Abstract
Eusocial wasps of the family Vespidae are thought to have derived their social behavior from a common ancestor that had a rudimentary caste-containing social system. In support of this behavioral scenario, the leading phylogenetic hypothesis of Vespidae places the eusocial wasps (subfamilies Stenogastrinae, Polistinae, and Vespinae) as a derived monophyletic clade, thus implying a single origin of eusocial behavior. This perspective has shaped the investigation and interpretation of vespid social evolution for more than two decades. Here we report a phylogeny of Vespidae based on data from four nuclear gene fragments (18S and 28S ribosomal DNA, abdominal-A and RNA polymerase II) and representatives from all six extant subfamilies. In contrast to the current phylogenetic perspective, our results indicate two independent origins of vespid eusociality, once in the clade Polistinae+Vespinae and once in the Stenogastrinae. The stenogastrines appear as an early diverging clade distantly related to the vespines and polistines and thus evolved their distinctive form of social behavior from a different ancestor than that of Polistinae+Vespinae. These results support earlier views based on life history and behavior and have important implications for interpreting transitional stages in vespid social evolution.
Keywords: Hymenoptera, molecular phylogenetics, social behavior, social insect evolution, Vespidae
Insect societies are among the most complex systems in nature. Although diverse in kind, the most distinctive and widely known insect societies are those characterized by a reproductive “queen” and an effectively sterile “worker” caste that assists in raising the next generation of reproductive offspring. These eusocial societies (1), which comprise a small but ecologically successful fraction of insect species, are believed to have evolved several times independently within the insect order Hymenoptera: once in ants (2), once in wasps of the family Sphecidae (3), once in wasps of the family Vespidae (4, 5), and several times within bees (6, 7). What were the circumstances that gave rise to eusociality (8–11), and is this rare state inevitably the result of a stepwise progression through the transitional stages, from solitary nesting through “primitive” sociality (12–14) to a derived state of eusocial complexity (15, 16)? To answer these intriguing questions requires, minimally, two conditions: (i) the existence of a socially diverse group of taxa, and (ii) a robust estimate of their phylogeny to provide the historical framework from which to investigate changes from solitary to social behavior (17).
Vespidae is one of the few groups that has retained the necessary transitional states to elucidate social evolution, encompassing solitary, presocial, facultatively eusocial (18), and eusocial taxa (17, 19). There are ≈4,200 described vespid species currently classified into 6 subfamilies based on morphological evidence (4, 5). Euparagiinae (9 species, found only in southwestern North America and northern Mexico) (20) and Masarinae (“pollen wasps,” ≈300 species) (21) are solitary; Eumeninae (“potter and mason wasps,” ≈3,000 species worldwide) (22, 23) exhibit both solitary and presocial behavior (23); Stenogastrinae (“hover wasps,” ≈50 species found in the Indo-Pacific tropics) (24) are facultatively eusocial (25); and Polistinae (“paper wasps,” ≈800 species) (5) and Vespinae (“hornets and yellowjackets,” ≈60 species) (26) are eusocial (27). However, relationships among the vespid subfamilies have been controversial (28–35), and the alternative proposed relationships have strikingly different implications for the evolution of social behavior.
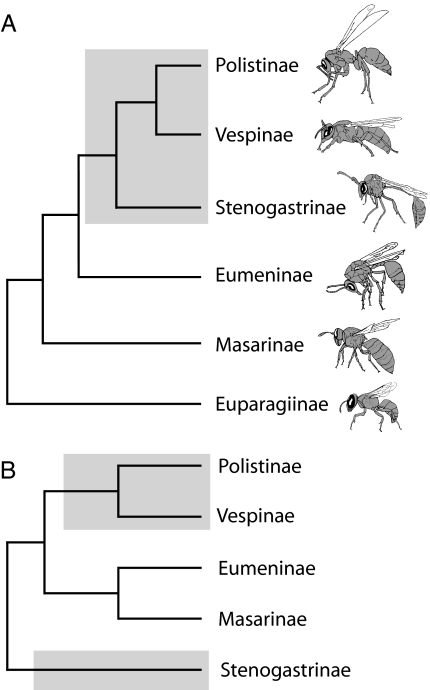
The current leading phylogenetic hypothesis for Vespidae (Fig. 1A) (4, 5, 31) implies a single origin of social behavior in the common ancestor of a clade comprising Stenogastrinae + (Polistinae+Vespinae). This hypothesis was initially based on parsimony analysis of morphological and behavioral characters, but the three putative synapomorphies found to support the clade exhibit homoplasy (4) and may not reflect common ancestry. Additional behavioral characters proposed as shared, derived traits (synapomorphies) for the clade (5, 35) appear to be ambiguously characterized (19, 37). Indeed, earlier investigators of vespid social behavior argued that the often dramatic trait differences between stenogastrines and polistines+vespines in behavior (egg-laying, larval provisioning, nest architecture) and morphology (wing folding, pupal posture) reflect separate origins of sociality (19, 28, 29, 32–34). Moreover, a separate origin of stenogastrine sociality was inferred from a preliminary molecular analysis (Fig. 1B) of <600 nucleotides from 16S and 28S rDNA (30), an analysis that remains controversial because of the absence of some ingroup subfamilies and the inappropriate selection of outgroups, which resulted in uncertainties in rooting the phylogeny (31).
Fig. 1.
Previous hypotheses of subfamily-level relationships of Vespidae. (A) The conventional hypothesis based on morphological and behavioral characters (4). (B) An alternative hypothesis based on limited molecular data (30), including the placement of Masarinae from a later study (36). Eusocial clades are indicated with gray shading.
Although interest in vespid relationships has expanded our understanding of these wasps, the proliferation of multiple phylogenies has placed a burden on students of social behavior to select the one that best reflects the true history. An incorrect phylogeny hampers a realistic understanding of hymenopteran social evolution, for example, by incorrectly inferring the number of social origins (6, 7, 38), misinterpreting homologies and transitional stages among social traits (6, 7, 38), and providing an inappropriate test of behavioral models of eusocial evolution (5, 14). Here we present a multigene phylogeny of Vespidae, including representatives of all currently recognized extant subfamilies. This phylogeny provides robust support for two independent origins of social behavior in these wasps.
Results
A data set comprising 3,002 aligned nucleotide sites was assembled from four gene fragments: 18S rDNA [≈735 amplified bp/29 parsimony informative (PI) sites]; 28S rDNA (≈790 bp/142 PI sites); abdominal-A (abd-A) (≈550 bp/158 PI sites); and RNA polymerase II (RNA pol II) (841 bp/250 PI sites). Bayesian and parsimony analyses of individual gene fragments resulted in highly resolved and well supported phylogenies with similar patterns of relationship (see supporting information (SI) Fig. 3), with the exception of 18S rDNA, which was too conserved to resolve many relationships. The conventional subfamilies (4) were monophyletic, with two exceptions: Masarinae was paraphyletic with respect to Euparagiinae for abd-A and RNA pol II, and Eumeninae was consistently paraphyletic with respect to Polistinae+Vespinae. Each gene fragment (again excluding 18S) provided strong support for a subset of Eumeninae as the sister group to Polistinae+Vespinae.
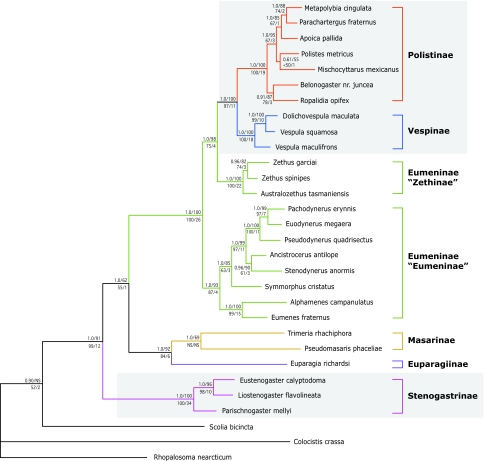
Bayesian, maximum likelihood (ML), and maximum parsimony (MP) analyses of the combined sequence data resulted in a highly resolved vespid phylogeny with strong support (Fig. 2). All subfamilies except Eumeninae were monophyletic, although Masarinae was supported only under Bayesian and ML inference. Stenogastrinae attached as the sister group to the remaining Vespidae for all inference methods, and thus is distantly related to Polistinae+Vespinae. Masarinae was strongly supported as the sister group to Euparagiinae. Eumeninae was resolved as a paraphyletic sister clade to Vespinae+Polistinae with maximum support (posterior probability = 1.0, ML and MP bootstrap values = 100). Eumeninae divided into two separate monophyletic clades: “Zethinae” (39) was sister to Polistinae+Vespinae, and the remaining “Eumeninae” were sister to Zethinae+(Polistinae+Vespinae). “Eumeninae” divided into three principal groups: Pachodynerus-Stenodynerus, Symmorphus, and Alphamenes+Eumenes. Polistinae tribes (40) were recovered as monophyletic, with Ropalidiini (Ropalidia and Belonogaster) as sister to the remaining Polistinae, rather than the currently recognized Polistini (5).
Fig. 2.
Bayesian consensus phylogeny of all genes combined. Node support values are Bayesian posterior probabilities followed by ML bootstrap values on top and parsimony bootstrap followed by Bremer support values on the bottom. Clade values represented by NS are not supported for that analysis. Color-coded branches represent subfamilies, following the classificatory system of Carpenter (4). The eusocial clades are represented by gray shading.
Discussion
Our result that eusocial behavior evolved twice in two distantly related lineages is incongruent with the conventional single-origin hypothesis (4). Given our phylogeny, a dual-origin scenario is more parsimonious than a single origin of eusociality for Vespidae, which would require one gain of sociality along the branch leading to Vespidae, and three independent losses of sociality. Potential sources of error not otherwise addressed that could have misled these analyses include base composition bias and long-branch attraction. Base composition bias is an unlikely explanation for the grouping of Eumeninae with Vespinae+Polistinae, to the exclusion of Stenogastrinae, because eumenine and stenogastrine base composition is similar across genes. For instance, AT base composition of parsimony informative characters from the combined data set is 52.0% for Eumeninae and 52.1% for Stenogastrinae, and differs between the two subfamilies by only 1.5% for abd-A, the most informative gene for uniting Eumeninae with Polistinae+Vespinae. Long-branch attraction among clades is an unlikely source of error given that ingroup clades exhibit similar rates of substitutional change.
From this robust phylogenetic perspective, we can more confidently explore the hierarchical framework in which eusociality evolved independently from solitary ancestors and elucidate traits that may be driving vespid social evolution and their remarkable behavioral variation. A key group in which to search for early stages in the evolution of eusocial behavior is the solitary and presocial Eumeninae. The perspective that eumenines are the nearest relatives of eusocial Vespinae+Polistinae is not new. Earlier observers argued for this relationship on the basis of behavioral and morphological traits (41–45), such as the longitudinal folding of wings, a commonly used diagnostic feature of Vespidae that occurs only in eumenines, polistines, and vespines. Examining patterns of trait evolution within the morphologically and behaviorally diverse Eumeninae, which comprise the vast majority of vespid species, can shed light on whether and under what conditions a stepwise progression toward increasingly social traits occurred en route to the highly eusocial Polistinae+Vespinae. Our finding that Eumeninae is paraphyletic accords well with the distribution of their trait variation and supports an earlier taxonomic classification (39) of two subfamilies, Zethinae and Eumeninae. Eumenine relationships similar to our results were obtained in morphological analyses of Eumeninae by Vernier (46) and Carpenter (22), both of whom found a zethine taxon to be sister to the remaining Eumeninae.
“Zethinae,” the sister group to the eusocial taxa, exhibits traits that may be transitional between those of the ancestral eumenines and the eusocial Vespinae+Polistinae. For instance, rather than the typical eumenine nest construction with mud, the zethine genera Zethus and Calligaster are known to construct nests of plant material, a behavior that could precede the construction of nests from long-fiber wood pulp in the manner of Vespinae and Polistinae (41, 47, 48). Furthermore, Zethus miniatus will oviposit into an incompletely constructed nest cell (41), a behavior otherwise unique to polistines and vespines. Z. miniatus (41) progressively provisions larvae with intact prey, a behavior intermediate between the solitary vespid condition of mass provisioning with intact prey and the eusocial condition of progressive provisioning with macerated prey (17). Z. miniatus also exhibits communal behavior on occasions, with multiple adult females present on a multicelled aggregate nest. Females on these communal nests show a high degree of plasticity in whether they build and oviposit in a new cell, oviposit in a vacant cell, or usurp and oviposit in the cell of a less aggressive female; usurped females may depart and initiate a new nest (23). A similar reproductive flexibility characterizes the eusocial Polistinae. Indeed, Z. miniatus is believed to exemplify a social system from which polistine eusociality could have evolved (14, 49, 50). Cowan (ref. 23, p. 73) notes that Zethus and Calligaster “are regularly cited as exemplifying the critical evolutionary stages of subsocial and communal behavior that connect solitary and eusocial wasps,” a perspective that dates to de Saussure (51). The phylogeny presented here strongly supports this line of thinking and lends strong support to Cowan's (ref. 23, p. 73) plea that “thorough reinvestigations of these insects, including careful attention to their life histories, nesting and mating behaviors, and population structures, are badly needed.” A more comprehensive eumenine phylogeny is also needed, both to investigate other potential instances of eumenine paraphyly and to identify basal character states that may pertain to the acquisition of social behavior in Zethinae and the higher grouping (Eumeninae+(Zethinae+(Polistinae+Vespinae))).
Our results highlight the distinct origin of social behavior in Stenogastrinae, a group that provides an independent source of information concerning traits that promote eusociality. Stenogastrine sociality is flexible and rudimentary, with nests that usually contain <10 adults and eusocial traits that are facultatively expressed (52, 53). Numerous authors have noted that the eusociality of Stenogastrinae differs in key aspects from that of Polistinae+Vespinae (24, 28, 29, 33, 34, 54). For example, stenogastrines lack the true gynes and workers found in Vespinae and Polistinae (19, 24). All three groups exhibit progressive provisioning, but unlike Vespinae+Polistinae, which provision larvae frequently throughout the day, stenogastrines provision larvae once per day or less frequently. Earlier interpretation that this stenogastrine condition represents an intermediate stage between mass provisioning and progressive provisioning (33) must be reassessed in light of our results, which indicate that they acquired their particular form of larval provisioning independently. Another unique trait in Stenogastrinae is the application of a Dufour's (abdominal) gland secretion to eggs, which serves as a tool for oviposition and a substrate for larval adhesion and food deposition. In contrast, Vespinae+Polistinae oviposit directly into cells, use the chorion for early larval adhesion, and feed young larvae mouth to mouth. Stenogastrines are further differentiated behaviorally from Vespinae+Polistinae by licking larval oral secretions from the cuticle, rather than engaging in mouth-to-mouth larval to adult trophallaxis, and by using secretions from the Dufour's gland in place of the van der Vecht's gland to repel ants (24). Furthermore, stenogastrines use a wider diversity of construction materials (mud, masticated vegetation, or wood fibers) and nest design than polistines and vespines, possibly reflecting a more labile ancestral condition. As with Eumeninae, a better understanding of social evolution in Stenogastrinae can be gained with a robust and comprehensive species phylogeny and further study of their diverse natural history.
The dual acquisition of sociality in vespid wasps highlights the evolutionary importance of various traits, now seen as convergent, that play significant roles in the evolution of their sociality (17). Although the traits differ in detail, adult females of both Stenogastrinae and Vespinae+Polistinae construct aerial nests of multiple open cells that house uneven-aged brood, progressively provision multiple larvae simultaneously, receive nourishment while processing larval provisions, and drink larval saliva (17). These attributes involve larval–adult interaction and brood care, which may have been central to both origins of eusocial behavior (17, 55, 56). Another trait shared by Stenogastrinae and Vespinae+Polistinae is the partitioning of reproduction through dominance hierarchies. In Stenogastrinae and some Polistes (52), this involves age-based queues, whereas in other Polistinae and Vespinae dominance is either aggressively or pheromonally established. The various convergent forms of eusocial and presocial traits, such as progressive provisioning, exhibited in Vespinae+Polistinae, Stenogastrinae, some Eumeninae, and even some Masarinae (57), suggest that a basic set of groundplan traits in Vespidae has made multiple routes to sociality probable. Our phylogeny constitutes a firm foundation on which to further compare and contrast traits that may have led to colonial life in Vespidae.
Materials and Methods
Taxon Sampling and Sequencing.
We selected a broad sample of 27 species of Vespidae representing all 6 currently recognized subfamilies (4), including taxa from the 4 known tribes of Polistinae (40), the historically recognized eumenine subfamilies Zethinae and Eumeninae (4), and 3 of the 7 described genera of Stenogastrinae (58) (SI Table 1). Outgroup taxa included exemplars from 3 other vespoid families: Rhopalosomatidae, Scoliidae, and Tiphiidae. Voucher specimens are retained at the Illinois Natural History Survey (Urbana, IL).
We generated DNA sequences from four gene fragments: the highly conserved nuclear 18S rDNA (variable regions V3–5 and related core elements), nuclear 28S rDNA (D2-D3 expansion regions and related core elements), and two intron-free protein-encoding nuclear gene fragments, RNA pol II and abd-A. We extracted DNA from thoracic muscle or legs using the Dneasy Tissue Kit (Qiagen, Valencia, CA) and PCR-amplified each gene fragment with standard protocols by using Eppendorf HotMaster Taq and the following PCR conditions: initial denaturation at 94°C for 5 min; 35 repetitions of 94°C denaturation for 1 min, 48–58°C annealing for 1 min, and 72°C elongation for 1 min, 10 sec; and final elongation at 72°C for 5 min. Primers for each gene and their annealing temperatures are provided in SI Table 2. We purified the resulting products by using either the QIAquick PCR Purification Kit (Qiagen) or the QIAquick Gel Extraction Kit (Qiagen). We carried out sequencing reactions for both forward and reverse strands using BigDye v3.1 (ABI PRISM; Applied Biosystems, Foster City, CA). Automated sequencing was implemented at the W. C. Keck Center for Comparative and Functional Genomics (University of Illinois).
Alignment.
We initially aligned sequences from the protein-encoding genes using the default parameters of Clustal W in Bioedit (59). We further refined the alignment of a few amino acid indels in abd-A (RNA pol II contained no indels) using protein translation. We aligned 28S and 18S rDNA sequences to secondary structure (60), following the methods of Gillespie (61), based on recent structural models for arthropod rRNA (62) (alignments are available in SI Materials and Methods). We excluded ≈95 bp across regions of rDNA alignments where positional homology could not be established using structural criteria, including regions of alignment ambiguity, expansion and contraction, and slipped-strand compensation.
Phylogenetic Analyses.
We estimated phylogenies for each gene region individually and for a combined-gene data set using both MP implemented in PAUP* v.4.0b10 (63) (heuristic search, 1,000 random additions, TBR branch swapping) and Bayesian inference implemented in MrBayes v3.1.2 (64) (4 independent runs at 3,000,000 generations, 4 chains, saving trees every 100 generations; consensus trees and posterior probability values obtained after removing trees from the first 300,000 generations, a point of convergence across all analyses). For the combined data set, we also performed an ML analysis in PAUP* [heuristic search, neighbor-joining starting tree, TBR branch swapping, SYM+I+G model and parameter values selected by using Modeltest v3.7 (65)].
For estimating clade support under parsimony, we implemented nonparametric bootstrapping in PAUP* (400 replicates, 10 random additions per replicate, TBR branch swapping). For ML bootstrap estimates, we applied the same conditions used for obtaining the phylogeny, repeated for 100 replicates. We also calculated Bremer support values for the nodes in the combined-gene tree by using the strict consensus of the two most parsimonious trees as an input tree in the program TreeRot.v2 (66). Bremer support values represent the number of extra steps required under parsimony if a given node is collapsed.
All Bayesian analyses were run with flat priors and different models for partitions, including variable rates and unlinked model parameters across partitions. For abd-A and RNA pol II, we combined codon positions 1 and 2 into a single partition and treated third positions separately. We partitioned 28S and 18S by stem and nonstem regions. Nucleotides that form pairs in 18S and 28S rRNA stems were treated as nonindependent units by using the doublet secondary structure model in MrBayes (64). Nucleotide substitution models were selected based on the Akaike information criterion in Modeltest and included GTR+I+G for 28S rRNA stem and nonstem regions, 18S rRNA stems, and abd-A position 3; GTR+I for 18S rRNA stems, RNA pol II positions 1 + 2, and abd-A positions 1 + 2; and HKY+I+G for RNA pol II position 3.
Supplementary Material
Acknowledgments
We especially thank R. Wallon, who assisted with the 28S rDNA sequencing, and C. K. Starr, J. Neff, and S. Turillazzi, who assisted with important specimen collections, along with A. R. Deans, E. J. Hernandez, M. G. Keeping, J. P. Pitts, J. N. Seal, and A. Toth. We also thank M. J. Yoder for assistance with RNA-related Perl scripting and R. Gadagkar, A. Toth, J. Whitfield, and two anonymous reviewers for insightful comments on the manuscript. This research was supported by a U.S. Department of Agriculture (National Research Initiative CSREES 2004-35302-15077) grant (awarded to S.A.C.). J.H.H. was supported by a University of Missouri-St. Louis Research Leave Grant and the University of Illinois Sociogenomics Initiative (G. Robinson, Principal Investigator). J.J.G. was supported by National Institute of Allergy and Infectious Diseases Contract HHSN266200400035C (awarded to Bruno Sobral, Virginia Bioinformatics Institute at Virginia Tech, Blacksburg, VA).
Abbreviations
- abd-A
abdominal-A
- RNA pol II
RNA polymerase II
- ML
maximum likelihood
- MP
maximum parsimony
- PI
parsimony informative.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in GenBank (accession nos. EF190706–EF190821; also listed in SI Table 1).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610140104/DC1.
References
- 1.Michener CD. Annu Rev Entomol. 1969;14:299–342. [Google Scholar]
- 2.Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Science. 2006;321:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- 3.Matthews RW. In: The Social Biology of Wasps. Ross KG, Matthews RW, editors. Ithaca, NY: Comstock, Cornell Univ Press; 1991. pp. 570–602. [Google Scholar]
- 4.Carpenter JM. Syst Entomol. 1982;7:11–38. [Google Scholar]
- 5.Carpenter JM. In: The Social Biology of Wasps. Ross KG, Matthews RW, editors. Ithaca, NY: Comstock, Cornell Univ Press; 1991. pp. 7–32. [Google Scholar]
- 6.Cameron SA, Mardulyn P. Syst Biol. 2001;50:194–214. [PubMed] [Google Scholar]
- 7.Danforth BN. Proc Natl Acad Sci USA. 2002;99:286–290. doi: 10.1073/pnas.012387999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler WM. Proc Am Philos Soc. 1918;57:293–343. [Google Scholar]
- 9.Lin N, Michener CD. Q Rev Biol. 1972;47:131–159. [Google Scholar]
- 10.Hamilton WD. J Theor Biol. 1964b;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- 11.Hunt JH, Amdam GV. Science. 2005;308:264–267. doi: 10.1126/science.1109724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batra SWT. Indian J Entomol. 1966;28:375–393. [Google Scholar]
- 13.Michener CD. The Social Behavior of the Bees: A Comparative Study. Cambridge, MA: Belknap, Harvard Univ Press; 1974. [Google Scholar]
- 14.West-Eberhard MJ. J Kans Entomol Soc. 1978;51:832–856. [Google Scholar]
- 15.Wheeler WM. Social Life Among the Insects. New York: Harcourt Brace; 1923. [Google Scholar]
- 16.Evans HE. Proc Int Congr Entomol 10th 1956. 1958. pp. 449–457.
- 17.Hunt JH. Evolution (Lawrence, Kans) 1999;53:225–237. doi: 10.1111/j.1558-5646.1999.tb05348.x. [DOI] [PubMed] [Google Scholar]
- 18.Crespi BJ, Yanega D. Behav Ecol. 1995;6:109–115. [Google Scholar]
- 19.Hunt JH. The Evolution of Social Wasps. New York: Oxford Univ Press; 2007. in press. [Google Scholar]
- 20.Bohart RM. J Kans Entomol Soc. 1989;62:462–467. [Google Scholar]
- 21.Gess SK. The Pollen Wasps: Ecology and Natural History of the Masarinae. Cambridge, MA: Harvard Univ Press; 1996. [Google Scholar]
- 22.Carpenter JM, Cumming JM. J Nat Hist. 1985;19:877–916. [Google Scholar]
- 23.Cowan DP. In: The Social Biology of Wasps. Ross KG, Matthews RW, editors. Ithaca, NY: Comstock, Cornell Univ Press; 1991. pp. 33–73. [Google Scholar]
- 24.Turillazzi S. In: The Social Biology of Wasps. Ross KG, Matthews RW, editors. Ithaca, NY: Comstock, Cornell Univ Press; 1991. pp. 74–98. [Google Scholar]
- 25.Field J, Foster W, Shreeves G, Sumner S. Proc R Soc London Ser B. 1998;265:973–977. [Google Scholar]
- 26.Matsuura M, Yamane S. Biology of the Vespine Wasps. Berlin: Springer; 1990. [Google Scholar]
- 27.Wilson EO. The Insect Societies. Cambridge, MA: Belknap, Harvard Univ Press; 1971. [Google Scholar]
- 28.Turillazzi S. J Insect Behav. 1989;2:649–661. [Google Scholar]
- 29.Pardi L, Turillazzi S. Atti Accad Naz Ital Entomol Rendic. 1982;30:3–21. [Google Scholar]
- 30.Schmitz J, Moritz RFA. Mol Phylogenet Evol. 1998;9:183–191. doi: 10.1006/mpev.1997.0460. [DOI] [PubMed] [Google Scholar]
- 31.Carpenter JM. Am Mus Novit. 2003;3389:1–20. [Google Scholar]
- 32.Richards OW. Biol Rev Cambridge Philos Soc. 1971;46:483–528. [Google Scholar]
- 33.Spradbery JP. J Aust Entomol Soc. 1975;14:309–318. [Google Scholar]
- 34.Vecht J van der. Tijdschr Entomol. 1977;120:55–75. [Google Scholar]
- 35.Carpenter JM. J N Y Entomol Soc. 1988;96:140–175. [Google Scholar]
- 36.Schmitz J, Moritz RFA. In: Hymenoptera: Evolution, Biodiversity and Biological Control. Austin AD, Dowton M, editors. Victoria, Australia: CSIRO, Collingwood; 2000. pp. 84–89. [Google Scholar]
- 37.Hunt JH. Ann Zool Fenn. 2006;43:407–422. [Google Scholar]
- 38.Lockhart PJ, Cameron SA. Trends Ecol Evol. 2001;16:84–88. doi: 10.1016/s0169-5347(00)02054-1. [DOI] [PubMed] [Google Scholar]
- 39.Bequaert J. Ann Mag Nat Hist. 1928;10:138–176. [Google Scholar]
- 40.Carpenter JM. In: Biological Relationships Between Africa and South America. Goldblatt P, editor. New Haven, CT: Yale Univ Press; 1993. pp. 139–155. [Google Scholar]
- 41.Ducke A. Zool Jahr Abt Syst Geogr Biol Tiere. 1914;36:303–330. [Google Scholar]
- 42.Bequaert J. Bull Am Mus Nat Hist. 1918;39:1–384. [Google Scholar]
- 43.Roubaud E. Ann Sci Nat Zool. 1916;1:1–160. [Google Scholar]
- 44.Roubaud E. Smithson Inst Annu Rep. 1911;1910:507–525. [Google Scholar]
- 45.Williams FX. Experiment Station of the Hawaiian Sugar Planters Association, Entomology Series. 1919;14:19–186. [Google Scholar]
- 46.Vernier R. Bull Soc Neuchatel Sci Nat. 1997;120:87–98. [Google Scholar]
- 47.Rau P. The Jungle Bees and Wasps of Barro Colorado Island. Kirkwood, MO: Phil Rau; 1933. [Google Scholar]
- 48.Rau P. Ann Entomol Soc Am. 1943;36:516–536. [Google Scholar]
- 49.West-Eberhard MJ. In: Chemistry and Biology of Social Insects. Eder J, Rembold H, editors. Munich: J Peperny; 1987. [Google Scholar]
- 50.West-Eberhard MJ. In: Animal Societies: Theories and Facts. Itô Y, Brown JL, Kikkawa J, editors. Tokyo: Scientific Societies; 1987. pp. 35–51. [Google Scholar]
- 51.de Saussure H. Smithson Misc Collect. 1875;254 [Google Scholar]
- 52.Strassmann JE, Hughes CR, Turillazzi S, Solís CR, Queller DC. Anim Behav. 1994;48:813–821. [Google Scholar]
- 53.Field J, Foster W, Shreeves G, Sumner S. Proc R Soc London Ser B. 1998;265:973–977. [Google Scholar]
- 54.Hansell MH. Anim Behav. 1987;35:131–141. [Google Scholar]
- 55.Hunt JH. In: The Social Biology of Wasps. Ross KG, Matthews RW, editors. Ithaca, NY: Comstock, Cornell Univ Press; 1991. pp. 426–450. [Google Scholar]
- 56.Hunt JH. In: Nourishment and Evolution in Insect Societies. Hunt JH, Nalepa CA, editors. Boulder, CO: Westview; 1994. [Google Scholar]
- 57.Brauns H. Z Wiss Insektbiol. 1910;6:348–387. 445–447. [Google Scholar]
- 58.Carpenter JM, Starr CK. Am Mus Novit. 2000;3291:1–12. [Google Scholar]
- 59.Hall TA. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 60.Kjer KM. Mol Phylogenet Evol. 1995;4:314–330. doi: 10.1006/mpev.1995.1028. [DOI] [PubMed] [Google Scholar]
- 61.Gillespie JJ. Mol Phylogenet Evol. 2004;33:936–943. doi: 10.1016/j.ympev.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR. Insect Mol Biol. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2001. Version 4. [Google Scholar]
- 64.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 65.Posada D, Crandall KA. Bioinformatics. 1998;9:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 66.Sorensen MD. TreeRot. Boston: Boston Univz; 1999. Version 2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.