Abstract
Transport proteins constitute ≈10% of most proteomes and play vital roles in the translocation of solutes across membranes of all organisms. Their (dys)function is implicated in many disorders, making them frequent targets for pharmacotherapy. The identification of substrates for members of this large protein family, still replete with many orphans of unknown function, has proven difficult, in part because high-throughput screening is greatly complicated by endogenous transporters present in many expression systems. In addition, direct structural studies require that transporters be extracted from the membrane with detergent, thereby precluding transport measurements because of the lack of a vectorial environment and necessitating reconstitution into proteoliposomes for activity measurements. Here, we describe a direct scintillation proximity-based radioligand-binding assay for determining transport protein function in crude cell extracts and in purified form. This rapid and universally applicable assay with advantages over cell-based platforms will greatly facilitate the identification of substrates for many orphan transporters and allows monitoring the function of transport proteins in a nonmembranous environment.
Keywords: membrane protein, neurotransmitter:sodium symporter, scintillation proximity, substrate binding
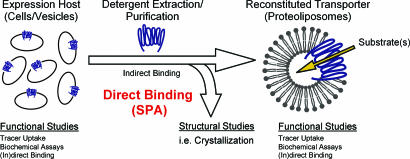
Membrane transport proteins fulfill an essential function in every living cell by catalyzing the translocation of solutes, including ions, nutrients, neurotransmitters, and numerous drugs, across biological membranes. Their (mal)function is directly implicated in many diseases including autism, epilepsy, migraine, depression, drug abuse, and cystic fibrosis, and they play an important role in the success or lack of success of cancer chemotherapy. Hence, they are of primary medical/pharmacological interest for target-oriented drug discovery and delivery. Their function is routinely studied in suitable expression hosts and, if feasible, after reconstitution into proteoliposomes. Direct biophysical and structural studies, including crystallization, however, require that these integral membrane proteins be solubilized and purified (Fig. 1). Between their extraction from the membrane and reconstitution, transport activity cannot be measured because of the lack of a vectorial environment. As a consequence, the determination of their function in detergent has been limited to indirect binding studies, such as substrate protection against cysteine modification (1–3) or substrate-induced changes in tryptophan fluorescence (3, 4), approaches that require the fortuitous or engineered localization of cysteines or tryptophans as well as laborious development and implementation.
Fig. 1.
Monitoring the function of membrane transport proteins outside of the membrane. Schematic representation of the analyses of membrane transport proteins in different states. See Introduction for details.
To overcome this significant bottleneck in membrane transporter proteomics, we developed a direct radiotracer-binding assay using scintillation proximity (5–8) to monitor the function of transport proteins in crude membrane extracts and in purified form. Because this enormous family of membrane transport proteins is still replete with many hypothetical proteins for which the substrates are not known, this rapid, simple, and universally applicable format will also allow the identification of substrates for orphan transporters by simple screening in crude membrane extracts without the need for testing or engineering of expression hosts in which endogenous transporters have been eliminated.
Results
Functional Assays in Crude Membrane Extracts.
As a model system for this study, we used the recombinant His10-tagged Na+/tyrosine transporter (Tyt1) of Fusobacterium nucleatum in conjunction with the copper chelate affinity-based scintillation proximity assay (SPA) to optimize the expression host, the choice of detergent and purification technique, and efficient reconstitution conditions of fully functional purified transporter in proteoliposomes. Tyt1 is a member of the neurotransmitter:sodium symporter (NSS) family (9–11) that, analogous to the majority of heterologously expressed transport proteins, was expressed and characterized in a suitable Escherichia coli host (9). However, our extensive attempts to reconstitute purified Tyt1 isolated from a number of E. coli hosts were unsuccessful. Because it is not possible by using only transport assays to determine the amount of functional transporter in the cytoplasmic membrane, we hypothesized that most of the Tyt1 detected by immunoblotting or protein staining was nonfunctional and that the small fraction of functional transporter in E. coli accounted for the robust transport activity. This hypothesis was supported by the observation that when the Tyt1 mutant was expressed in E. coli C41(DE3) (12) or MQ614 (9), the substrate tyrosine failed to significantly protect from reaction with the thiol reagent 2-(aminoethyl)methane thiosulfonate (MTSEA) [see supporting information (SI) Text] a single cysteine substituted for Ile-104 (index position 3.46 (see refs. 13 and 14 for indexing scheme), which lines the substrate-binding site of the homologous tryptophan transporter TnaT (N. R. Goldberg and J.A.J., unpublished work) and of LeuTAa (11). Thus, the predominant species of Tyt1 expressed in the E. coli inner membrane appears to be nonfunctional, suggesting that the expressed protein is not suitable for biochemical studies of the detergent-solubilized and/or purified protein.
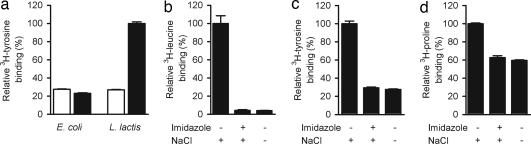
In contrast, when the same Tyt1 mutant was expressed in Lactococus lactis NZ9000 (15), substantial tyrosine protection against reaction of the single cysteine at position 104 with MTSEA was detectable (data not shown.), suggesting that a much higher fraction of protein is functional in this expression system. Consistent with this hypothesis, using the metal chelate-based SPA, we measured robust Na+-dependent tyrosine binding to recombinant (His10-tagged) Tyt1 expressed in L. lactis, whereas the same recombinant Tyt1 variant expressed in E. coli C41(DE3) (12) lacked this binding activity (Fig. 2a). Of note, by using the SPA, we were able to detect binding of 3H compounds in crude membrane extracts of E. coli and L. lactis hosts expressing other members of the NSS family, LeuTAa (11) and an uncharacterized homologue, MhsT of Bacillus halodurans, as well as a member of the sodium:solute symporter family (16), the Na+/proline transporter (PutP) (17) (Fig. 2 b, c, and d, respectively).
Fig. 2.
Detection of transporter function in nonmembranous environment. (a) Optimization of the expression host. The activity of Tyt1 solubilized from membrane vesicles of L. lactis NZ9000 or E. coli CD41(DE3) was assayed for binding of 1 μM l-[3H]tyrosine by means of scintillation proximity (filled bars; n = 3). Membrane vesicles of L. lactis NZ9000/pNZ8048 or E. coli CD41(DE3)/pQE60 served as control (open bars; n = 3). Binding of (b) 0.1 μM l-[3H]leucine to crude membranes extracts of E. coli C41(DE3) containing LeuTAa, (c) 1 μM l-[3H]tyrosine to membrane extracts of L. lactis NZ9000 harboring MhsTBh, or (d) 1 μM l-[3H]proline to solubilized membranes of E. coli WG170 harboring PutP was detected outside of a membranous environment (n = 3). (b–d) Binding was assayed in the presence of 150 mM NaCl (NaCl), 150 mM NaCl plus 150 mM imidazole (Imidazole), or in the absence of NaCl. The results of the latter two conditions were not significantly different from those observed with sample originating from control vesicles (data not shown). Data were expressed as percentage of the highest signal without background correction. Specific 3H-substrate binding activity (after background correction) was (in pmol/μg solubilized membrane protein) 1.04 ± 0.09 [3H]leucine for LeuTAa (b), 0.074 ± 0.004 [3H]tyrosine for MhsTBh (c), and 0.033 ± 0.002 [3H]proline for PutP (d). All scintillation proximity binding assays shown in this report were performed in the T = 0 assay format (simultaneous incubation of protein sample, radiotracer, and SPA beads; see manufacturer's instructions for details). Performing the radiotracer binding step with the respective vesicles before solubilization (i.e., delayed SPA format) led to similar levels of binding (data not shown).
Optimization of Functional Assays.
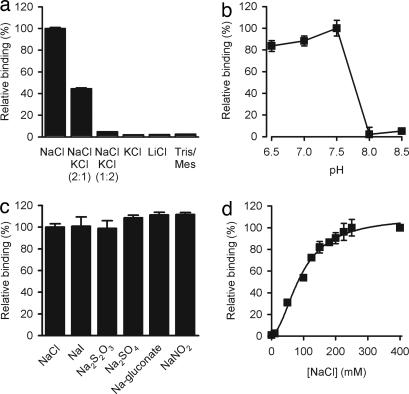
The versatility of the SPA format allowed further characterization of radiotracer binding. Fig. 3a shows the effect of the cationic composition of the assay buffer on tyrosine binding to Tyt1. Among the cations tested, only Na+ promoted robust [3H]tyrosine binding, consistent with previous results with Na+-dependent tyrosine cotransport (9). Whereas the pH-dependence of Tyt1 tyrosine transport activity in E. coli exhibited an optimum at 8.5 (9), Tyt1-mediated tyrosine binding was optimal between pH 6.5 and 7.5 but dropped dramatically (>90% reduction of binding activity) at pH ≥8.0 (Fig. 3b). However, upon reconstitution of the transporter in liposomes, the pH optimum of Na+-dependent tyrosine transport was 8.5 (Y. Zhao, M.Q., and J.A.J., unpublished work), similar to that observed when the transporter was situated in the cytoplasmic membrane of intact E. coli cells. Tyrosine binding to Tyt1 was not chloride-dependent, because replacement of NaCl with other sodium salts maintained specific binding (Fig. 3c), consistent with the lack of chloride-dependence of tyrosine transport (9). Whereas the apparent Na+ affinity for tyrosine transport in intact E. coli MQ614 cells (9) or in Tyt1-containing proteoliposomes (Fig. 5d) was ≈1 mM, Na+ activation of Tyt1-mediated tyrosine binding was half-maximal at ≈92 mM NaCl, with a Hill coefficient of 1.94 ± 0.28 (Fig. 3d).
Fig. 3.
Tyrosine binding to Tyt1 is strictly Na+- and pH-dependent. (a) Binding of l-[3H]tyrosine to solubilized Tyt1 was performed in assay buffer composed of 50 mM Tris/Mes, pH 7.5/20% glycerol/2 mM TCEP/0.05% N-dodecyl-β-d-maltopyranoside/150 mM of the indicated salts (at the indicated ratios) (n = 6). (b) The effect of protons on the binding activity was performed by varying the pH of the assay buffer in the presence of 150 mM NaCl (n = 6). (c) Tyrosine binding to solubilized Tyt1 is not chloride-dependent as determined by the replacement of NaCl with other sodium salts. (d) Tyt1 exhibits an apparent 2 Na+:1 tyrosine binding stoichiometry. The apparent Na+-activation constant of l-tyrosine binding (KDNa+) was determined to be 91.5 ± 4 mM with a Hill coefficient of 1.94 ± 0.28. Binding of 1 μM l-[3H]tyrosine was performed by varying the NaCl concentration from 0–400 mM (equimolar replacement with Tris/Mes; n = 3).
Fig. 5.
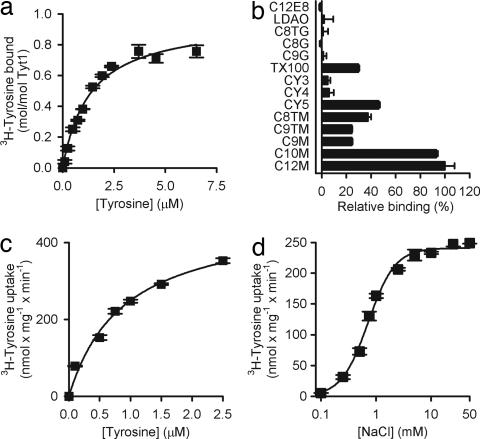
Optimization of reconstitution conditions for Tyt1. (a) l-[3H]tyrosine binding to Tyt1 is saturable with a KDTyr of 1.35 ± 0.17 μM (n = 3). Tyt1 function is maintained during the solubilization process with a molar binding ratio of ≈1 (0.96 ± 0.04 mol of tyrosine per mol of Tyt1; n = 3). (b) Effect of detergents on the binding of 1 μM l-[3H]tyrosine to purified and desalted Tyt1 (n = 3). Detergents used were: n-dodecyl-β-d-maltopyranoside (C12M), n-decyl-β-d-maltopyranoside (C10M), n-nonyl-β-d-maltopyranoside (C9M), n-nonyl-β-d-thiomaltopyranoside (C9TM), n-octyl-β-d-thiomaltopyranoside (C8TM), CYMAL-5 (CY5), CYMAL-4 (CY4), CYMAL-3 (CY3), Triton X-100 (TX100), n-nonyl-β-d-glucopyranoside (C9G), n-octyl-β-d-glucopyranoside (C8G), n-octyl-β-d-thioglucopyranoside (C8TG), n-dodecyl-N,N-dimethylamine-N-oxide (LDAO), and polyoxyethylene(8)dodecyl ether (C12E8). Data in a and b represent the background-corrected binding activity. Nonspecific background binding activity was determined in the presence of 150 mM imidazole and was not significantly different from that observed with sample originating from control vesicles. (c) Upon the functional reconstitution of Tyt1 in proteoliposomes tyrosine transport by Tyt1-containing proteoliposomes is saturable with a K0.5Tyr of 0.9 ± 0.19 μM and a VmaxTyr of 470 ± 42 nmol × mg Tyt1−1× min−1 (n = 3), resulting in a catalytic turnover number (kcat) of 0.4 ± 0.04 × s−1. (d) Membrane-inserted Tyt1 exhibits an apparent K0.5Na+ of 0.72 ± 0.05 mM with a Hill coefficient of 1.8 ± 0.21 (n = 3).
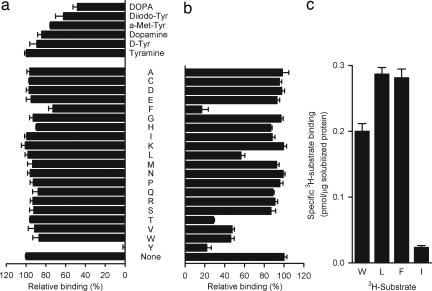
The substrate specificity of Tyt1 was tested by a competition SPA assay (Fig. 4a). Among the naturally occurring 20 amino acids, 10 μM l-tyrosine inhibited binding of 0.1 μM l-[3H]tyrosine to Tyt1 almost completely. Of the other amino acids and tyrosine analogues tested, only α-methyl-l-tyrosine, phenylalanine, 5-diiodo-l-tyrosine, or 3-(3,4-dihydrophenyl)-l-alanine (DOPA) inhibited l-[3H]tyrosine binding ≥25% at a concentration of 10 μM, demonstrating the high substrate specificity of Tyt1 for tyrosine. In contrast, a similar application of the SPA identified MhsTBh as a multisubstrate transporter (Fig. 4 b and c; M.Q., H. Yano, and J.A.J., unpublished work).
Fig. 4.
Determination of the substrate specificity of Tyt1 and MhsT. (a) Tyt1 binding is highly specific. The substrate specificity of solubilized Tyt1 was measured by a competition assay. The binding of 0.1 μM (final concentration) l-[3H]tyrosine was assayed in the presence or absence of 10 μM nonradioactive amino acid (or tyrosine analogue) as indicated (a–c, in the one-letter amino acid code used; the tyrosine analogues shown in a are: tyramine, d-tyrosine (D-Tyr), dopamine, α-methyl-l-tyrosine (a-Met-Tyr), 5-diiodo-l-tyrosine (Diiodo-Tyr), and 3-(3,4-dihydrophenyl)-l-alanine (DOPA). (b) MshT of B. halodurans is a multisubstrate transporter. Binding of 0.1 μM l-[3H]tyrosine was tested in the presence or absence of 10 μM nonradioactive amino acid as indicated (n = 3). l-[3H]tyrosine binding to MshTBh was inhibited ≥50% by 10 μM l-tyrosine, l-phenylalanine, l-tryptophan, l-valine, l-isoleucine, or l-threonine. (c) This specificity pattern was confirmed by assessing the binding activity of a 1 μM concentration of the indicated l-[3H]amino acids (n = 3) as well as by transport assays (data not shown; M.Q., Hideaki Yano, and J.A.J., manuscript in preparation). Data represent the background-corrected binding activity measured in the presence of 150 mM imidazole, which was not significantly different from that observed with sample originating from control vesicles.
Monitoring the Purification and Reconstitution Conditions.
To monitor the (potential) activity loss of Tyt1 during the purification process, an SPA was performed with detergent-solubilized Tyt1 at a Na+ concentration of 150 mM NaCl in which the [3H]tyrosine concentration was increased from 0.1 to 6.5 μM (Fig. 5a). l-[3H]tyrosine binding is saturable, with a half-maximum saturation constant (KDTyr) of 1.35 ± 0.17 μM, a value comparable with the apparent K0.5Tyr for tyrosine transport in intact E. coli cells (9) (Table 1). Determination of the molar binding ratio of Tyt1 revealed a binding coefficient of ≈1, indicating (i) a 1 tyrosine:1 Tyt1 binding stoichiometry and (ii) ≈100% activity (i.e., no significant activity loss) of Tyt1 upon solubilization with n-dodecyl-β-d-maltopyranoside. The same binding assay was performed after the purification of Tyt1 by immobilized metal chelate affinity chromatography (IMAC) (Y. Zhao, M.Q., and J.A.J., unpublished work), and similar results were obtained (Table 1). This result indicates that Tyt1 retained its functionality during the purification procedure when n-dodecyl-β-d-maltopyranoside was used as detergent.
Table 1.
Kinetic parameters of Tyt1 in different states
| E. coli(9) | Solubilized | IMAC | Proteoliposomes | |
|---|---|---|---|---|
| Tyrosine affinity (K0.5Tyr,KDTyr) (μM) | 0.34 ± 0.08 | 1.35 ± 0.06 | 1.33 ± 0.24 | 0.9 ± 0.19 |
| Maximum velocity (VmaxTyr)(nmol × mg−1 × min−1)* | 6.9 ± 0.3 | – | – | 470 ± 42 |
| Molar binding ratio | – | 0.93 ± 0.04 | 0.98 ± 0.03 | – |
| App. Na+ affinity (K0.5Na+,KDNa+) (mM) | 0.75 ± 0.03 | 95.2 ± 6.7 | 86.4 ± 12.3 | 0.72 ± 0.05 |
| Hill coefficient | 2 ± 0.2 | 2.01 ± 0.08 | 1.93 ± 0.16 | 1.8 ± 0.21 |
| Turnover no. (kcat) | – | – | – | 0.4 ± 0.04 s−1 |
Kinetic parameters were determined as described in the legend to Figs. 3 or 5 or taken from ref. 9. Binding data for Tyt1 purified by immobilized metal affinity chromatography (IMAC) were determined as described for the detergent-solubilized fraction of Tyt1 containing membrane vesicles with ≈0.5 μg Tyt1 per mg of YSi beads (see SI Fig. 6 and SI Text). Kinetic parameters for solubilized or IMAC-purified Tyt1 were determined in three individual experiments, and the constants represent the average ± SD of the individual determinations. The kinetic parameters for Tyt1 reconstituted into proteoliposomes are based on the data shown in Fig. 5 c and d, with errors representing the SEM of the fit. Note that the variation of these parameters is [ltequ]20% when compared with independent experiments (Y. Zhao, M.Q., and J.A.J., unpublished work. App., apparent.
*Note that the maximum velocity refers to the total amount of membrane protein in E. coli cells or the amount of purified Tyt1 reconstituted in proteoliposomes.
The effect of different detergents on the binding activity of purified Tyt1 was assayed (Fig. 5b). In the presence of detergents with maltopyranoside head groups, Tyt1 exhibited the highest activity with a reduction of the activity pattern generally reflecting the chain length of the detergent. However, detergents with a glucopyranoside moiety or n-dodecyl-N,N-dimethylamine-N-oxide- or polyoxyethylene(8)dodecyl ether-impaired Tyt1 binding activity. Triton X-100 was the only nonmaltoside detergent tested that preserved ≈30% residual binding activity of Tyt1 compared with that in n-dodecyl-β-d-maltopyranoside.
Tyt1-containing proteoliposomes exhibited saturable tyrosine transport, which depended strictly on the Na+ concentration (Fig. 5 c and d). The (apparent) K0.5 for tyrosine and Na+ were comparable with those reported for Na+-dependent tyrosine cotransport measured in intact E. coli cells expressing Tyt1 (9) (Table 1). After the determination of the actual Tyt1 amount used per transport assay, the turnover number (kcat) of Na+/tyrosine cotransport was determined to be 0.4 ± 0.04 × s−1. This value is in perfect agreement with the turnover number calculated for the human dopamine transporter (18), consistent with the functional homology of Tyt1 and other NSS family members.
Discussion
This study introduces an approach to transporter proteomics by directly monitoring the function of recombinant membrane transport proteins in detergent-solubilized crude membrane extracts and in purified form by using the scintillation proximity assay format. Although SPA approaches have been widely used to study ligand binding to receptors (19, 20), to our knowledge, they have not been used previously to study substrate binding to transporters, despite the many advantages that we demonstrate here, including the suitability of the assay for relatively low-affinity substrates, the binding of which is difficult or impossible to study by using standard filtration methods and has necessitated the development of complex indirect binding assays.
By using Tyt1, we were able to evaluate this method on a recently characterized NSS transporter (9). In addition, this approach was used to (i) identify a previously uncharacterized member of the NSS family, MhsT of B. halodurans (Fig. 2c and 4 b and c), (ii) measure leucine binding to LeuTAa (11) (Fig. 2b), (iii) measure substrate binding activity of detergent-solubilized PutP of E. coli, a member of the sodium:solute symporters family (Fig. 2d), and (iv) successfully establish conditions for substrate binding studies on the human dopamine transporter (hDAT) expressed in a mammalian cell line (data not shown).
This article describes the copper-chelate scintillation proximity-based assay format in conjunction with suitable radiotracers as a rapid and simple technique to assess the function of a given membrane transport protein in a nonmembranous environment, because this method conserves the mechanistic features, i.e., substrate affinity and specificity and ion dependence, of the transporter. The differences in the pH and Na+-dependence observed for Tyt1 in a membranous and nonmembranous environment (Table 1) may reflect the impact of the electrochemical gradient on transport kinetics. Indeed, whereas the apparent K0.5Na+ for tyrosine transport by Tyt1 in proteoliposomes was ≈1 mM (see Fig. 5d), the apparent KDNa+ for tyrosine binding in proteoliposomes, which were deenergized by the addition of gramicidin, was ≈100 mM. This result shows that the differences in the apparent Na+ affinities for tyrosine transport in intact cells and proteoliposomes and for binding in the SPA cannot be attributed to the presence or absence of lipid but, rather, reflect the lack of the electrochemical gradient across the membrane. Detailed kinetic analyses and mathematical simulations for the human Na+/glucose transporter (hSGLT1) (21) reveal that the transition of the unloaded transporter from the “inward-facing” conformation (C6) to the “outward-facing” conformation (C1) [the conformational state that can bind two Na+ ions leading to the Na+-loaded conformation (C2)] is the major voltage-dependent step in the transport cycle. Previous studies with hSGLT1 (22) showed that the voltage-dependence of charge movement is modulated by the Na+ concentration, indicating the synergistic action of membrane potential and activating cation on the distribution of conformational states of the transporter. Thus, it is tempting to speculate that because of the lack of a vectorial environment in our binding assay, i.e., the solubilization of Tyt1 from the membrane, higher Na+ concentrations are required to promote substrate (tyrosine) binding to Na+-loaded Tyt1 (C2 → C3 transition (see ref. 9 for the proposed Tyt1 kinetic scheme). The Hill coefficient of ≈2 for Na+-dependent tyrosine binding to Tyt1 is consistent with an apparent 2 Na+:1 tyrosine binding stoichiometry in an ordered kinetic binding model. Note that in the crystal structure of LeuTAa (11) two Na+ ions were identified near or in contact with the bound substrate, leucine. The functional studies presented in Fig. 3 a and d reveal the strict Na+ requirement of Tyt1-catalyzed tyrosine binding and hence provide further support for an ordered binding mechanism in which the binding of two Na+ ions organizes the substrate-binding site of the transporter before the binding of the substrate.
This study emphasizes the power of the scintillation proximity assay as (i) a generally applicable direct assay to monitor the activity and integrity of membrane transport proteins in detergent and (ii) a valuable tool to enhance the effectiveness of the expression system and initial characterization of the protein. This information is crucial for the optimization of conditions for the successful purification and reconstitution of transport proteins in their physiological (functional) state. It is also directly applicable to optimization of crystallization conditions in which the protein is maintained in detergent and not reconstituted; particularly because information on substrate and inhibitor binding will be useful for designing cocrystallization experiments.
The requirement that the substrate used in the SPA be radiolabeled is a limitation to the universal applicability of the assay. Nonetheless, many potential substrates, including all of the amino acids, are readily available in tritiated form. It also remains to be determined whether binding of substrates for which a given transporter possesses a lower affinity (i.e., in the mM range) can be detected readily with this assay. Despite these potential limitations, in a proteomic approach, this rapid and simple assay enables the screening of different substrates/inhibitors in crude cell extracts without the need for laborious protein purification. Because this enormous family of membrane transport proteins is still replete with many orphan transporters for which the substrates are not known, the scintillation proximity binding assay will be a valuable tool toward the elucidation of their structure–function relationships.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli MQ614 [SVS1144 mtr aroP tnaB271::Tn5 tyrP1 pheP::cat] (9) or C41(DE3) [F− ompT gal hsdSB (rB−mB−) dcm lon λDE3 plus an uncharacterized mutation] (12) (Imaxio formerly Avidis, Saint-Beauzire, France) harboring plasmid pQ2 was used for the overproduction of Tyt1 (9). L. lactis NZ9000 (15) and expression plasmid pNZ8048 (23) were obtained from W. Konings (University of Gronigen, Haren, The Netherlands) through H. R. Kaback (Universtiy of California, Los Angeles, CA). Recombinant tyt1 [containing a His10 coding region before the tyt1 start codon (9)] was subcloned into pNZ8048 by using the BstBI/HindIII restriction enzyme combination, followed by transformation of electrocompetent L. lactis NZ9000 with the ligation mix. The resulting plasmid, designated pNZ2, was isolated by plasmid miniprep (Qiagen, Valencia, CA) after lysozyme (10 mg/ml) treatment at 55°C for 10 min. The gene of an uncharacterized NSS homologue of B. halodurans (accession no. NP_241994), designated mhsT (mhsTBh) hereafter, was PCR-amplified from B. halodurans genomic DNA (BAA-125D; American Type Culture Collection, Manassas, VA). Unique NcoI and NheI sites were introduced at the haaT 5′ and 3′ termini, respectively. mhsTBh was subcloned into pQ2 by means of NcoI/NheI restriction digest, replacing the entire tyt1 gene. To generate an N-terminally His10-tagged mhsTBh gene product, NcoI/NheI-digested mhsTBh was ligated into similarly cut pNZ2N (in pNZ2N the HindIII site flanking the tyt1 stop codon was replaced with an unique NheI site after HindIII-digestion of pNZ2, followed by Klenow treatment and relegation of the vector) generating pNZmhsT. Similarly, the leuT gene of Aquifex aeolicus was PCR-amplified with genomic DNA of A. aeolicus (A. aeolicus cells were a generous gift from H. Huber, University of Regensburg, Germany) as template. Unique NcoI and HindIII sites were introduced at the 5′ and 3′ termini, and the resulting gene product was cloned into pQ2 by using this enzyme combination. The fidelity of all plasmids was confirmed by DNA sequencing (Columbia University Sequencing Facility).
Gene Expression.
L. lactis NZ9000 harboring pNZ8048 or its derivatives were cultivated in M17 medium supplemented with 0.5% glucose and 5 μg of chloramphenicol per ml at 30°C. Gene expression in exponentially growing cells was induced for 4 h by the addition of 5 ng of nisin per ml. Cells were harvested by centrifugation at 7,500 × g for 10 min at 4°C, followed by a 1-h treatment with 10 mg of lysozyme per ml in 100 mM potassium phosphate, pH 7.0, at 30°C. L. lactis protoplasts (≈1 g wet weight per ml) were disrupted by a 3-fold passage through an EmulsiFlex-C5 cell homogenizer (Avestin, Ottawa, Canada) at 30,000 psi. Unbroken cells and cell debris were removed by centrifugation (10,000 × g for 10 min at 4°C) before membrane vesicles were collected at 262,000 × g for 45 min at 4°C. Expression of the tyt1 gene in E. coli MQ614 or C41(DE3) was performed as described (9). Alternatively, cells were incubated in Terrific broth, supplemented with 100 μg of ampicillin per ml at 37°C until the cultures reached an absorbance at 600 nm of 0.6. Tyt1 expression was induced by the addition of 0.15 mM isopropyl-β-d-thiogalactopyranoside for 16 h at 20°C. E. coli MQ614 or C41(DE3) harboring pQE60 (9) served as control. LeuT and PutP were expressed in E. coli C41(DE3) or WG170 as described (11, 17). E. coli membrane vesicles were prepared by passing the cell suspension through an Avestin EmulsiFlex-C5 cell homogenizer at 15,000 psi. For SPA-based substrate-binding experiments, membrane vesicles of E. coli C41(DE3) harboring pQE60 (9), or WG170 transformed with pTrc99a (17) served as control.
Solubilization and Purification.
Membrane vesicles were resuspended in ice-cold 50 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol (Tris)/2-(N-morpholino)-ethanesulfonic acid (Mes), pH 7.5/20% glycerol/150 mM NaCl/2 mM Tris(2-carboxyethyl) phosphine (TCEP) (unless otherwise noted) at a protein concentration of 10 mg/ml and solubilized by the addition of 1% (wt/vol) n-dodecyl-β-d-maltopyranoside for 1 h at 4°C. Solubilized protein and insoluble matter were separated by centrifugation at 262,000 × g for 45 min at 4°C. The supernatant was either used for binding assays (see below) or incubated with preequilibrated Ni2+-Sepahrose Fastflow (GE Healthcare, Piscataway, NJ) (1 ml of resin/200 mg of membrane protein) for 1.5 h with gentle shaking at 4°C in the presence of 20 mM imidazole. The protein–resin complex was then loaded into a column. Unbound protein was removed by washing with 50 mM Tris/Mes, pH 7.5/20% glycerol/150 mM NaCl/2 mM TCEP/60 mM imidazole/0.05% (wt/vol) n-dodecyl-β-d-maltopyranoside until the absorbance at 280 nm returned to the baseline. Tyt1 was eluted by a step increase of the imidazole concentration in the buffer to 250 mM. Removal of imidazole or desalting/buffer exchange of the eluted fraction before the SPA was achieved with Zeba Desalt Spin Columns (Pierce, Rockford, IL). To assess the effect of various detergents on Tyt1 activity, the purified and desalted sample was incubated in the presence of various detergents as indicated at a concentration of ≈2.5 times their critical micellar concentration (CMC) for 16 h at 4°C. The binding step was performed by diluting the samples in assay buffer in which 0.05% n-dodecyl-β-d-maltopyranoside was replaced with the respective detergent at ≈1 times their CMC in the presence of 1 μM (final concentration) l-[3H]tyrosine.
Reconstitution/Transport Assay.
Preformed liposomes were prepared as described (24) by using E. coli total lipid extract (Avanti Polar Lipids, Alabaster, AL). The liposome suspension was suspended in 100 mM potassium phosphate, pH 7.5/2 mM TCEP and frozen in liquid N2. Before reconstitution, thawed liposomes were extruded through a 400-nm membrane filter (Whatman, Clifton, NJ). Liposomes (5 mg lipid per ml) were destabilized by the addition of 0.12% (wt/vol) Triton X-100 (onset of solubilization) and mixed with purified Tyt1 in a 250:1 ratio (wt/wt) for 10 min at 23°C. Detergent was removed by adding Bio-Beads SM-2 (Bio-Rad, Hercules, CA). Tyt1-containing proteoliposomes (or control liposomes) were concentrated by centrifugation at 320,000 × g for 45 min, resuspended in 100 mM potassium phosphate, pH 6.5/2 mM TCEP (≈100 mg of lipids per ml), and stored in liquid N2. Proteoliposomes were thawed at 23°C and extruded through a 400-nm filter, followed by extrusion through a 100-nm filter (Avanti Polar Lipids). After a 100-fold dilution of the proteoliposomes in 50 mM Tris/Mes, pH 8.5/2 mM TCEP/25 mM NaCl (unless otherwise indicated) an artificial membrane potential (inside negative) was generated by the addition of 2 μM valinomycin 30 seconds before the addition of l-[3H]tyrosine [53 Ci/mmol (1 Ci = 37 GBq); concentration as indicated] (GE Healthcare). Transport was assayed as described (24) at 23°C. The protein content of each batch of proteoliposomes was assayed as described (25).
SPA.
Cu2+ chelate YSi scintillation SPA beads (GE Healthcare) were diluted in 50 mM Tris/Mes, pH 7.5/20% glycerol/150 mM NaCl/2 mM TCEP/0.05% n-dodecyl-β-d-maltopyranoside (unless indicated otherwise) at a concentration of 2.5 mg/ml. For Tyt1 SPA binding experiments l-[3H]tyrosine (at a specific radioactivity of 53 Ci/mmol or, for routine experiments, at 10 Ci/mmol; GE Healthcare) was added to the bead solution at the indicated concentration. One hundred microliters of this suspension was mixed with the supernatant of the solubilized membrane vesicles or purified and desalted Tyt1 (at a protein amount of ≈50% of the maximum binding capacity of the SPA beads; (SI Fig. 6) in individual wells of a 96-well white-wall clear-bottom plate in the absence or presence of 150 mM imidazole (as background correction) or compounds as indicated. The plate was vigorously shaken on a vibrating platform for 15 min at 23°C (unless indicated otherwise) before counting in a photomultiplier tube MicroBeta counter (Wallac) (SPA cpm mode). Afterward, 200 μl of OptiPhase SuperMix scintillation mixture (PerkinElmer, Wellesley, MA) were added per well, and the plate was counted again, this time in the regular cpm mode. This number was taken for the accurate determination of the cpm-to-mol substrate coefficient rather than rely on the theoretical dpm value of the added radiotracer at its specific activity. Based on this coefficient, the cpm from the reading in the SPA mode were transformed into the molar amount of the radiotracer bound in the scintillation proximity assay. The SPA for LeuTAa, MhsTBh, and PutP was performed as described above [with 0.2 μM or 1 μM (final concentration) l-[3H]leucine (40 Ci/mmol; (American Radiolabeled Chemicals, St. Louis, MO), 1 μM (final concentration) of l-[3H]tyrosine (53 Ci/mmol; GE Healthcare), l-[3H]tryptophan (20 Ci/mmol; GE Healthcare), l-[3H]phenylalanine (55 Ci/mmol; Moravek Radiochemicals, Brea, CA), l-[3H]isoleucine (20 Ci/mmol; GE Healthcare), or l-[3H]proline (80 Ci/mmol; Moravek Radiochemicals, Brea, CA) as indicated], with the exception that the binding assay for PutP was performed at 4°C, followed by immediate counting of the plate at 23°C. This was necessary because of the lower affinity of the His6-tagged transporter for the Cu2+ chelating YSi SPA scintillation beads (note that the other recombinant transport proteins described in this study contain a His10 tag). A similar tendency was observed during purification of PutP; that is, the solubilized protein did not bind to the Ni2+–NTA resin when the purification was performed at room temperature (M.Q. and H. Jung, unpublished results).
Data Analysis.
All experiments were repeated at least in duplicate starting with membrane vesicles prepared form different batches of cells. Figures represent a typical experiment, and, unless otherwise noted, data points represent the mean of a triplicate determination (n = 3) ± SD. Data fits of kinetic analyses were performed by using nonlinear regression algorithms in Sigmaplot (Systat Software, San Jose, CA), and errors represent the SEM of the fit.
Supplementary Material
Acknowledgments
We thank Dr. Myles H. Akabas for his inspiration regarding the potential use of the SPA format for studying binding to a transport protein, and Hideaki Yano for preparation of L. lactis membrane vesicles and expert technical assistance. This work was supported by National Institutes of Health Grants MH57324 and DA17293 (to J.A.J.).
Abbreviations
- IMAC
immobilized metal affinity chromatography
- LeuTAa
leucine transporter of A. aeolicus
- Mes
2-(N-morpholino)-ethanesulfonic acid
- MTSEA
2-(aminoethyl)methane thiosulfonate
- NSS
neurotransmitter:sodium symporter
- PutP
Na+/proline transporter
- SPA
scintillation proximity assay
- TCEP
Tris(2-carboxyethyl) phosphine
- Tris
2-amino-2-(hydroxymethyl)-1,3-propanediol
- Tyt1
Na+/tyrosine transporter of F. nucleatum.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609573104/DC1.
References
- 1.Wu J, Kaback HR. Biochemistry. 1994;33:12166–12171. doi: 10.1021/bi00206a020. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Frillingos S, Voss J, Kaback HR. Protein Sci. 1994;3:2294–2301. doi: 10.1002/pro.5560031214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weitzman C, Consler TG, Kaback HR. Protein Sci. 1995;4:2310–2318. doi: 10.1002/pro.5560041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li XD, Villa A, Gownley C, Kim MJ, Song J, Auer M, Wang DN. FEBS Lett. 2001;494:165–169. doi: 10.1016/s0014-5793(01)02334-1. [DOI] [PubMed] [Google Scholar]
- 5.Gruner SM, Kirk G, Patel L, Kaback HR. Biochemistry. 1982;21:3239–3243. doi: 10.1021/bi00256a033. [DOI] [PubMed] [Google Scholar]
- 6.Nelson N. Anal Biochem. 1987;165:287–293. doi: 10.1016/0003-2697(87)90271-5. [DOI] [PubMed] [Google Scholar]
- 7.Udenfriend S, Gerber L, Nelson N. Anal Biochem. 1987;161:494–500. doi: 10.1016/0003-2697(87)90479-9. [DOI] [PubMed] [Google Scholar]
- 8.Bosworth N, Towers P. Nature. 1989;341:167–168. doi: 10.1038/341167a0. [DOI] [PubMed] [Google Scholar]
- 9.Quick M, Yano H, Goldberg NR, Duan L, Beuming T, Shi L, Weinstein H, Javitch JA. J Biol Chem. 2006;281:26444–26454. doi: 10.1074/jbc.M602438200. [DOI] [PubMed] [Google Scholar]
- 10.Androutsellis-Theotokis A, Goldberg NR, Ueda K, Beppu T, Beckman ML, Das S, Javitch JA, Rudnick G. J Biol Chem. 2003;278:12703–12709. doi: 10.1074/jbc.M206563200. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 12.Miroux B, Walker JE. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg NR, Beuming T, Soyer OS, Goldstein RA, Weinstein H, Javitch JA. Eur J Pharmacol. 2003;479:3–12. doi: 10.1016/j.ejphar.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 14.Beuming T, Shi L, Javitch JA, Weinstein H. Mol Pharmacol. 2006;70:1630–1642. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- 15.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 16.Saier MH., Jr J Cell Biochem. 1999;(Suppl32/33):84–94. doi: 10.1002/(sici)1097-4644(1999)75:32+<84::aid-jcb11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 17.Quick M, Jung H. Biochemistry. 1998;37:13800–13806. doi: 10.1021/bi980562j. [DOI] [PubMed] [Google Scholar]
- 18.Sonders MS, Zhu SJ, Zahniser NR, Kavanaugh MP, Amara SG. J Neurosci. 1997;17:960–974. doi: 10.1523/JNEUROSCI.17-03-00960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, Grandy DK, Langen H, Monsma FJ, Jr, Civelli O. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- 20.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo DD, Hirayama BA, Cha A, Bezanilla F, Wright EM. J Gen Physiol. 2005;125:13–36. doi: 10.1085/jgp.200409150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quick M, Loo DD, Wright EM. J Biol Chem. 2001;276:1728–1734. doi: 10.1074/jbc.M005521200. [DOI] [PubMed] [Google Scholar]
- 23.de Ruyter PG, Kuipers OP, de Vos WM. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quick M, Wright EM. Proc Natl Acad Sci USA. 2002;99:8597–8601. doi: 10.1073/pnas.132266599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffner W, Weissmann C. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.