ciência e a Tecnologia Grant POCI/BIA-PRO/59118/2004 and the Biotechnology and Biological Sciences Research Council. F.M.V.D. is supported by Fundação para a Ciência e a Tecnologia (PraxisXXI/BD/21250/1999). E.A.B. acknowledges financial support from Israel Science Foundation Grant 442/05.
Abbreviations
- Coh
cohesin
- Doc
dockerin
- ITC
isothermal titration calorimetry.
Footnotes
The authors declare no conflict of interest.
Data deposition: Coordinates and observed structure factor amplitudes for the Coh-Doc S45A-T46A mutant complex, to 2 Å resolution, have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 2CCL).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611173104/DC1.
References
- 1.Warren RA. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Himmel ME, Ruth MF, Wyman CE. Curr Opin Biotechnol. 1999;10:358–364. doi: 10.1016/S0958-1669(99)80065-2. [DOI] [PubMed] [Google Scholar]
- 3.Bhat MK. Biotechnol Adv. 2000;18:355–383. doi: 10.1016/s0734-9750(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 4.Boudet AM, Kajita S, Grima-Pettenati J, Goffner D. Trends Plants Sci. 2003;8:576–581. doi: 10.1016/j.tplants.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Jr, Hallett JP, Leak DJ, Liotta CL, et al. Science. 2006;11:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 6.Lamed R, Bayer EA. Adv Appl Microbiol. 1988;33:1–46. [Google Scholar]
- 7.Lamed R, Setter E, Kenig R, Bayer EA. Biotechnol Bioeng Symp. 1983;13:163–181. [Google Scholar]
- 8.Bayer EA, Belaich JP, Shoham Y, Lamed R. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 9.Beguin P, Lemaire M. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 10.Shoham Y, Lamed R, Bayer EA. Trends Microbiol. 1999;7:275–281. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 11.Demain AL, Newcomb M, Wu JH. Microbiol Mol Biol Rev. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi RH, Kosugi A. Nat Rev Microbiol. 2004;2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- 13.Gerngross UT, Romaniec MP, Kobayashi T, Huskisson NS, Demain AL. Mol Microbiol. 1993;8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 14.Salamitou S, Raynaud O, Lemaire M, Coughlan M, Beguin P, Aubert JP. J Bacteriol. 1994;176:2822–2827. doi: 10.1128/jb.176.10.2822-2827.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibovitz E, Beguin P. J Bacteriol. 1996;178:3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciruela A, Gilbert HJ, Ali BR, Hazlewood GP. FEBS Lett. 1998;422:221–224. doi: 10.1016/s0014-5793(97)01590-1. [DOI] [PubMed] [Google Scholar]
- 17.Yaron S, Morag E, Bayer EA, Lamed R, Shoham Y. FEBS Lett. 1995;360:121–124. doi: 10.1016/0014-5793(95)00074-j. [DOI] [PubMed] [Google Scholar]
- 18.Carvalho AL, Dias FM, Prates JA, Nagy T, Gilbert HJ, Davies GJ, Ferreira LMA, Romao MJ, Fontes CMG. Proc Natl Acad Sci USA. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams JJ, Pal G, Jia Z, Smith SP. Proc Natl Acad Sci USA. 2006;103:305–310. doi: 10.1073/pnas.0507109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho AL, Pires VM, Gloster TM, Turkenburg JP, Prates JA, Ferreira LMA, Romao MJ, Davies GJ, Fontes CM, Gilbert HJ. J Mol Biol. 2005;349:909–915. doi: 10.1016/j.jmb.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 21.Spinelli S, Fierobe HP, Belaich A, Belaich JP, Henrissat B, Cambillau C. J Mol Biol. 2000;304:189–200. doi: 10.1006/jmbi.2000.4191. [DOI] [PubMed] [Google Scholar]
- 22.Tavares GA, Beguin P, Alzari PM. J Mol Biol. 1997;273:701–713. doi: 10.1006/jmbi.1997.1326. [DOI] [PubMed] [Google Scholar]
- 23.Noach I, Frolow F, Jakoby H, Rosenheck S, Shimon LW, Lamed R, Bayer EA. J Mol Biol. 2005;348:1–12. doi: 10.1016/j.jmb.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Noach I, Lamed R, Xu Q, Rosenheck S, Shimon LJ, Bayer EA, Frolow F. Acta Crystallogr D Biol Crystallogr. 2003;59:1670–1673. doi: 10.1107/s0907444903014094. [DOI] [PubMed] [Google Scholar]
- 25.Shimon LJ, Bayer EA, Morag E, Lamed R, Yaron S, Shoham Y, Frolow F. Structure (London) 1997;5:381–390. doi: 10.1016/s0969-2126(97)00195-0. [DOI] [PubMed] [Google Scholar]
- 26.Handelsman T, Barak Y, Nakar D, Mechaly A, Lamed R, Shoham Y, Bayer EA. FEBS Lett. 2004;572:195–200. doi: 10.1016/j.febslet.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 27.Schaeffer F, Matuschek M, Guglielmi G, Miras I, Alzari PM, Beguin P. Biochemistry. 2002;41:2106–2114. doi: 10.1021/bi011853m. [DOI] [PubMed] [Google Scholar]
- 28.Flint J, Bolam DN, Nurizzo D, Taylor EJ, Williamson MP, Walters C, Davies GJ, Gilbert HJ. J Biol Chem. 2005;280:23718–23726. doi: 10.1074/jbc.M501551200. [DOI] [PubMed] [Google Scholar]
- 29.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 30.Mechaly A, Fierobe HP, Belaich A, Belaich JP, Lamed R, Shoham Y, Bayer EA. J Biol Chem. 2001;276:9883–9888. doi: 10.1074/jbc.M009237200. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz WH, Zverlov VV, Bahl H. Adv Appl Microbiol. 2004;56:215–261. doi: 10.1016/S0065-2164(04)56007-0. [DOI] [PubMed] [Google Scholar]
- 32.Lytle B, Myers C, Kruus K, Wu JH. J Bacteriol. 1996;178:1200–1203. doi: 10.1128/jb.178.4.1200-1203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fierobe HP, Bayer EA, Tardif C, Czjzek M, Mechaly A, Belaich A, Lamed R, Shoham Y, Belaich JP. J Biol Chem. 2002;277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 34.Fierobe HP, Mechaly A, Tardif C, Belaich A, Lamed R, Shoham Y, Belaich JP, Bayer EA. J Biol Chem. 2001;276:21257–21261. doi: 10.1074/jbc.M102082200. [DOI] [PubMed] [Google Scholar]
- 35.Fierobe HP, Mingardon F, Mechaly A, Belaich A, Rincon MT, Pages S, Lamed R, Tardif C, Belaich JP, Bayer EA. J Biol Chem. 2005;280:16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 36.Leslie AGW. Protein Crystallogr. 1992;26:27–33. [Google Scholar]
- 37.Kabsch W. J Appl Crystallogr. 1988;21:67–71. [Google Scholar]
- 38.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 39.Vagin A, Tepliakov A. J Applied Cryst. 1997;30:1022–1025. [Google Scholar]
- 40.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. Acta Crystallogr D. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.