Abstract
Plasmacytoid dendritic cells (pDCs) play a central role in innate and adaptive immune responses against viral infections. pDCs secrete type I IFNs and proinflammatory cytokines upon stimulation by either TLR7 or TLR9. Throughout the course of HIV infection, the production of type-I IFNs is profoundly impaired, and total pDC cell counts in peripheral blood correlates inversely with viral load and positively with CD4+ T cell count. The origin of these defects is unclear. pDCs express CD4, CCR5, and CXCR4, the primary receptor and coreceptors, respectively, for the HIV envelope; yet little is known concerning the effects of the viral envelope on these cells. Here, we show that exposure of pDCs to gp120 results in the suppression of activation of these cells. This suppression is specific for TLR9-mediated responses, because TLR7-mediated responses are unaffected by gp120. gp120 also suppressed TLR9-mediated induction of proinflammatory cytokines and expression of CD83, a marker of DC activation. Finally, gp120 suppressed pDC-induced cytolytic activity of natural killer cells. Taken together, these data demonstrate that the direct interaction of HIV-1 gp120 with pDCs interferes with TLR9 activation resulting in a decreased ability of pDCs to secrete antiviral and inflammatory factors that play a central role in initiating host immune responses against invading pathogens.
Keywords: CpG, interferon α
Plasmacytoid dendritic cell (pDCs), which are found in both peripheral blood and in T cell-rich areas of secondary lymphoid tissues (1, 2), play a central role in immune responses against viral infections. pDCs are the principal producers of Type 1 interferons (IFN-α/β) (3). These cytokines exhibit potent antiviral activity insofar as they regulate the responses of numerous cell subsets involved in both innate and adaptive immune responses against viral pathogens. pDCs respond to viruses and other pathogens primarily through the recognition of pathogen-associated molecular patterns (PAMPs), by two intracellular Toll-like receptors (TLRs), TLR7 and TLR9. The former recognizes single stranded RNA (4), whereas the latter recognizes unmethylated DNA motifs (5). The engagement of TLR7 and TLR9 by PAMPS activates pDCs to rapidly produce high levels of type 1 IFNs and moderate amounts of inflammatory cytokines, including TNF-α and IL-6 (6). Through TLR-activation, human pDCs promote B cell-, T cell- and natural killer (NK) cell-mediated immune responses. The interaction between pDCs and NK cells has a particularly profound effect on innate immune responses. Activation of pDCs, after engagement of TLR-9, activates autologous NK cells and enhances their cytolytic activity (7, 8). In this regard, pDCs have been identified as the non-T, non-B, nonmonocytic cell type required for NK cell-mediated killing of virus-infected cells or tumor cell lines (9), and pDC-derived IFN-α exerts a dominant role in this interaction (10). The role of pDCs in HIV disease is a subject of great interest. HIV infection results in a decrease in the number of circulating pDCs (11, 12). Levels of circulating pDCs correlate inversely with plasma viral load and directly with CD4+ T cell counts (12, 13). These effects can be partially reversed by antiretroviral therapy (14). Additionally, pDCs isolated from individuals acutely infected with HIV exhibit an impaired ability to secrete type 1 IFNs (15, 16). The underlying cause(s) of these alterations in pDC numbers and activity, as a result of HIV infection, are unknown. However, given the central role that these cells play in both innate and adaptive antiviral immune responses, understanding the mechanisms whereby pDCs interact with and respond to HIV may provide fundamental insights into HIV pathogenesis. pDCs can be infected with HIV in vitro, but productive infection is achieved only by using high-titer stocks (17, 18), and it is unlikely that in vivo direct infection accounts for the altered pDC activity described above. Incubation of pDCs with HIV-1 virions induces their maturation and the production of IFNα. This response is driven by TLR7 recognition of HIV ssRNA (19). However, additional interactions between pDCs and virion components, including the viral envelope, may also impact pDC maturation and function. In this regard, pDCs express CD4, CCR5, and CXCR4, all of which are signal-transducing ligands for the viral envelope protein gp120. gp120 has been shown to transduce intracellular signals in CD4+ T cells and macrophages (20, 21) and can have a profound effect on the function and viability of these cells (22, 23). Yet, little is known about gp120-mediated effects on pDCs, although one report indicates that HIV envelope protein promotes IFN-α secretion (24).
In this report, we examined the effect of gp120 on pDCs function. We found that gp120 disrupts TLR9-mediated activation in a relatively specific manner. gp120 treatment suppressed TLR-9-induced secretion of type-1 IFNs and of other inflammatory cytokines. Functionally, pDCs exposed to gp120 exhibited a reduced capacity to induce cytotoxic activity in NK cells. As a possible mechanism of gp120-mediated interference with the IFN pathway, we describe its binding to BDCA-2, a C-type lectin receptor expressed on the surface of pDCs.
Results
gp120 Inhibits TLR9-, but Not TLR7-. Mediated Secretion of IFN-α from pDCs.
Upon endocytosis of HIV-1 virions, pDCs are activated and secrete IFN-α (19). Although this response requires virus capture by CD4 receptors on the surface of pDCs, several reports suggest that activation and IFN-α secretion occur as a consequence of intracellular TLR7 recognition of viral ssRNA (19). However, one study reports that gp120 alone, in the absence of RNA, can trigger pDC activation and IFN-α secretion (24). To better understand the effect of exposure of pDCs to HIV gp120, we cultured freshly isolated pDCs for 18 h in the presence or absence of either an R5 or an X4 gp120 recombinant protein. Using a sensitive multisubtype INF-α ELISA, we were unable to detect gp120-induced IFN-α (Fig. 1a). In contrast, cultures treated with CpGs induced high levels of IFN-α (Fig. 1a). Considering that gp120 disrupts the maturation of monocyte-derived DC (25), we sought to determine whether gp120 would also disrupt TLR-mediated activation and maturation of pDCs. As described above, freshly isolated pDCs were treated with CpGs, which activate pDCs through TLR9 (26), or alternatively with Imiquimod, an imidazoquinoline compound that activates pDCs through TLR7 (27), in the presence or absence of gp120. As expected, overnight exposure to CpG alone induced significant levels of IFN-α; however, concomitant treatment with an R5 gp120 reduced the CpG-induced-IFN-α levels by >50% (Fig. 1b). Similar effects were observed with an X4 gp120 (data not shown). This reduction was significant relative to CpG-treated cells in five experiments (P = 0.006). gp120 is presented as a trimer on HIV virions (28). To determine whether this form of gp120 induced similar effects, pDCs were exposed to gp120 trimer in the presence of CpG, and an even greater suppression of IFN-α secretion was observed (Fig. 1b). Additionally, we evaluated a recombinant simian immunodeficiency virus (SIV) gp120 derived from the sooty mangabey virus SIVsmm/PBj (29). This envelope protein suppressed IFN-α levels to a greater degree than any of the HIV-1 gp120s tested (Fig. 1b). Finally, as a control, we treated cells with CpG and a recombinant control protein, in this instance sCD4, to demonstrate that the suppression we observed was not a general phenomenon resulting from the exposure to any recombinant protein; as expected, we observed no significant effect on IFN-α secretion (Fig. 1b). Subsequently we evaluated several concentrations of gp120 ranging from 200 nM to 1 nM, and determined that its effect on IFN-α secretion was concentration-dependent and that, in the majority of the experiments, 10 nM gp120 consistently reduced IFN-α by at least 50% (data not shown). In marked contrast, gp120 exerted no effect on TLR7-mediated induction of IFN-α (Fig. 1c). pDCs treated overnight with Imiquimod induced significant levels of IFN-α, and neither monomeric nor trimeric gp120 suppressed this induction [the differences in the amount of the IFN-α secreted were not significant (P > 0.05)]. Thus, the gp120-mediated suppression of TLR9-IFN-α secretion was not the result of a generalized effect on pDCs. Indeed, because there is a substantial overlap in TLR7 and TLR9 signal transduction pathways (30), gp120 disruption of TLR9 stimulation would appear to reflect a relatively specific effect. In addition to CpG, we tested the effect of gp120 on the stimulation of pDCs by HSV-2, a naturally occurring TLR9 agonist (5). Although virus alone induced INF-α, inclusion of gp120 in cultures together with HSV-2 reduced IFN-α secretion to an undetectable level (Fig. 1d). Finally, we determined whether IFN-β secretion was also suppressed by gp120. Both CpG and HSV-2 induced IFN-β, and the addition of gp120 to cultures suppressed IFN-β secretion by both of these TLR9 agonists (Fig. 1e).
Fig. 1.
IFN-α production induced by TLR9-ligands, but not by a TLR7-ligand is inhibited by both monomeric and trimeric gp120. (a) pDCs were treated for with CpG (5 μg/ml), R5-gp120 (100 nM), or mock-treated with PBS. IFN-α in the supernatants was detected by using human IFN-α multisubtype ELISA. (b) IFN-α produced from freshly isolated pDC treated with CpGs (500 ng/ml) in presence or absence of R5-gp120 monomer or trimer (10 nM), SIV monomer (10 nM), or control recombinant protein (10 nM). (c) IFN-α produced from pDCs treated with the TLR 7 ligand Imiquimod (1 μg/ml) in the presence or absence of R5-gp120 monomer or trimer (gp120tr) (10 nM). (d) IFN-α produced from pDCs exposed to HSV-2 for 18 h in the presence or absence of HIV gp120. (e) IFN-β produced from pDCs exposed to HSV-2, or treated with CpG, in the presence or absence of gp120. Error bars represent SDs calculated from replicates. Results shown are representative of at least five independent experiments
gp120 Inhibits TLR9-Mediated Up-Regulation of CD83 and the Secretion of Inflammatory Cytokines.
Both CD83 and CD86 are membrane receptors that are expressed at very low (CD86) or undetectable (CD83) levels on immature pDCs but are up-regulated upon activation of these cells by TLR ligands (6). CD86 is one of the principal costimulatory receptors on APCs (31), whereas the function of CD83 is largely unknown (32). Of note, CD83 up-regulation after culture of DC isolated from HIV-infected individuals is reduced relative to DCs isolated from healthy donors (33). We tested the effect of gp120 on TLR-mediated induction of both CD83 and CD86. R5 gp120 suppressed TLR9-mediated induction of CD83 (Fig. 2a) (P = 0.007). A recombinant R5 trimer exerted a stronger suppressive effect, whereas X4 gp120 suppressed CD83 in a manner comparable with an R5 gp120 (data not shown). In contrast, none of the recombinant envelopes tested suppressed TLR9-mediated induction of CD86 (Fig. 2b) (P > 0.05). TLR7-mediated induction of CD83 and CD86 were unaffected by exposure of pDCs to gp120 (data not shown), consistent with the inhibitory activity of gp120 on TLR9-, but not TLR7-, mediated induction of IFN-α. Along with IFN-α, activated pDCs secrete moderate amounts of other inflammatory cytokines, including TNF-α, IL1β, IL4, IL6, IL12p70, and IP10. Using a flow cytometry-based bead array assay (detection limit 3.9 pg/ml), we were able to detect only TNF-α, IL6, and the chemokine IP10 after TLR9-mediated stimulation. As with IFN-α, gp120 significantly suppressed the production of all three cytokines (Fig. 3) (P < 0.05). The gp120 trimer was notably more potent, suppressing TNF-α secretion by >70%.
Fig. 2.
gp120 suppresses TLR9-mediated up-regulation of CD83, but not CD86, on pDCs. pDCs were treated with CpGs in the presence or absence of R5-gp120 monomer or trimer and stained with PE anti-CD83 (a) or CD86 (b) mAbs and analyzed by flow-cytometry. Mean fluorescence intensities (MFI) were averaged from three different experiments. Error bars represent SDs calculated on them.
Fig. 3.
Secretion of inflammatory cytokines induced by TLR9 stimulation in pDCs is inhibited by gp120. Concentrations of TNF-α, IL6, and IP10 in the supernatants of pDC cultures with CpGs in the presence or absence of R5-gp120 monomer or trimer were detected by using BD Cytometric Bead Array (CBA) cell signaling flex sets. SDs were calculated from replicates. Results shown are representative of at least five independent experiments.
gp120 Interferes with pDC-Driven NK Cell Cytotoxicity.
NK cells respond to viral infections by producing IFN-γ and lysing infected cells (10). This antiviral response requires NK cell activation, and pDCs play an important role in this activation. Viruses stimulate pDCs through either TLR7 or TLR9, and they respond by secreting IFN-α and TNF-α, which, in turn, contribute to the activation of NK cells (7, 8). In light of our observations that gp120 can suppress TLR9-mediated secretion of IFN-α and TNF-α (Figs. 1 and 3), we sought to determine whether these effects were of sufficient magnitude to impact pDC-driven NK cell cytolytic activity. We activated pDCs overnight with CpG in the presence or absence of gp120. Cells were then washed to remove gp120 from the culture medium. Freshly isolated autologous NK cells were then added for an additional 24 h, after which an NK sensitive cell line, K562, was added to the culture. Cell killing was measured after 2 hours by using a flow cytometry-based assay. In good agreement with previous reports, cytolytic activity was clearly enhanced when NK cells were cocultured with CpG-stimulated pDCs relative to control cultures lacking pDCs (Fig. 4). Of note, we observed a significant reduction in the cytolytic activity of NK cells activated with pDCs that were treated with CpG in the presence of gp120 relative to pDCs that were treated with CpG in the absence of gp120 (P < 0.05). Both monomeric (Fig. 4a) and trimeric (Fig. 4b) gp120 mediated this suppression, consistent with the capacity of both proteins to suppress the secretion of IFN-α and TNF-α. We cannot rule out the possibility that additional pDC activation factors were dysregulated by gp120 treatment and also played a role in the reduced activation of NK cells. Nevertheless the reduced capacity of NK cells to kill targets when CpG-treated pDC are exposed to gp120 underscores the potential of gp120 to disrupt the network of antiviral immune responses in which pDCs are involved.
Fig. 4.
NK cell cytotoxicity induced by TLR9-stimulated pDCs is inhibited by gp120. pDCs were treated with CpGs in the presence or absence of an R5 gp120 monomer (a) or trimer (b). Stimulated pDCs were added to homologous NK cell cultures. NK cells cultured in the absence of pDCs were included as a control. PKH67-K562 target cells were added to cultures at ratios: 1:2, 1:1, and 2:1 (NK:K562). Frequency of cell killing was determined by flow cytometric measurement of the number of K562 cells staining positively for PKH67-green and propidium iodide. NK cytotoxicity is reported as the percentage of dead K562 cells calculated by subtracting in each condition the percentage of dead cells of the untreated control (K562 cultured without NK cells). SDs were calculated on the replicates.
gp120 Interacts with pDCs Through CD4-Dependent and -Independent Mechanisms.
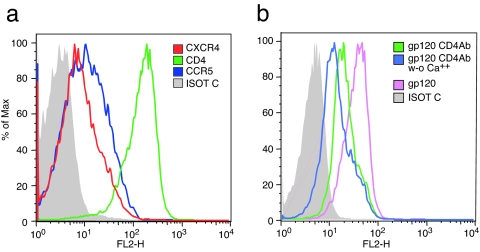
To better understand the mechanisms underlying gp120-mediated disruption of pDC function, we investigated the interactions between gp120 and pDC surface receptors. Most DCs subtypes express CD4 and the HIV coreceptors CXCR4 and CCR5. In addition, gp120 is also susceptible to capture by several antigen-capturing C-type lectin receptors that are expressed on various DCs and APCs. These include DC-SIGN, expressed on myeloid DCs, langerin on Langerhans cells, and the macrophage mannose receptor expressed on macrophages (34). All of these receptors bind to gp120 carbohydrate residues, but none of the above-mentioned receptors is present on pDCs. We measured both CD4-dependent and -independent binding on pDCs. Consistent with previous reports, flow-cytometric analysis of these cells demonstrated expression of CD4 as well as CCR5 and CXCR4 (Fig. 5a). pDCs were also stained with a biotin-conjugated R5-tropic gp120, followed by a fluoresceine conjugate. High levels of gp120 binding were detected on pDCs (Fig. 5b). To determine how much of this binding was CD4-mediated, cells were preincubated with a blocking anti-CD4 mAb, Leu3a. Although binding was reduced in the presence of Leu3A, a significant level of residual, non CD4-mediated binding remained (Fig. 5b), indicating that gp120 binds other unidentified receptors on pDCs. Because binding to C-type lectin receptors is calcium-dependent (35), we then compared CD4-independent binding in the presence or absence of Ca2+ (Fig. 5b) and noted a significant reduction in CD4-independent binding in the absence of Ca2+. We conclude that gp120 binds pDCs in both CD4-dependent and -independent mechanisms.
Fig. 5.
gp120 binds pDCs through both CD4-dependent and Ca2+-dependent interactions. (a) Freshly isolated pDC were stained with anti-CD4, anti-CCR5, and anti-CXCR4 mAbs or an isotype control mAb. (b) Freshly isolated pDC were preincubated with an unlabeled anti-CD4, gp120-blocking mAb (Leu3a), and stained with biotin-gp120- streptavidin-PE in the presence or absence of Ca2+.
gp120 Binds to the C-Type Lectin Receptor BDCA-2.
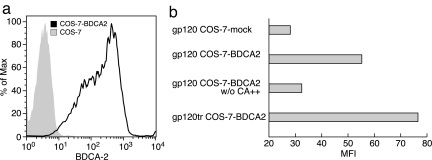
BDCA-2 is a C-type lectin receptor expressed on pDCs, which functions primarily as an antigen-capturing receptor (36). It has been reported that BDCA-2 ligation and cross-linking results in the inhibition of CpG-mediated induction of IFN-α/β (36, 37). Given the results presented above, we investigated the possibility that HIV-1 gp120 binds this C-type lectin receptor. In preliminary experiments, we consistently observed a partial reduction of the gp120 binding to the surface of pDCs in the presence of an anti-BDCA-2 mAb (data not shown). To determine directly whether gp120 binds BDCA-2, we transiently transfected COS-7 cells with a BDCA-2 expression vector. After 48 h, a high level of BDCA-2 expression was achieved (Fig. 6a). We reproducibly observed low but significant levels of gp120 binding to the BDCA-2-transfected cells (P = 0.03), but not mock-transfected cells, and this binding was inhibited by the removal of Ca2+ from the staining buffer (Fig. 6b). Trimeric gp120 bound to BDCA-2 transfected cells to a greater degree (Fig. 6b), suggesting that the affinity of gp120–BDCA-2 interactions are facilitated by avidity effects associated with envelope trimers.
Fig. 6.
gp120 binds BDCA-2 COS-7-transfected cells. (a) BDCA-2 expression on BDCA-2-transfected COS-7 cells vs. mock-transfected cells. gp120 binding to mock-transfected and BDCA-2-transfected cells in the presence or absence of Ca2+. (b) gp120-trimer binding to BDCA-2-transfected cells. Results are representative of three independent experiments.
BDCA-2-mediated inhibition of IFN-α/β secretion has been linked to tyrosine phosphorylation (36). We asked whether gp120 treatment of pDCs also promoted tyrosine phosphorylation of intracellular substrates. Overnight-cultured pDCs were treated with either gp120 or an irrelevant control protein (sCD4) for 30 min, after which cells were lysed, and the tyrosine phosphorylation of two substrates, p38 and ERK1/2, was monitored with phosphospecific antibodies by flow cytometry. CpG DNA, which is known to induce the phosphorylation of these two substrates, was used as a positive control. Both gp120 and CpG DNA mediated p38 and ERK1/2 tyrosine phosphorylation, whereas the control protein did not [supporting information (SI) Fig. 7). This observation is consistent with BDCA-2-mediated signaling. Although we cannot rule out the possibility that gp120 is also signaling through CD4, under similar conditions, CD4 ligation does not induce tyrosine phosphorylation of intracellular substrates on pDCs (38).
Discussion
As the principal producers of IFN-α, pDCs play a central role in antiviral immune responses (3). Type I interferons regulate a broad range of responses involving T, B, and NK cells. In HIV disease, these cell populations exhibit profound functional defects (39). The potential role of pDCs in immune dysfunction in HIV has therefore generated substantial interest. As Bhardwaj and colleagues have elegantly shown, pDCs respond to HIV RNA through TLR7 (19). To better understand the specific effects of gp120 ligation, in the absence of other viral components, including viral RNA, we treated freshly isolated pDCs with several recombinant R5 and X4 gp120 proteins, including a trimeric R5 envelope. Under the conditions we used, none of these gp120s induced IFN-α (Fig. 1a), consistent with the observations of Beignon et al. (19). These results differ from a report by Gessani and colleagues (24) who did observe gp120-induced IFN-α. This discrepancy may reflect the fact that those experiments we carried out in the presence of IL3, which can predispose pDCs to secrete IFN-α (40). gp120 treatment did however produce specific alterations in pDC function. When pDCs were treated simultaneously with gp120 and either a TLR9 agonist (CpG) or HSV-2, TLR9-mediated responses were blunted. We observed reduced secretion of IFNs, TNF-α, and IL6 and reduced up-regulation of CD83. We further demonstrated that NK cells, which are activated by pDCs to kill target cells, exhibited reduced cytolytic activity after exposure of CpG-treated pDCs to gp120. These effects did not result from reduced pDC viability, as evidenced by our observation that CD86 up-regulation in untreated vs. gp120-treated pDCs was unchanged. Moreover, gp120 treatment had no discernible effect on pDC responses to TLR7-induced secretion of cytokines, or the up-regulation of cell surface markers of activation. These dichotomous effects on TLR9- vs. TLR7-mediated activation are surprising insofar as the downstream signaling pathways associated with these two types of stimulation are largely overlapping. Of note, TLR7 and TLR9 pathways differ in their sensitivity to chloroquine (41). Additional studies are required to understand the mechanism by which gp120 disrupts TLR9-, but not TLR7-, mediated activation of pDCs.
Because pDCs express high levels of CD4 and significant levels of CCR5 and CXCR4, gp120-mediated signal transduction through these receptors may contribute to the responses described in this report. Although gp120 signaling through these receptors is well documented in lymphocytes (20, 21), little is know about gp120-mediated signaling through these receptors on pDCs. In addition, in this report, we demonstrate that BDCA-2, a C-type lectin, which is expressed on pDCs, binds HIV-1 gp120 in a Ca2+-dependent manner, raising the possibility that this interaction contributes to suppression of IFN-α production. Two lines of investigation suggest this to be the case. First, two recent reports show that BDCA-2 ligation with an anti-BDCA-2 antibody inhibits CpG DNA-induced IFN-α production (36, 37). Second, we observed gp120-mediated tyrosine phosphorylation of intracellular substrates, which occurs as a consequence of BDCA-2 ligation, but not CD4 ligation (38). Thus, gp120-mediated suppression of CpG-induced IFN-α secretion appears to result directly from ligation of BDCA-2. Interestingly, signaling through BDCA-2 interferes with the nuclear translocation of IRF-7, one of the principal transcription factors involved in the regulation of IFN-α production (42). Although we could not identify CD4-mediated signaling in pDCs, cross-linking CD4 has also been reported to suppress CpG-mediated IFN-α secretion (38). Thus, the unique capacity of gp120 to bind both CD4 and BDCA-2 may contribute to the suppression of IFN-α secretion in response to TLR9, but not TLR7, stimulation.
In conclusion, pDCs respond to HIV in a complex manner that includes effects mediated by both viral RNA and the HIV envelope. HIV virions induce pDCs to secrete IFN-α through TLR7 recognition of viral RNA (19). In this report, we show that, in addition to viral RNA, pDCs also respond to the viral envelope protein in a manner that renders them less able to respond to DNA viruses and other TLR9-stimulating pathogens, including bacteria. Thus, direct interactions between HIV and pDCs may contribute to chronic activation of the immune system and simultaneously suppress responses to specific opportunistic infections. In light of the central role that these cells play in regulating the immune system, the results reported herein may provide insight into the role of HIV–pDC interactions in HIV-driven dysfunction of the immune system.
Materials and Methods
Cells and Reagents.
pDCs were obtained from PBMCs derived from healthy donors by using a BDCA-4 Miltenyi selection kit (Miltenyi Biotec, Auburn CA). Cell purities routinely exceeded 95% by flow cytometric analysis using a BDCA-2 mAb. Cells were used immediately after selection, without preculture. Homologous NK cells were isolated from elutriated lymphocytes by using a negative selection enrichment kit (Stemcell Technologies, Vancouver BC, Canada). NK purity was >98%. The K-562 cell line was obtained from the American Type Culture Collection (Manassas, VA) (43). The TLR9-ligands (TLR9L), CpG A (ODN2216), and CpG C (ODN2395) were obtained from Invivogen (San Diego, CA) and were used interchangeably (unless otherwise specified) at 500 ng/ml. The TLR7-ligand (TLR7L) Imiquimod (Invivogen) was used at 1 μg/ml. Recombinant gp120 proteins (JR-FL, 92Ug037, 92Ug021, and SIV PBj1.9) were produced and purified as described (44). The trimeric recombinant gp120 92Ug037 was constructed by using the fibritin domain of phage T7 (45). Purified envelope proteins underwent three successive Triton X-114 extractions to remove trace endotoxins (46) and were tested for endotoxin with the Limulus amoebocyte lysate (LAL) assay (BioWhittaker, Walkersville, MD) (<0.1 unit/μg) gp120 was biotinylated by using NHS-EZ link Biotin Reagents (Pierce, Rockford IL). Anti-CD4 PE, anti-CCR5 PE, anti-CXCR4 PE, anti-CD83 PE, anti-CD86 PE, or Streptavidin FITC and PE were from BD Biosciences (San Diego, CA), anti-BDCA-2 PE and APC were from Miltenyi Biotec. CD4 mAb and Leu3A were from BD Biosciences (San Jose, CA). FACS staining buffer: 10 mM Hepes, 150 mM NaCl, 10 mM CaCl, 0.09% Na Azide, 2% Fetal Bovine Serum (FBS). Fluorescence was measured on a BD FACSCalibur (BD Biosciences, San Jose).
Cytokine Analysis.
Supernatants were collected 18 h after stimulation. IFN-α and IFN-β were determined by ELISA using multisubtype IFN-α and IFN-β ELISAs (PBL Biomedical, Piscataway NJ) per the manufacturers instructions. TNF-α, IL1β, IL4, IL6, IL10, IL12p70 and IP10 were measured via Cytometric Bead Array (CBA) cell signaling flex sets (BD Biosciences San Jose CA).
NK Cytotoxicity Assay.
NK cell cytotoxicity assay was carried out as previously described with minor modifications (47, 48). Briefly, freshly isolated pDCs were stimulated with TLR agonists with or without gp120 as described above. Autologous NK cells were kept overnight at 4C°. After 18 h, pDCs were washed to remove gp120, and NK cells were added at a ratio of 1 pDC per 2 NK cells for an additional 18 h. Cells were then washed, and PKH72-green labeled K562 cells were added to the culture for an additional 2 h. Cells were washed and resuspended in staining buffer containing propidium iodide (PI). The frequency of killed K562 cells was calculated as the number of (PI+/PKH72+ cells)/(PI+/PKH72+ cells in the absence of NK cells).
Transfection of Cells with BDCA-2 Plasmid.
COS-7 cells were transiently transfected by using Profectin (Qiagen, Valencia CA) following the manufacturer's instructions. The human BDCA-2 coding sequence was synthesized (DNA 2.0, Menlo Park CA) and inserted into pCMV 3.0 (Promega, Madison, WI). Surface expression of BDCA-2 was checked with BDCA-2-PE (Miltenyi Biotech) and gp120 binding was assessed with biotinylated gp120 after 24 and 48 h by using BD FACSCalibur (BD Biosciences).
Statistical Analysis.
Data were analyzed by using Prism 4 Biostatistic software comparing all of the experiment performed in absence of gp120 with the ones performed in presence of gp120. Significance was evaluated by using Student's or paired t test at P < 0.05. The analysis on the NK cell cytotoxicity assays was performed on replicates in each experiment.
Supplementary Material
Acknowledgments
We acknowledge the National Institute of Allergy and Infectious Diseases AIDS reagent repository for supplying numerous reagents used in this study.
Abbreviations
- pDC
plasmacytoid dendritic cell
- gp120tr
gp120 trimer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611353104/DC1.
References
- 1.Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. J Exp Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Penna G, Vulcano M, Sozzani S, Adorini L. Hum Immunol. 2002;63:1164–1171. doi: 10.1016/s0198-8859(02)00755-3. [DOI] [PubMed] [Google Scholar]
- 3.Asselin-Paturel C, Brizard G, Chemin K, Boonstra A, O'Garra A, Vicari A, Trinchieri G. J Exp Med. 2005;201:1157–1167. doi: 10.1084/jem.20041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YJ. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 7.Romagnani C, Della Chiesa M, Kohler S, Moewes B, Radbruch A, Moretta L, Moretta A, Thiel A. Eur J Immunol. 2005;35:2452–2458. doi: 10.1002/eji.200526069. [DOI] [PubMed] [Google Scholar]
- 8.Gerosa F, Gobbi A, Zorzi P, Burg S, Briere F, Carra G, Trinchieri G. J Immunol. 2005;174:727–734. doi: 10.4049/jimmunol.174.2.727. [DOI] [PubMed] [Google Scholar]
- 9.Trinchieri G, Santoli D, Dee RR, Knowles BB. J Exp Med. 1978;147:1299–1313. doi: 10.1084/jem.147.5.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall JD, Heeke DS, Abbate C, Yee P, Van Nest G. Immunology. 2006;117:38–46. doi: 10.1111/j.1365-2567.2005.02261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacanowski J, Kahi S, Baillet M, Lebon P, Deveau C, Goujard C, Meyer L, Oksenhendler E, Sinet M, Hosmalin A. Blood. 2001;98:3016–3021. doi: 10.1182/blood.v98.10.3016. [DOI] [PubMed] [Google Scholar]
- 12.Soumelis V, Scott I, Gheyas F, Bouhour D, Cozon G, Cotte L, Huang L, Levy JA, Liu YJ. Blood. 2001;98:906–912. doi: 10.1182/blood.v98.4.906. [DOI] [PubMed] [Google Scholar]
- 13.Pacanowski J, Develioglu L, Kamga I, Sinet M, Desvarieux M, Girard PM, Hosmalin A. J Infect Dis. 2004;190:1889–1892. doi: 10.1086/425020. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt B, Fujimura SH, Martin JN, Levy JA. J Clin Immunol. 2006;26:55–64. doi: 10.1007/s10875-006-8401-3. [DOI] [PubMed] [Google Scholar]
- 15.Kamga I, Kahi S, Develioglu L, Lichtner M, Maranon C, Deveau C, Meyer L, Goujard C, Lebon P, Sinet M, et al. J Infect Dis. 2005;192:303–310. doi: 10.1086/430931. [DOI] [PubMed] [Google Scholar]
- 16.Muller-Trutwin M, Hosmalin A. Immunol Cell Biol. 2005;83:578–583. doi: 10.1111/j.1440-1711.2005.01394.x. [DOI] [PubMed] [Google Scholar]
- 17.Patterson S, Rae A, Hockey N, Gilmour J, Gotch F. J Virol. 2001;75:6710–6713. doi: 10.1128/JVI.75.14.6710-6713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt B, Scott I, Whitmore RG, Foster H, Fujimura S, Schmitz J, Levy JA. Virology. 2004;329:280–288. doi: 10.1016/j.virol.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. J Clin Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman F, Jensen WA, Johnson GL, Heasley L, Cambier JC. J Immunol. 1994;153:2905–2917. [PubMed] [Google Scholar]
- 21.Hivroz C, Mazerolles F, Soula M, Fagard R, Graton S, Meloche S, Sekaly RP, Fischer A. Eur J Immunol. 1993;23:600–607. doi: 10.1002/eji.1830230303. [DOI] [PubMed] [Google Scholar]
- 22.Kinter M, Spitz DR, Roberts RJ. J Nutr. 1996;126:2952–2959. doi: 10.1093/jn/126.12.2952. [DOI] [PubMed] [Google Scholar]
- 23.Masci AM, Galgani M, Cassano S, De Simone S, Gallo A, De Rosa V, Zappacosta S, Racioppi L. J Leukoc Biol. 2003;74:1117–1124. doi: 10.1189/jlb.0503239. [DOI] [PubMed] [Google Scholar]
- 24.Del Corno M, Gauzzi MC, Penna G, Belardelli F, Adorini L, Gessani S. J Virol. 2005;79:12597–12601. doi: 10.1128/JVI.79.19.12597-12601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fantuzzi L, Purificato C, Donato K, Belardelli F, Gessani S. J Virol. 2004;78:9763–9772. doi: 10.1128/JVI.78.18.9763-9772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. J Exp Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 28.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 29.Fultz PN, McClure HM, Anderson DC, Switzer WM. AIDS Res Hum Retroviruses. 1989;5:397–409. doi: 10.1089/aid.1989.5.397. [DOI] [PubMed] [Google Scholar]
- 30.Perry AK, Chen G, Zheng D, Tang H, Cheng G. Cell Res. 2005;15:407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- 31.Harris NL, Ronchese F. Immunol Cell Biol. 1999;77:304–311. doi: 10.1046/j.1440-1711.1999.00835.x. [DOI] [PubMed] [Google Scholar]
- 32.Lechmann M, Berchtold S, Hauber J, Steinkasserer A. Trends Immunol. 2002;23:273–275. doi: 10.1016/s1471-4906(02)02214-7. [DOI] [PubMed] [Google Scholar]
- 33.McIlroy D, Autran B, Clauvel JP, Oksenhendler E, Debre P, Hosmalin A. AIDS Res Hum Retroviruses. 1998;14:505–513. doi: 10.1089/aid.1998.14.505. [DOI] [PubMed] [Google Scholar]
- 34.Turville S, Wilkinson J, Cameron P, Dable J, Cunningham AL. J Leukoc Biol. 2003;74:710–718. doi: 10.1189/jlb.0503208. [DOI] [PubMed] [Google Scholar]
- 35.Weis WI, Taylor ME, Drickamer K. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 36.Dzionek A, Sohma Y, Nagafune J, Cella M, Colonna M, Facchetti F, Gunther G, Johnston I, Lanzavecchia A, Nagasaka T, et al. J Exp Med. 2001;194:1823–1834. doi: 10.1084/jem.194.12.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerkmann M, Rothenfusser S, Hornung V, Towarowski A, Wagner M, Sarris A, Giese T, Endres S, Hartmann G. J Immunol. 2003;170:4465–4474. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- 38.Fanning SL, George TC, Feng D, Feldman SB, Megjugorac NJ, Izaguirre AG, Fitzgerald-Bocarsly P. J Immunol. 2006;177:5829–5839. doi: 10.4049/jimmunol.177.9.5829. [DOI] [PubMed] [Google Scholar]
- 39.Fauci AS. J Acquir Immune Defic Syndr. 1993;6:655–662. [PubMed] [Google Scholar]
- 40.Fuchs A, Cella M, Kondo T, Colonna M. Blood. 2005;106:2076–2082. doi: 10.1182/blood-2004-12-4802. [DOI] [PubMed] [Google Scholar]
- 41.Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, Raz E, Cottam HB. Proc Natl Acad Sci USA. 2003;100:6646–6651. doi: 10.1073/pnas.0631696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 43.Lozzio BB, Lozzio CB. Int J Cancer. 1979;24:513. doi: 10.1002/ijc.2910240421. [DOI] [PubMed] [Google Scholar]
- 44.Cicala C, Arthos J, Martinelli E, Censoplano N, Cruz CC, Chung E, Selig SM, Van Ryk D, Yang J, Jagannatha S, et al. Proc Natl Acad Sci USA. 2006;103:3746–3751. doi: 10.1073/pnas.0511237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letarov AV, Londer YY, Boudko SP, Mesyanzhinov VV. Biochemistry (Moscow) 1999;64:817–823. [PubMed] [Google Scholar]
- 46.Aida Y, Pabst MJ. J Immunol Methods. 1990;132:191–195. doi: 10.1016/0022-1759(90)90029-u. [DOI] [PubMed] [Google Scholar]
- 47.Gupta N, Arthos J, Khazanie P, Steenbeke TD, Censoplano NM, Chung EA, Cruz CC, Chaikin MA, Daucher M, Kottilil S, et al. Virology. 2005;332:491–497. doi: 10.1016/j.virol.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Kottilil S, Shin K, Jackson JO, Reitano KN, O'Shea MA, Yang J, Hallahan CW, Lempicki R, Arthos J, Fauci AS. J Immunol. 2006;176:1107–1114. doi: 10.4049/jimmunol.176.2.1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.