Abstract
During cell division, cessation of transcription is coupled with mitotic chromosome condensation. A fundamental biological question is how gene expression patterns are retained during mitosis to ensure the phenotype of progeny cells. We suggest that cell fate-determining transcription factors provide an epigenetic mechanism for the retention of gene expression patterns during cell division. Runx proteins are lineage-specific transcription factors that are essential for hematopoietic, neuronal, gastrointestinal, and osteogenic cell fates. Here we show that Runx2 protein is stable during cell division and remains associated with chromosomes during mitosis through sequence-specific DNA binding. Using siRNA-mediated silencing, mitotic cell synchronization, and expression profiling, we identify Runx2-regulated genes that are modulated postmitotically. Novel target genes involved in cell growth and differentiation were validated by chromatin immunoprecipitation. Importantly, we find that during mitosis, when transcription is shut down, Runx2 selectively occupies target gene promoters, and Runx2 deficiency alters mitotic histone modifications. We conclude that Runx proteins have an active role in retaining phenotype during cell division to support lineage-specific control of gene expression in progeny cells.
Keywords: chromatin, epigenetic control, mitosis, cell division
Lineage commitment and cell proliferation are critical for normal tissue development. Preservation of phenotype during clonal expansion of committed cells necessitates the faithful segregation of chromosomes and the conveyance of lineage-specific gene regulatory machinery to progeny cells. Mitosis involves nuclear reorganization, global chromosome condensation, and transcription silencing and occurs concomitant with protein degradation and/or displacement of many regulatory factors from chromosomes (1–4). One fundamental question is how cells are programmed to sustain phenotypic gene expression patterns after cell division when transcriptional competency is restored in progeny cells.
Cell fate is determined in response to extracellular cues by lineage-specific master regulators that include the Runx family of transcription factors. In mammals, these proteins are required for development of hematopoietic (Runx1), osteogenic (Runx2), and gastrointestinal and neuronal (Runx3) cell lineages (5–11). Runx factors integrate cell signaling pathways (e.g., TGF-β/bone morphogenetic protein and Yes/Src) and recruit chromatin-modifying enzymes (e.g., histone deacetylases, histone acetyltransferases, SWI/SNF, SuVar139) to modulate promoter accessibility within a nucleosomal context (10, 12–17). Runx proteins function as promoter-bound scaffolds that organize the regulatory machinery for gene expression within punctate subnuclear domains during interphase (18, 19). Pathological perturbations in the organization of these domains are linked with altered development and tumorigenesis (10, 20–29). Temporal and spatial changes of these architecturally organized Runx domains occur during mitosis (13).
Osteogenic cell fate decisions and subsequent proliferation of osteoprogenitor cells are controlled by Runx2 (10, 30–33). A mechanism must be operative that ensures Runx2-dependent regulation of this osteogenic identity through multiple mitotic cell divisions. Here we have combined mitotic cell synchronization, expression profiling, chromatin immunoprecipitation (ChIP), and RNA interference to investigate this mechanism. During mitosis, Runx2 interacts directly with a novel set of cell fate- and cell cycle-related target genes that exhibit distinct Runx2-dependent modifications in histone acetylation and methylation. Our results indicate that Runx transcription factors reinforce cell fate through an epigenetic mechanism that retains phenotypic gene expression patterns after cell division.
Results/Discussion
Runx2 Protein Is Stable During Mitosis and Associated with Mitotic Chromosomes.
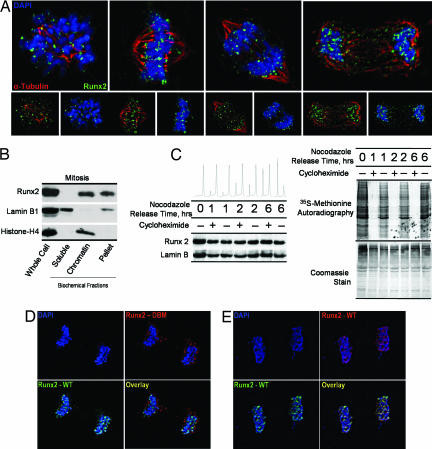
Runx2, a tissue-specific transcription factor that confers cell fate and lineage commitment, is organized in multiple distinct subnuclear foci during interphase (12). Here we show that Runx2 is localized to chromosomes at all stages of mitosis, including metaphase, as determined by three-dimensional deconvolution immunofluorescence microscopy (Fig. 1A). Subcellular fractionation validates the association of Runx2 with mitotic chromatin (Fig. 1B). We also observe mitotic association of endogenous Runx2 with chromosomes in multiple cell lines, including normal calvarial osteoblasts and osteosarcomas (data not shown). Although Runx2 is localized with mitotic chromosomes, a fraction of Runx2 is extrachromosomal, perhaps in association with microtubules (ref. 34 and our unpublished observations). Our conclusion that Runx2 associates with mitotic chromosomes is based on both microscopic evidence (Fig. 1) and biochemical data obtained by ChIP (see below).
Fig. 1.
Runx2 is stable and associated with chromosomes during mitosis. (A) Asynchronously growing Saos-2 cells were fixed and stained for DNA by using DAPI and for Runx2 by using a rabbit polyclonal antibody. Mitotic cells were identified by chromosome morphology. High-resolution images obtained by three-dimensional deconvolution algorithms reveal that Runx2 (green) is localized in mitotic chromosomes. A subset of Runx2 colocalizes with the microtubules, labeled by α-tubulin staining (red). (B) Localization of Runx2 by biochemical fractionation using standard techniques to generate soluble, chromatin-associated, and insoluble protein fractions compared with whole-cell protein levels. Each fraction was analyzed by Western blotting using antibodies against Runx2, as well as lamin B1 and histone H4 as controls. (C) Stability of Runx2 protein in mitosis. Saos cells were arrested at the G2/M boundary by nocodazole treatment and released through mitosis into G1 in the presence or absence of the protein translation inhibitor cycloheximide (50 μg/ml). At the indicated times, protein synthesis was assayed by pulse labeling with [35S]methionine. In parallel, protein samples were isolated for Western blot analysis. (D and E) Colocalization studies of wild-type Runx2 and a DNA-binding mutant Runx2. HeLa cells were cotransfected with wild-type Runx2 and the R182Q mutant Runx2 (GenBank accession no. D14637), which were N-terminally tagged with HA and Xpress epitopes, respectively. In situ immunofluorescence microscopy was performed with DNA staining by DAPI and indirect immunolabeling with antibodies directed against HA and Xpress epitopes with appropriate secondary antibodies. Mitotic cells were identified by DNA morphology. Control colocalization experiments were performed by using HA- and Xpress-tagged wild-type proteins.
To assess whether Runx2 is metabolically stable during mitosis, we examined protein levels in synchronized cells. Mitotic cells were released into G1 in the presence or absence of the protein translation inhibitor cycloheximide. Progression into G1 was monitored by microscopy and FACS analysis, and inhibition of translation was verified in parallel by metabolic labeling with [35S]methionine. As cells exit mitosis and enter G1, levels of Runx2 or lamin B1 protein are unaffected by inhibition of translation (Fig. 1C). Thus, Runx2 protein synthesized before division is not turned over and is retained at the onset of the next G1 phase. The stability of Runx2 during mitosis and its association with mitotic chromosomes indicate a potentially novel regulatory function for this cell fate determinant.
Mitotic Chromosome Association of Runx2 Is Abrogated by a Specific Point Mutation.
Loss-of-function mutations that abrogate the sequence-specific DNA binding of Runx proteins alter cell phenotype and result in cancer (e.g., acute myelogenous leukemia) and other human disorders [e.g., cleidocranial dysplasia (CCD) and familial platelet disorder] (35, 36). We hypothesized that the chromosome association of Runx2 in mitosis involves protein–DNA interactions through its conserved Runt homology domain. To test this concept, we used site-directed mutagenesis to generate a CCD-related point mutation in the Runt homology domain, R182Q, that abrogates DNA binding (35, 36). Runt homology domain mutations do not affect nuclear localization during interphase. Electrophoretic mobility shift assays confirmed that the R182Q mutation renders Runx2 defective in DNA binding, as expected (data not shown). Colocalization studies using indirect immunofluorescence microscopy reveal that the Runx2 DNA-binding mutant is excluded from chromosomes during mitosis (Fig. 1 D and E). Thus, association of Runx2 protein with mitotic chromosomes requires an intact Runt homology DNA-binding domain and provides evidence that Runx2 remains bound to its cognate regulatory elements within target genes.
Identification of Mitotically Regulated Runx2 Target Genes Related to Cell Cycle Control and Osteogenesis.
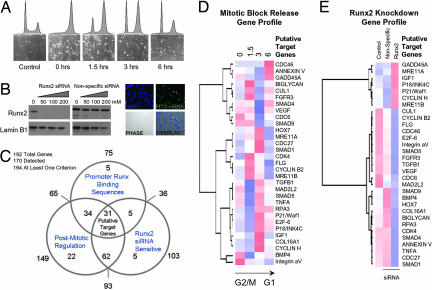
We applied a focused functional genomics strategy to identify mitotic targets of Runx2 related to proliferation and phenotype commitment. Genes were selected that are sensitive to Runx2 siRNA, are mitotically controlled, and have Runx consensus motifs in their promoters. Using cDNA arrays comprising a total of 192 osteogenic and/or cell cycle regulatory genes, we discovered 31 genes that satisfy these three biological criteria (Fig. 2A–E). Efficacy of siRNA oligonucleotides at multiple doses was established (Fig. 2B). Using ChIP assays, we confirmed that at least 14 of these genes are direct Runx2 targets (Fig. 3B). Two of these genes have previously been established as Runx2-responsive (e.g., p21 and VEGF) and thus validate our approach (37, 38). Independent siRNA experiments, in which the knockdown of Runx2 was shown by Western blotting, were analyzed by RT-PCR and directly confirmed that Runx2 controls expression of selected genes (Fig. 3A). Analysis of the target genes identified in our candidate screen [supporting information (SI) Table 1] indicates that Runx2 exerts phenotype control at the transcriptional level to mediate cell cycle progression and signaling pathways that establish competence for lineage commitment.
Fig. 2.
Runx2 target gene identification. To identify mitotic target genes of Runx2, we applied a functional genomics strategy. Genes were selected that exhibit alterations in steady-state mRNA during progress from mitosis into G1, that are sensitive to Runx2 siRNA, and that have promoters with Runx consensus motifs. The first two criteria were assessed by cDNA array-based gene profiling (SuperArray Bioscience Corporation), and the final criterion was assessed through a bioinformatics analysis by using TFSEARCH (48). (A) Mitotic cells were released into G1, and RNA was taken at 0, 1.5, 3, and 6 h for analysis. (B) siRNA knockdown of Runx2 was monitored by Western blot analysis at concentrations of 50, 100, and 200 nM. Fluorophore-conjugated siRNA oligonucleotides were transfected in parallel to determine transfection efficiency. Micrographs show localization of siRNA oligonucleotides in cells with ≈95% efficiency at 100 nM. (C) Venn diagram indicates the number of genes in each of the three functional groups. Thirty-one target genes satisfying all three criteria were analyzed by hierarchical clustering based on cell cycle expression data (D) and expression in the Runx2-knockdown experiment (E) (see SI Data Set 1 for primer information). Color maps are applied to standardized gene expression data: pure blue, −3; pure white, 0; and pure red, 3.
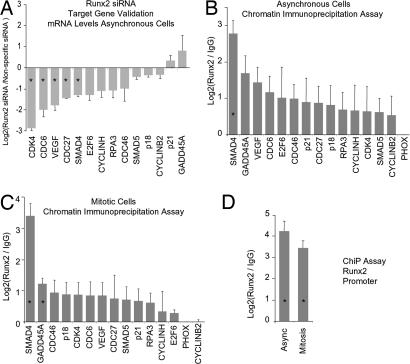
Fig. 3.
Target gene validation. (A) Fourteen putative Runx2 target genes identified in a primary screen (see Fig. 2) were tested for responsiveness to depletion of Runx2 by RNA interference. Independent siRNA experiments were analyzed in duplicate by RT-qPCR with primer sets for each of the target genes (SI Data Set 1). Expression data are normalized and displayed as the log2 difference between Runx2 and nonspecific siRNAs. Error bars reflect SEM. (B) Runx2 target genes were validated by ChIP in two independent experiments (SI Data Set 1). Samples were quantified by qPCR relative to input and normalized to nonspecific immunoprecipitation of the PHOX gene promoter. Values represent the log2 difference between Runx2-specific and control IgG signals. (C) Interaction of Runx2 with its novel target genes during mitosis was assessed by ChIP assays. Mitotic cells were isolated by nocodazole synchronization and mitotic shakeoff in two independent experiments, and samples were assayed in duplicate by qPCR. Data analysis is described in B. (D) Interaction of Runx2 with its promoter was assessed by ChIP on asynchronous and mitotic cells. Asterisks indicate statistical significance at the 0.05 level based on a t test.
Because our immunofluorescence microscopy studies indicate that Runx2 binds sequence-specifically to mitotic chromosomes, we tested whether Runx2 associates with target genes at mitosis. ChIP assays were performed on prometaphase cells. Analysis by quantitative PCR (qPCR) and radiolabeled PCR of ChIP products from multiple independent experiments reveals that Runx2 retains association with the promoters of nearly all target genes examined within the condensed mitotic chromosomes (Fig. 3C and SI Fig. 7). Furthermore, we have established that Runx2 protein binds to the Runx2 promoter during mitosis, suggesting a mitotic autoregulatory function (Fig. 3D). Interestingly, association of Runx2 with the promoter of the cyclin B2 gene, which is involved in control of mitotic progression, is reduced in mitosis. Our findings suggest that Runx2 provides a critical regulatory function in progeny cells for postmitotic gene expression in G1.
Runx2 Target Genes Exhibit Mitotic-Specific Histone Modifications.
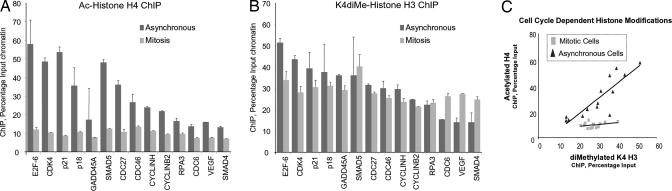
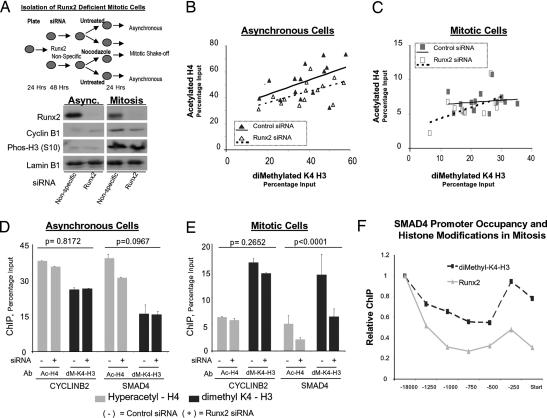
Recent work indicates that specific histone modifications may mark active genes in mitosis (39). We investigated whether promoters of Runx2 target genes exhibit mitotic-specific epigenetic changes that could be indicative of postmitotic transcriptional state. Acetylation of histone H4 and dimethylation of histone H3 on lysine 4 (K4), which are both linked with active gene expression, were examined in both asynchronous and mitotically synchronized cells (Fig. 4). The majority of Runx2-responsive genes exhibit substantially decreased H4 acetylation in mitosis (Fig. 4A). In contrast, we observe a retention or selective increase in mitotic histone H3–K4 dimethylation compared with asynchronous cells (Fig. 4B). Our data show that in general, histone H4 acetylation is positively correlated with histone H3–K4 dimethylation in asynchronous cells, as would be expected for actively transcribed genes. However, in mitosis we observe a basal level of H4 acetylation that is no longer tightly linked with H3–K4 methylation (Fig. 4C). The reduction of histone H4 acetylation may be coupled with the general shutdown of transcription in mitosis. We propose that the persistence of basal levels of H4 acetylation and constitutive histone H3–K4 dimethylation at Runx target gene promoters during mitosis reflects a transcriptionally poised chromatin structure.
Fig. 4.
Runx2 is associated with epigenetically modified target genes in mitosis. Histone modifications at the 14 target genes were assayed by ChIP analyses of asynchronous (dark gray bars) and pure mitotic cells (light gray bars) synchronized as described in Fig. 3. Duplicate samples were analyzed by qPCR, quantified as a percentage of input, and normalized for comparison with subsequent functional experiments in Fig. 5. (A) Histone H4 acetylation and (B) histone H3–K4 dimethylation of gene promoters. (C) Scatterplot of H4 acetylation (ordinate) versus H3–K4 dimethylation (abscissa) for all 14 genes in asynchronous cells (black triangles) and mitotic cells (gray squares) is depicted. A least-squares regression line is shown for each population.
Runx2 Controls Posttranslational Histone Modifications at Target Gene Promoters During Mitosis.
Runx2 mediates activation and repression of gene transcription through interactions with a diverse set of chromatin-modifying enzymes. Because Runx2 associates during mitosis with target gene promoters that exhibit distinct histone modifications, we mechanistically addressed whether Runx2 mediates these epigenetic alterations. We generated Runx2-deficient mitotic cells by using RNA interference and confirmed reduction in Runx2 levels by Western blot analysis (Fig. 5A). The effect on H3–K4 dimethylation and H4 acetylation at Runx2-regulated promoters was then determined. Quantitative ChIP analyses for the 14 target genes revealed that depletion of Runx2 protein alters promoter histone modifications (P = 0.0005) and that these effects are gene-specific (P = 0.0001). In asynchronous cells, we find an overall reduction in levels of H4 acetylation but not H3–K4 dimethylation at Runx2 target gene promoters (Fig. 5B). In contrast, during mitosis we find that loss of Runx2 significantly diminishes both H3–K4 dimethylation and H4 acetylation at target gene promoters (Fig. 5C). We observe the greatest effect on histone modifications at the SMAD4 gene, which in response to Runx2 knockdown exhibits decreased H3–K4 dimethylation and H4 acetylation during mitosis (P < 0.0001) but not in asynchronous cells (Fig. 5 D and E). These observations are consistent with high levels of Runx2 interaction with the SMAD4 promoter in mitosis and sensitivity of SMAD4 expression to Runx2 siRNA (Fig. 3 A and B). For comparison, we do not detect Runx2-dependent histone modifications at the cyclin B2 promoter, which does not bind Runx2 during mitosis (Fig. 5 D and E). Consistent with these molecular observations, we find that Runx2 binding at multiple sites across the SMAD4 locus mechanistically correlates with dimethylation at K4 of histone H3 (Fig. 5F). Taken together, our findings indicate that Runx2 contributes to epigenetic regulation by supporting histone modification at target gene promoters during mitosis.
Fig. 5.
Runx2 affects mitotic histone modifications at target gene promoters. The effects of Runx2 on promoter histone modifications were assessed by combining siRNA gene knockdown with mitotic cell synchronization. (A) Experimental strategy to obtain Runx2-depleted mitotic cells. Histone modification levels at target gene promoters in mitotic and asynchronous cells were assayed by ChIP and analyzed by qPCR. Protein was extracted from parallel plates to validate Runx2 knockdown. Cyclin B1 levels and histone H3 (S10) phosphorylation status serve as markers of mitosis and lamin B1 as a loading control. Efficiency of siRNA transfection (>90%) was determined in parallel (data not shown). (B and C) Levels of hyperacetylated histone H4 and dimethylated histone H3 (K4) in control and Runx2 siRNA-treated cells were determined in two ChIP assays, each analyzed in duplicate by qPCR. Scatterplot of H4 acetylation versus H3 K4-dimethylation is shown for all 14 target genes in asynchronous (B) and mitotic cells (C) treated with control (black symbols) or Runx2 siRNA (open symbols). A least-squares regression line is shown for each population: control (solid line) or Runx2 siRNA (broken line). (D and E) A mixed-model ANOVA was used to assess significance of Runx2 siRNA effects for all target genes. Multiple pairwise comparisons (Tukey's HSD) were evaluated to determine which effects differ at a 0.05 level and to establish P values; error bars are SE (n = 4). Plots show ChIP results for SMAD4 and CYCLINB2 in Runx2 and control siRNA-treated asynchronous (D) and mitotic cells (E). (F) Binding of Runx2 across the SMAD4 promoter was compared with dimethyl-K4 histone H3 modifications. Primer sets encompassed positions in the proximal and distal SMAD4 promoter, as well as the transcription start site (see SI Data Set 1). Within-group (Runx2 or K4–H3 dimethylated) ratios as calculated in D and E are plotted.
Runx2 Is a Lineage-Specific Regulator for Mitotic Retention of Gene Expression Patterns.
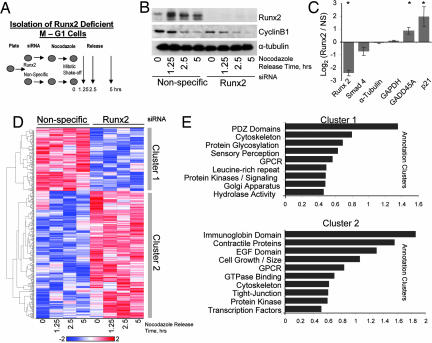
Because the lineage-specific Runx2 protein controls cell fate, binds to mitotic chromosomes, and modifies chromatin of its target genes, we sought to determine on a genome-wide scale the breadth of gene classes modulated by Runx2 during and immediately after mitosis. We performed genome-wide expression profiling of mitotically synchronized cells (Fig. 6A and SI Data Sets 2 and 3). Runx2 siRNA or nonsilencing control siRNA-treated cells were blocked in mitosis and allowed to release synchronously into G1. Depletion of Runx2 protein was confirmed by Western blot analysis, and cyclin B1 protein levels were monitored to confirm egress from mitotic arrest (Fig. 6B). RNA samples from both treatment groups were analyzed by using Affymetrix microarrays (Hu-U133Plus2 chips) at four different time points after release of cells into G1 (Fig. 6 C and D). The microarray data show that SMAD4, GADD5A, and p21, which are Runx2 target genes identified in our focused screens and ChIP analyses (see), exhibit the expected modulations in mRNA levels upon Runx2 siRNA treatment (Fig. 6C). Down-regulation of Runx2 RNA levels further confirms efficacy of Runx2 siRNA.
Fig. 6.
Genome-wide identification of Runx2-sensitive gene expression patterns during the mitosis to G1 transition. (A) Runx2 and control siRNA-treated cells were synchronized by nocodazole and mitotic shakeoff. Mitotic cells were isolated at shakeoff (0 h), and remaining cells were replated and released for progression into G1 (1.25, 2.5, and 5 h). (B) Total cellular protein was isolated for Western blot analysis to confirm Runx2 knockdown by siRNA. Cyclin B1 levels confirm release from mitosis. (C) Time-averaged expression of Runx2 and target genes detected on Affymetrix microarrays. Asterisks indicate significance at the 0.05 level. (D) Using an empirical Bayes linear modeling approach, we identified 500 genes significantly altered by siRNA treatment. A heat map illustrating hierarchical cluster analysis is shown. Two main clusters reflect genes that are repressed (Cluster 1) and activated (Cluster 2) by Runx2 knockdown. (E) Gene annotation enrichment analysis was performed to elucidate the biological processes and pathways associated with each gene cluster. The top annotation clusters are shown in bar plots; the value of the abscissa reflects the annotation enrichment score. Group names on the ordinate were based on interpretation of the underlying annotations.
To identify genes within our microarray data set that are regulated differentially between Runx2 and nonspecific siRNA-treated cells progressing through the M to G1 transition, we used an empirical Bayes linear modeling approach (40). Our analysis of the time course data revealed a large set of genes that are significantly altered in the mitosis to G1 transition by depletion of Runx2 protein. Hierarchical cluster analysis of the top 500 genes defined groups of genes that are either down-regulated (Cluster 1) or up-regulated (Cluster 2) by treatment with Runx2 siRNA when cells progress from mitosis into G1 (Fig. 6D). Cluster 1, which represents genes activated by Runx2, contains a preponderance of genes that encode proteins involved in cell signaling and distinct nonnuclear functions including cytoskeletal proteins (Fig. 6E). Cluster 2, which encompasses proteins whose expression is normally suppressed by Runx2, contains prominent subsets involved in cell signaling, growth, and cell size, as well as factors involved in transcriptional regulation. Our results define a mitosis/early G1 Runx2-dependent gene expression signature and indicate that Runx2 mediates continuity of expression between parental and progeny cells.
Conclusions
We have investigated the Runx2 transcription factor as a paradigm for understanding mechanisms by which phenotypic control of gene expression is sustained during mitotic division. Our results show that Runx2 interacts sequence-specifically with mitotic chromosomes at target gene promoters and controls histone H4 acetylation and histone H3–K4 dimethylation during mitosis. Runx2 is thought to function as a promoter-bound scaffold for the temporal recruitment of coactivators or repressors and associated chromatin-modifying factors that are capable of establishing histone modification patterns in mitosis. Loss-of-function DNA-binding mutations in Runx proteins eliminate mitotic chromosome association and are linked with alterations in cell phenotype in multiple human disorders. We propose that chromosomal association of Runx2 in mitosis supports epigenetic retention of phenotype during cell division to maintain lineage identity of progeny cells.
Materials and Methods
Cell Culture and Cell Synchronization.
Cell synchronization was accomplished by nocodazole block and mitotic shakeoff. Protocols are provided in SI Materials and Methods.
Gene Expression Analysis.
Constructs and methods for EMSA have been reported previously (12). Western blot analysis was performed by using commercially available or reported antibodies (41). Total RNA was subjected to real-time PCR using SYBR chemistry (Applied Biosystems, Inc., Foster City, CA). Primers for gene validation studies span exons that are contained in the transcripts. For siRNA-knockdown experiments, Saos-2 cells were transfected by using Oligofectamine (Invitrogen, Carlsbad, CA) with siRNA duplexes specific for human Runx2 (Qiagen, Inc., Valencia, CA). Gene profiling and histone modification studies used oligonucleotides at 50 and 25 nM, respectively. Focused expression profiling was performed by using the osteogenic and cell cycle cDNA arrays according to the manufacturer's GEArray instructions (SuperArray Bioscience Corporation, Frederick, MD). Gene expression patterns were identified by using hierarchical cluster analysis of row-wise standardized data with dCHIP software (42). Affymetrix microarrays (Hu-U133Plus2 chips) and ChIP assays were performed as described (43–45).
In Situ Immunofluorescence Microscopy.
Cells were grown on gelatin-coated coverslips and processed for in situ immunofluorescence by using standard techniques. Immunostaining of cell preparations was recorded by using an CCD camera attached to a epifluorescence Zeiss Axioplan 2 (Zeiss, Inc., Thorwood, NY) microscope.
Statistical Analysis.
t tests were performed to assess the significance of observed changes in gene expression associated with siRNA knockdown of Runx2 protein and to determine the significance of observed Runx2 binding to target gene promoters. ANOVA was carried out to assess the significance of Runx2 knockdown on histone modifications at target genes. A mixed-model analysis was performed on log-transformed ChIP data by using SAS/Analyst (Sas Institute, Inc., Cary, NC) with genes (14), antibodies (2), and cell cycle stage (2) incorporated as fixed effects and qPCR well position as a random effect. The complete microarray data analysis approach is detailed in SI Materials and Methods. Runx2-responsive genes identified in the microarray analysis were subjected to functional annotation clustering by using DAVID 2006 found at http://david.abcc.ncifcrf.gov (46, 47).
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey Nickerson for consultation on microscopy, Stephen Baker for statistical consutations, and Judy Rask and Elizabeth Bronstein for editorial assistance. This work was supported in part by National Institutes of Health Grants P01AR48818 (to G.S.S.), P01CA82834 (to G.S.S.), AR049069 (to A.J.W.), AR39588 (to G.S.S.), T32 AR07572, and P30 DK32520.
Abbreviation
- qPCR
quantitative PCR.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611419104/DC1.
References
- 1.Gottesfeld JM, Forbes DJ. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 3.Muchardt C, Reyes J-C, Bourachot B, Legouy E, Yaniv M. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 4.Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J-Y, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, et al. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao Y-H, Inada M, et al. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, et al. Nat Neurosci. 2002;5:946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- 9.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 10.Westendorf JJ, Hiebert SW. J Cell Biochem Suppl. 1999;32-33:51–58. doi: 10.1002/(sici)1097-4644(1999)75:32+<51::aid-jcb7>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- 11.Blyth K, Cameron ER, Neil JC. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 12.Zaidi SK, Javed A, Choi J-Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- 13.Zaidi SK, Young DW, Pockwinse SH, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Proc Natl Acad Sci USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniuchi I, Littman DR. Oncogene. 2004;23:4341–4345. doi: 10.1038/sj.onc.1207671. [DOI] [PubMed] [Google Scholar]
- 15.Vradii D, Doan DN, Wagner S, Nickerson JA, Lian JB, Stein JL, van Wijnen AJ, Imbalzano AN, Stein GS. J Cell Physiol. 2006;206:112–118. doi: 10.1002/jcp.20432. [DOI] [PubMed] [Google Scholar]
- 16.Young DW, Pratap J, Javed A, Weiner B, Ohkawa Y, van Wijnen A, Montecino M, Stein GS, Stein JL, Imbalzano AN, et al. J Cell Biochem. 2005;94:720–730. doi: 10.1002/jcb.20332. [DOI] [PubMed] [Google Scholar]
- 17.Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, Arriagada G, Olate J, Imschenetzky M, van Wijnen AJ, et al. Mol Cell Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young DW, Zaidi SK, Furcinitti PS, Javed A, van Wijnen AJ, Stein JL, Lian JB, Stein GS. J Cell Sci. 2004;117:4889–4896. doi: 10.1242/jcs.01229. [DOI] [PubMed] [Google Scholar]
- 20.Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Proc Natl Acad Sci USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes GL, Javed A, Waller SM, Kamal MH, Hebert KE, Hassan MQ, Bellahcene A, van Wijnen AJ, Young MF, Lian JB, et al. Cancer Res. 2003;63:2631–2637. [PubMed] [Google Scholar]
- 22.Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Cancer Res. 2004;64:4506–4513. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- 23.Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER, Neil JC, Stewart M. Oncogene. 2001;20:295–302. doi: 10.1038/sj.onc.1204090. [DOI] [PubMed] [Google Scholar]
- 24.Brubaker KD, Vessella RL, Brown LG, Corey E. Prostate. 2003;56:13–22. doi: 10.1002/pros.10233. [DOI] [PubMed] [Google Scholar]
- 25.Cameron ER, Neil JC. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- 26.Ito Y. Oncogene. 2004;23:4198–4208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- 27.Neil J, Stewart M, Terry A, O'Hara M, Hu M, Blyth K, Baxter E, Onions D, Cameron E. Leukemia. 1999;13(Suppl 1):S83–S86. doi: 10.1038/sj.leu.2401295. [DOI] [PubMed] [Google Scholar]
- 28.Vaillant F, Blyth K, Terry A, Bell M, Cameron ER, Neil J, Stewart M. Oncogene. 1999;18:7124–7134. doi: 10.1038/sj.onc.1203202. [DOI] [PubMed] [Google Scholar]
- 29.Otto F, Kanegane H, Mundlos S. Hum Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- 30.Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 31.Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 33.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- 34.Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nickerson J, Javed A, Redick S, Lian JB, van Wijnen AJ, Stein JL, et al. J Cell Physiol. 2006;206:354–362. doi: 10.1002/jcp.20469. [DOI] [PubMed] [Google Scholar]
- 35.Osato M. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 36.Zhou G, Chen Y, Zhou L, Thirunavukkarasu K, Hecht J, Chitayat D, Gelb BD, Pirinen S, Berry SA, et al. Hum Mol Genet. 1999;8:2311–2316. doi: 10.1093/hmg/8.12.2311. [DOI] [PubMed] [Google Scholar]
- 37.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zelzer E, Glotzer DJ, Hartmann C, Thomas D, Fukai N, Soker S, Olsen BR. Mech Dev. 2001;106:97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 39.Kouskouti A, Talianidis I. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth GK. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. Proc Natl Acad Sci USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Wong WH. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Meijden CM, Lapointe DS, Luong MX, Peric-Hupkes D, Cho B, Stein JL, van Wijnen AJ, Stein GS. Cancer Res. 2002;62:3233–3243. [PubMed] [Google Scholar]
- 44.Balint E, Lapointe D, Drissi H, van der Meijden C, Young DW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. J Cell Biochem. 2003;89:401–426. doi: 10.1002/jcb.10515. [DOI] [PubMed] [Google Scholar]
- 45.Hovhannisyan H, Cho B, Mitra P, Montecino M, Stein GS, van Wijnen AJ, Stein JL. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 48.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.