Abstract
Fusicoccins are a class of diterpene glucosides produced by the plant-pathogenic fungus Phomopsis amygdali. As modulators of 14-3-3 proteins, fusicoccins function as potent activators of plasma membrane H+-ATPase in plants and also exhibit unique biological activity in animal cells. Despite their well studied biological activities, no genes encoding fusicoccin biosynthetic enzymes have been identified. Cyclic diterpenes are commonly synthesized via cyclization of a C20 precursor, geranylgeranyl diphosphate (GGDP), which is produced through condensation of the universal C5 isoprene units dimethylallyl diphosphate and isopentenyl diphosphate by prenyltransferases. We found that (+)-fusicocca-2,10 (14)-diene, a tricyclic hydrocarbon precursor for fusicoccins, is biosynthesized from the C5 isoprene units by an unusual multifunctional enzyme, P. amygdali fusicoccadiene synthase (PaFS), which shows both prenyltransferase and terpene cyclase activities. The functional analysis of truncated mutants and site-directed mutagenesis demonstrated that PaFS consists of two domains: a terpene cyclase domain at the N terminus and a prenyltransferase domain at the C terminus. These findings suggest that fusicoccadiene can be produced efficiently in the fungus by using the C5 precursors, irrespective of GGDP availability. In fact, heterologous expression of PaFS alone resulted in the accumulation of fusicocca-2,10 (14)-diene in Escherichia coli cells, whereas no product was detected in E. coli cells expressing Gibberella fujikuroi ent-kaurene synthase, another fungal diterpene cyclase that also uses GGDP as a substrate but does not contain a prenyltransferase domain. Genome walking suggested that fusicoccin biosynthetic enzymes are encoded as a gene cluster near the PaFS gene.
Keywords: biosynthesis, cyclase, prenyltransferase
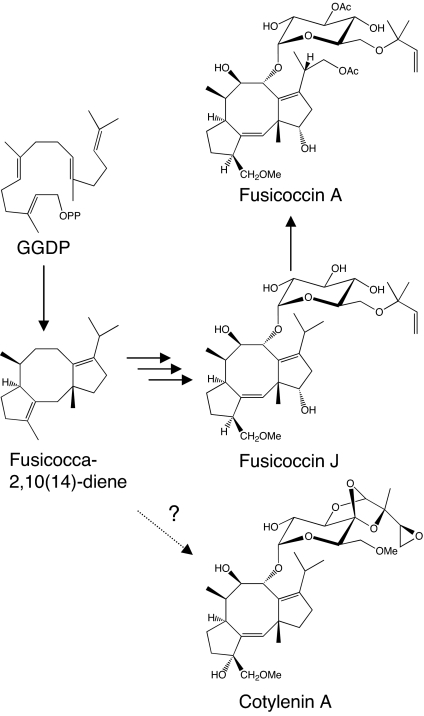
Fusicoccins (FC) A (1, 2) and J (3, 4) (Fig. 1) are the major metabolites produced by the plant-pathogenic fungus Phomopsis (Fusicoccum) amygdali (Del.) (5). Fusicoccin A and the structurally related cotylenins (CN) (6–8), e.g., cotylenin A (Fig. 1) isolated from Cladosporium sp. 501-7W, permanently activate plasma membrane H+-ATPases in all higher plants (9). Their mode of action has been detailed via x-ray crystallographic analysis of the ternary complex of a plant 14-3-3 protein, FC, and a phosphopeptide derived from the C terminus of H+-ATPase (10). FC is a positive modulator of the function of 14-3-3 proteins. Because 14-3-3 proteins form a highly conserved family of molecules in eukaryotes that regulate a wide array of cellular processes (11), the FC/CN class of diterpene glycosides was also anticipated to have effects in animal cells. Indeed, CN potently induces the differentiation of human myeloid leukemia cells (12–14) and acts synergistically with IFN-α to induce apoptosis in a wide array of cancer cells (15, 16). FC also induces the randomization of left–right patterning during amphibian embryogenesis through an interaction with 14-3-3 ε (17). FC/CNs are truly novel chemical genomic tools for modulating 14-3-3 activity. Investigating the diverse and important physiological roles of 14-3-3 proteins in intracellular signal transductions would be greatly enabled via access to a variety of FC/CN analogs. Because the complexity of these molecules precludes the total synthesis of FC/CN analogs, the identification of their biosynthetic genes is of great importance.
Fig. 1.
Proposed biosynthetic pathway of fusicoccin A and J.
The aglycon of FC is a cyclic diterpene derived from geranylgeranyl diphosphate (GGDP). Terpenoids are biosynthesized from the isoprene units isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which are derived through the mevalonate pathway, the methylerythritol pathway, or both (18). Cyclic terpenoids are constructed through the determination of prenyl chain length catalyzed by prenyltransferases, carbon skeleton formation by terpene cyclases, and chemical modification by oxidation, reduction, methylation, glucosylation, and other processes (19). Terpene cyclases are key branch-point enzymes catalyzing the complex intermolecular cyclizations of the linear prenyl diphosphates into cyclic hydrocarbons. Previously, we cloned three cDNAs encoding diterpene cyclase from fungi: PfCPS/KS (ent-kaurene synthase from Phaeosphaeria), GfCPS/KS (ent-kaurene synthase from Gibberella fujikuroi), and ACS (aphidicolan-16β-ol synthase from Phoma betae) (20–22). All of these enzymes initiated cyclization by protonation of the double bond of GGDP (type B), followed by second step cyclization initiated by ionization of the diphosphate (type A) (23). In contrast, FC was formed via fusicocca-2,10 (14)-diene (Fig. 1) based on corroborative experimental evidence (24–26), suggesting that the conversion of GGDP into fusicocca-2,10 (14)-diene would be initiated by ionization of the diphosphate (type A). However, no cDNA encoding type-A fungal diterpene cyclase has been isolated.
In this study, we isolated a cDNA, P. amygdali fusicoccadiene synthase (PaFS), encoding fusicocca-2,10 (14)-diene synthase from P. amygdali mycelia. This fungal enzyme catalyzes a cyclization of GGDP via a type-A mechanism. Surprisingly, the enzyme contains two domains, an N-terminal terpene cyclase domain and a C-terminal prenyltransferase domain, and converts isoprene units sequentially into GGDP and then into fusicocca-2,10 (14)-diene. An Escherichia coli strain harboring only the PaFS gene produced a high amount of fusicocca-2,10 (14)-diene, and a possible FC biosynthetic gene cluster occurs in the flanking regions of the PaFS. Taken together, efficient and high-production levels of FC and FC-related compounds can be achieved by using genetically engineered P. amygdali or heterologous cells such as yeast or E. coli.
Results
Isolation of cDNA Encoding an Unusual Chimera Terpene Synthase.
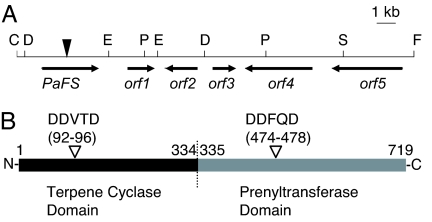
Diterpenoid biosynthetic genes, including a GGDP synthase (GGS) gene, have recently been shown to be clustered in the fungal genome, such as gibberellin and aphidicolin (27, 28). We used genome walking from GGS to find an unknown diterpene synthase gene. We isolated six cDNA fragments encoding putative GGS from mycelia of P. amygdali by RT-PCR with degenerate primers. By gene walking from the GGSs and homology-based RT-PCR, we found two terpene cyclase-like genes, PaDC3 and PaDC4, which were homologous to other fungal sesquiterpene cyclase genes. We determined a full-length PaDC4 cDNA sequence, which unexpectedly indicated that GGS is fused to PaDC4 at the 3′ end within a frame. We tentatively named this fused-type gene PaDC4:GGS (GenBank accession no. AB267396). This cDNA encodes 719-aa residues (81 kDa) and contains eight intron insertion sites based on a comparison with the corresponding genomic DNA sequence. The predicted peptide constitutes two possible domains, the terpene cyclase domain (position 1–334) at the N terminus and the prenyltransferase domain (position 335–719) at the C terminus (Fig. 2B). The terpene cyclase domain exhibited homology with aristolochene synthases, which are the fungal sesquiterpene cyclases (≈20% identities and 40% similarities). In contrast, the prenyltransferase domain was highly homologous with the GGSs in other species of fungi (≈40% identities and 60% similarities). The DDxxD motif, proposed to coordinate the Mg2+ ion (29), was conserved at both domains (Fig. 2B). A BLAST homology search using a full-length PaDC4:GGS amino acid sequence showed that homologs with two domains, annotated as hypothetical proteins or GGS, were registered in the database for Coccidioides immitis (EAS27885; 39% identity), Gibberella zeae (EAA68264; 36% identity), Chaetomium globosum (EAQ85668; 35% identity), and others. These results suggest that unusual chimera genes are ubiquitous in fungal species other than P. amygdali.
Fig. 2.
Chromosome map around PaFS and the primary structure of PaFS. (A) Chromosome map. A filled reverse triangle indicates the starting point of genome walking. The restriction sites for ScaI, DraI, EcoRV, FspI, PvuII, and StuI used in genome walking are represented by the letters C, D, E, F, P, and S, respectively. The direction and deduced region of transcription are represented by arrows. The deduced orf1, orf2, orf3, orf4, and orf5 represent 2-oxo-glutarate-dependent dioxygenase, P450 monooxygenase, short-chain dehydrogenase/reductase, α-mannosidase, and Nop14 like genes, respectively. Homology searches were performed by using BLAST (www.ncbi.nlm.nih.gov/BLAST/). (B) Schematic of two putative domains of PaFS. Black and gray bars indicate the terpene cyclase domain and the prenyltransferase domain, respectively. Reverse open triangles indicate the DDxxD motives of both domains. The borders between the terpene cyclase domain and the prenyltransferase domain were deduced by the homology with other fungal aristolochene synthases or GGSs.
PaFS Encodes Fusicocca-2,10 (14)-diene Synthase Also Possessing GGS Activity.
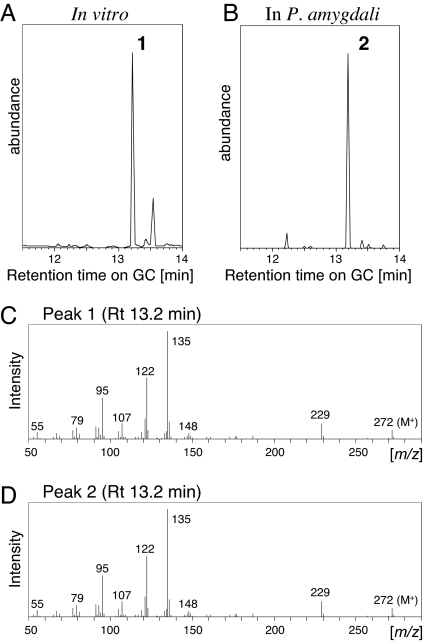
To study the function of PaDC4:GGS, we performed a terpene cyclase assay with the recombinant fusion protein with glutathione S-transferase (GST) at the N terminus, using geranyl diphosphate (GDP), farnesyl diphosphate (FDP), and GGDP as substrates. The recombinant GST-PaDC4:GGS primarily converted GGDP into a substance that peaked at a retention time of 13.2 min in GC (Fig. 3A, peak 1). The retention time and mass spectra of peak 1 (Fig. 3 A and C) were identical to those of authentic fusicocca-2,10 (14)-diene (Fig. 3 B, peak 2, and D) (30). Nine by-products related to fusicoccadiene were detected, of which eight were minor peaks. Among these, the retention time and mass spectra of the three minor peaks at 13.4, 13.8, and 16.9 min (not shown in Fig. 3A) were identical to those of fusicocca-3 (16),10 (14)-diene, δ–araneosene, and β-araneosene, respectively, the structures of which were determined in ref. 30. The structure of the by-product at 13.5 min, which was the most abundant by-product, could not be determined. These results strongly indicate that PaDC4:GGS encodes fusicocca-2,10 (14)-diene synthase; we therefore renamed this gene P. amygdali fusicoccadiene synthase (PaFS).
Fig. 3.
GC–MS results for products converted from GGDP by GST-PaFS. (A) Total ion chromatograms of the products after incubation of GGDP with the recombinant GST-PaFS. (B) Total ion chromatogram of the hydrocarbon fraction obtained from mycelia of P. amygdali N2. (C) Full-scan mass spectrum of peak 1. (D) Full-scan mass spectrum of authentic fusicocca-2,10 (14)-diene (peak 2).
PaFS contains two putative domains, i.e., a terpene cyclase domain at the N terminus and a GGS domain at the C terminus (Fig. 2B), prompting us to perform a terpene cyclase assay with DMAPP, GDP, or FDP with IPP. Surprisingly, the main product of recombinant GST-PaFS with DMAPP, GDP, or FDP in the presence of IPP was fusicocca-2,10 (14)-diene (data not shown), like with GGDP as a substrate. These results indicate that PaFS converted isoprene units into GGDP and successively converted GGDP into fusicocca-2,10 (14)-diene, suggesting that PaFS is an unusual multifunctional diterpene synthase possessing both prenyltransferase and terpene cyclase activities.
Identification of a Terpene Cyclase Domain and a Prenyltransferase Domain in PaFS.
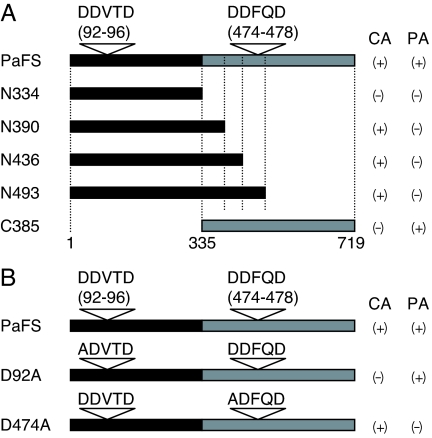
To demonstrate the function of both putative domains on the basis of homology of the primary structure, N- and C-terminal truncated mutants were prepared. The calculated molecular mass of N334 (position 1–334), N390 (position 1–390), N436 (position 1–436), N493 (position 1–493), and C385 (position 335–719) were 38, 44, 49, 56, and 43 kDa, respectively (Fig. 4A). N390, N436, and N493 exhibited cyclase activity, which was estimated by the production of fusicocca-2,10 (14)-diene from GGDP, as in full-length PaFS; N334 and C385 did not exhibit cyclase activity (Fig. 4A, CA). In contrast, C385 catalyzed the conversion of FDP and IPP into GGDP, whereas the four N-terminal peptides of course did not (Fig. 4A, PA). It was remarkable that N390 and N436 possessed cyclase activity even though its DDFQD motif in the GGS domain was omitted. To examine the role of the DDxxD motif in each domain, we prepared two mutants in which aspartate residues were substituted by the nonpolar residue, alanine: D92A and D474A (Fig. 4B). The cyclase assay showed that D474A converted GGDP into fusicocca-2,10 (14)-diene, whereas D92A did not (Fig. 4B, CA). The results of a prenyltransferase assay were complementary to those of the cyclase assay (Fig. 4B, PA). These results demonstrated that the N-terminal 44-kDa peptide catalyzed the cyclization of GGDP, and the C-terminal 43-kDa peptide showed GGS activity.
Fig. 4.
Cyclase and prenyltransferase activity of mutant proteins. Black and gray bars indicate the putative terpene cyclase domain and the prenyltransferase domain, respectively. Reverse open triangles indicate the aspartate-rich motif in each domain. CA, cyclase activity converting GGDP into fusicocca-2,10 (14)-diene; PA, prenyltransferase activity converting FDP and IPP into GGDP. (A) Truncated mutants. The calculated molecular mass of N334, N390, N436, N493, and C385 are 38, 44, 49, 56, and 43 kDa, respectively. The wild-type PaFS is a 719-aa polypeptide, and its molecular mass is 81 kDa. (B) Site-directed mutagenesis study. The aspartate residue in each domain was substituted by an alanine residue. (+), detected; (−), not detected. The activities of the mutant enzymes shown as (+) were almost the same as those of the wild type.
Accumulation of Fusicocca-2,10 (14)-diene in E. coli Expressing PaFS.
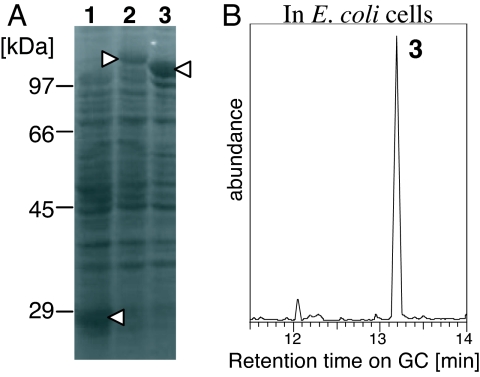
PaFS is responsible not only for forming the FC carbon skeleton but also for determining the chain length of the prenyldiphosphate substrate. We attempted to detect accumulated fusicocca-2,10 (14)-diene in E. coli cells harboring pGEX-PaFS. To check the limitation of GGDP, we also analyzed the hydrocarbon fraction from E. coli cells harboring pGEX-GfCPS/KS encoding ent-kaurene synthase from G. fujikuroi. Previously, we showed that this enzyme catalyzes the conversion of GGDP into ent-kaurene in vitro (21). E. coli cells incubated for 12 h at 17°C after induction of the recombinant protein production were used for extraction. We detected a large amount of fusicocca-2,10 (14)-diene in the hydrocarbon fraction of the extract from E. coli cells expressing GST-PaFS (Fig. 5B, peak 3). In contrast, no ent-kaurene was detected in cells harboring GST-GfCPS/KS (data not shown) in which sufficient amounts of the functional recombinant GST-GfCPS/KS was expressed (Fig. 5A, lane 2).
Fig. 5.
Accumulation of fusicocca-2,10 (14)-diene in E. coli cells carrying GST-PaFS. (A) CBB-stained SDS/PAGE gel to check the recombinant protein expression: lane 1, GST (vector control); lane 2, GST-GfCPS/KS (ent-kaurene synthase from G. fujikuroi, ≈130 kDa); lane 3, GST-PaFS (≈110 kDa). Open triangles show each recombinant protein. (B) Total ion chromatograms of the hydrocarbon fraction obtained from E. coli cells expressing GST-PaFS. The mass spectrum of peak 3 was identical to that of both peaks 1 and 2.
Fusicoccin Biosynthetic Genes Are Clustered in P. amygdali.
PaFS genome walking revealed a 2-oxoglutarate-dependent dioxygenase-like gene (PaDOL4-1), a cytochrome P450 monooxygenase-like gene (PaP450L4-1), a short-chain dehydrogenase/reductase-like gene (PaSDRL4-1), and an α-mannosidase-like gene (PaManL4-1) at the 3′ location downstream of PaFS (Fig. 2A; GenBank accession no. AB272062). Two types of oxidases are thought to be responsible for the oxidation of C8, C9, C12, C16, and C19. PaDOL4-1 belongs to the 2-oxo-glutarate-dependent dioxygenase family, which is specific to fungi (31). PaSDRL4-1 may catalyze the isomerization of the double bond at C2–C3 to C1–C2. The αmannosidase-like PaManL4-1 may be responsible for α-glucosylation at C9. The α-mannosidase/glucosidase could catalyze the translocation of mannose/glucose, in addition to cleavage. The glucosyltransferase family is generally considered responsible for the glucosylation of terpenoids; however, mannosidase has not been shown to catalyze the glucosylation of terpenoids, and we have yet to determine its exact function.
Discussion
We succeeded in cloning a gene for diterpene cyclase (PaFS), which catalyzes the conversion of GGDP into fusicocca-2,10 (14)-diene. PaFS also showed prenyl transferase activity, which determines the chain length of the direct substrate for cyclization reactions. The functional analysis of N- and C-terminal truncated mutants demonstrated that PaFS possesses two domains: a terpene cyclase domain at the N terminus and a prenyltransferase domain at the C terminus (Fig. 6). The site-directed mutagenesis of the DDxxD motif in each domain further indicated that the 92DDVTD motif and 474DDFQD motif were part of the active site for the terpene cyclase and prenyltransferase reactions, respectively. To date, several classes of multifunctional enzymes have been reported in the biosynthesis of natural products such as polyketides, terpenes, and primary metabolites (32–42). These enzymes mechanically catalyze distinct reactions by possessing multiple enzyme domains. Some fungal diterpene cyclases are bifunctional, having an N-terminal type-B cyclase domain and a C-terminal type-A cyclase domain and catalyzing two consecutive cyclization reactions from GGDP (43). However, the PaFS that we identified shows prenyltransferase activity, which provides the substrate GGDP for the cyclization reaction.
Fig. 6.
Synthesis of fusicocca-2,10 (14)-diene from isoprene units by PaFS. This enzyme is a diterpene hydrocarbon synthase possessing both prenyltransferase and terpene cyclase activity.
Previously, an artificially constructed chimeric enzyme containing an FDP synthase domain and a tobacco epi-aristolochene synthase domain was shown to produce the sesquiterpene epi-aristolochene from GDP and IPP (44). That report also suggested that the chimeric enzyme was more efficient than the mixture of two separate enzymes in converting GDP and IPP into epi-aristolochene in vitro (44). We therefore speculate that multifunctional fungal enzymes exhibiting both prenyltransferase and terpene cyclase activity may contribute to the efficient production of bioactive terpenes, regardless of the availability of GGDP in the cell. Our database survey suggested that such multifunctional terpene biosynthesis enzymes also occur in other fungal species, implying that this hypothetical mechanism for the efficient production of terpenes is not restricted to P. amygdali.
The 14-3-3 proteins are important regulators of cellular signaling pathways in eukaryotes and are targets of FC and CN. These terpenoids commonly function as activators of H+-ATPase in plants (9) by modifying the function of 14-3-3 proteins. However, FC and CN often have different effects in animal cells; some studies have reported anticancer activity by CNs (12–16), but none have described such activity by FC derivatives. These observations suggest that modifications of the aglycone of FCs and CNs may alter their biological effects in animal cells. The identification of FC biosynthetic enzymes, especially hydroxylases, will be important in providing enzymatic tools for the production of a new class of FC congeners having CN-like anticancer activity. The possible FC biosynthetic gene cluster that we identified by genome walking (Fig. 2A) will be useful in achieving this goal. We demonstrated that the heterologous expression of the PaFS gene alone is sufficient to produce a large amount of fusicocca-2,10 (14)-diene in E. coli. Because the C5 isoprene units are thought to be synthesized in all organisms as universal precursors to isoprenoids, PaFS will be a powerful and flexible tool for producing FCs in heterologous systems.
Materials and Methods
Chemicals.
IPP, DMAPP, GDP, FDP, and GGDP were purchased from Sigma–Aldrich, St. Louis, MO. 1-[14C]IPP (2.15 GBq/mmol) was obtained from Amersham Pharmacia, Piscataway, NJ.
RT-PCR.
P. amygdali N2 was cultured in shaken mode for 3 days at 25°C in a 500-ml flask (30), and the mycelia were obtained and frozen in liquid nitrogen. The poly(A)+ RNA was extracted from the frozen samples and was used to derive cDNA as described in ref. 45. With an Advantage 2 PCR system (Clontech, Mountain View, CA), RT-PCR was performed with the cDNA pool as a template and a set of degenerate primers: 5′-AAYAARACNGGNGGNYTNTT-3′ (sense) and 5′-CANARRTTCTARTARTCRTC-3′ (antisense) for isolation of GGS; and 5′-GCNATGGSNYTNACNATHCC-3′ (sense) and 5′-TARTCRTCNCKDATYTGRAA-3′ (antisense) for isolation of the chimera gene homolog. The PCR conditions were 40 cycles of denaturation at 94°C for 1 min, annealing at 48°C (GGS) or 50°C (chimera gene homolog) for 1 min, and elongation at 72°C for 1 min. RACE was performed by using gene-specific primers based on the corresponding genome DNA sequences, according to the methods described previously (45).
Genome Walking.
Genome walking was carried out by using a Universal Genome Walker kit (Clontech). Genomic DNA was extracted from the mycelia, as described above, with a Nucleon PhytoPure (Amersham Pharmacia). A genome walker library was constructed with the genomic DNA and was used for nested PCR according to the manufacturer's protocol. In addition to the enclosed restriction enzymes (DraI, EcoRV, PvuII, and StuI), four other enzymes (FspI, NruI, ScaI, and SspI) were used.
Functional Analysis.
To amplify the cDNA for functional analysis, RT-PCR was carried out with the primers 5′-GAATTCTATGGAGTTCAAATACTCGGA-3′ (F1, sense; EcoRI site underlined), 5′-GAATTCGACACAATTGGAATGGATGC-3′ (F2, sense; EcoRI site underlined), 5′-GCGGCCGCACAACCGTCAGAGTTACGAG-3′ (R1, antisense; NotI site underlined), 5′-GCGGCCGCTCAAAAGATGTTATGCGTCGAC-3′ (R2, antisense, NotI site underlined), 5′-GCGGCCGCTCATCCTTTAGATGGCATGGAA-3′ (R3, antisense; NotI site underlined), 5′-GCGGCCGCTCACAGCGAAGAATCCTGCCTA-3′ (R4, antisense; NotI site underlined), and 5′-GCGGCCGCTCACTTGTTGAATGAAACATCT-3′ (R5, antisense; NotI site underlined). The combinations of F1–R1, F1–R2, F1–R3, F1–R4, F1–R5, and F2–R1 were used to amplify cDNA encoding full-length PaFS, N493, N436, N390, N334, and C385, respectively. Site-directed mutagenesis was carried out by overlap extension PCR (46) by using two sets of mutagenic primers: 5′-TCTGCACGCTGATGTTACCGAC-3′ (sense, mutation underlined) and 5′-CGGTAACATCAGCGTGCAGAAAGG-3′ (antisense, mutation underlined) for D92A; 5′-GCTGCTCGCCGACTTCCAAGACA-3′ (sense, mutation underlined) and 5′-TCTTGGAAGTCGGCGAGCAGCAG-3′ (antisense, mutation underlined) for D474A. Each subcloned PCR product was digested with EcoRI and NotI and ligated into the pGEX-4T-3 vector (Amersham Pharmacia) for expression as a fusion protein with GST at the N terminus. The plasmid was transformed into E. coli XL1-Blue. Procedures for the growth of E. coli cells, induction of gene expression, extraction and purification of recombinant enzymes, and measurement of cyclase enzyme activity were identical to those described previously in ref. 47. A 20-μg aliquot of DMAPP, GDP, FDP, or GGDP was used as the substrate with or without 20 μg of IPP. Hydrocarbon products were analyzed by GC–MS. The estimation of prenyltransferase activity was carried out by using methods described in ref. 48.
Extraction and Purification of the Hydrocarbon Fraction.
The n-hexane extract was prepared in the usual way from the samples of P. amygdali mycelia or E. coli cells. This extract was loaded into a column packed with silica gel (BM-820MH; Fuji Silysia Chemical, Aichi, Japan). The column was eluted by n-hexane, and the eluate was evaporated and subjected to GC–MS analysis.
GC–MS.
GC–MS analysis was performed by using an Agilent 6890N GC-5973N mass selective detector system (Agilent Technologies, Palo Alto, CA): HP-1MS capillary column (0.25-mm width × 30-m length), 60°C (2 min)–150°C (30°C/min), 180°C (10°C/min), 210°C (2°C/min) (47).
Acknowledgments
We thank Dr. S. Kanematsu (Apple Research Center, National Institute of Fruit Tree Science, Morioka, Japan) for providing the fungus P. amygdali N2; Dr. S. Yamaguchi (RIKEN Plant Science Center, Yokohama, Japan) and Prof. H. Oikawa (Hokkaido University, Sappora, Japan) for their critical reading of earlier drafts of the manuscript; H. Kenmoku, Y. Kanno, S. Miura, T. Kagahara, and Y. Hirose (Yamagata University, Yamagata, Japan) and C. Ikeda and N. Tajima (Toyama Prefectural University, Toyama, Japan) for technical support. This work was supported in part by The Institute of Physical and Chemical Research (Japan) Grant-in-Aid for Science Research (A) 13306009 and 16208012.
Abbreviations
- CN
cotylenin
- DMAPP
dimethylallyl diphosphate
- FDP
farnesyl diphosphate
- FC
fusicoccin
- GDP
geranyl diphosphate
- GGDP
geranylgeranyl diphosphate
- GGS
geranylgeranyl diphosphate synthase
- GST
glutathione S-transferase
- IPP
isopentenyl diphosphate
- PaFS
P. amygdali fusicoccadiene synthase.
Footnotes
References
- 1.Ballio A, Brufani M, Casinovi CG, Cerrini S, Fedeli W, Pellicciari R, Santurbano B, Vaciago A. Experientia. 1968;24:631–635. doi: 10.1007/BF02153818. [DOI] [PubMed] [Google Scholar]
- 2.Barrow KD, Barton DHR, Chain EB, Ohnsorge UFW, Thomas R. Chem Commun. 1968:1198–1200. [Google Scholar]
- 3.Ballio A, Casinovi CG, D'Alessio V, Grandolini G, Randazzo G, Rossi C. Experientia. 1974;30:844–845. doi: 10.1007/BF01938313. [DOI] [PubMed] [Google Scholar]
- 4.Barrow KD, Barton DHR, Chain E, Bageenda-Kasujja D, Mellows G. J Chem Soc Perkin Trans 1. 1975:877–883. [Google Scholar]
- 5.Tuset JJ, Portilla T. Can J Bot. 1989;67:13275–13280. [Google Scholar]
- 6.Sassa T. Agric Biol Chem. 1971;35:1415–1418. [Google Scholar]
- 7.Sassa T, Togashi M, Shindo T. Agric Biol Chem. 1975;39:1735–1744. [Google Scholar]
- 8.Sassa T, Ooi T, Nukina M, Kato N. Biosci Biotechnol Biochem. 1998;62:1815–1818. doi: 10.1271/bbb.62.1815. [DOI] [PubMed] [Google Scholar]
- 9.Marré E. Ann Rev Plant Physiol. 1979;30:273–288. [Google Scholar]
- 10.Würtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C. EMBO J. 2003;22:987–994. doi: 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitken A. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Asahi K, Honma Y, Hazeki K, Sassa T, Kubohara Y, Sakurai A, Takahashi N. Biochem Biophys Res Commun. 1997;238:758–763. doi: 10.1006/bbrc.1997.7385. [DOI] [PubMed] [Google Scholar]
- 13.Honma Y. Leuk Lymphoma. 2002;43:1169–1178. doi: 10.1080/10428190290026222. [DOI] [PubMed] [Google Scholar]
- 14.Matsunawa M, Ishii Y, Kasukabe T, Tomoyasu S, Ota H, Honma Y. Leuk Lymphoma. 2006;47:733–740. doi: 10.1080/10428190500375839. [DOI] [PubMed] [Google Scholar]
- 15.Honma Y, Ishii Y, Yamamoto-Yamaguchi Y, Sassa T, Asahi K. Cancer Res. 2003;63:3659–3666. [PubMed] [Google Scholar]
- 16.Honma Y, Kasukabe T, Yamori T, Kato N, Sassa T. Gynecol Oncol. 2005;99:680–688. doi: 10.1016/j.ygyno.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Bunney TD, De Boer AH, Levin M. Development (Cambridge, UK) 2003;130:4847–4858. doi: 10.1242/dev.00698. [DOI] [PubMed] [Google Scholar]
- 18.Kuzuyama T. Biosci Biotechnol Biochem. 2002;66:1619–1627. doi: 10.1271/bbb.66.1619. [DOI] [PubMed] [Google Scholar]
- 19.Poulter CD, Rilling HC. In: Biosynthesis of Isoprenoid Compounds. Spurgeon SL, Porter JW, editors. New York: Wiley; 1982. pp. 161–224. [Google Scholar]
- 20.Kawaide H, Imai R, Sassa T, Kamiya Y. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 21.Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, Sassa T. Biosci Biotechnol Biochem. 2000;64:660–664. doi: 10.1271/bbb.64.660. [DOI] [PubMed] [Google Scholar]
- 22.Oikawa H, Toyomasu T, Toshima H, Ohashi S, Kawaide H, Kamiya Y, Ohtsuka M, Shinoda S, Mitsuhashi W, Sassa T. J Am Chem Soc. 2001;123:5154–5155. doi: 10.1021/ja015747j. [DOI] [PubMed] [Google Scholar]
- 23.MacMillan J, Beale MH. In: Comprehensive Natural Products Chemistry. Cane D, editor. Vol 2. Oxford: Elsevier Science; 1999. pp. 217–243. [Google Scholar]
- 24.Kato N, Zhang C-S, Matsui T, Iwabuchi H, Mori A, Ballio A, Sassa T. J Chem Soc Perkin Trans 1. 1998:2473–2474. [Google Scholar]
- 25.Kato N, Zhang C-S, Tajima N, Mori A, Graniti A, Sassa T. J Chem Soc Chem Commun. 1999:367–368. [Google Scholar]
- 26.Sassa T, Zhang C-S, Sato M, Tajima N, Kato N, Mori A. Tetrahedron Lett. 2000;41:2401–2404. [Google Scholar]
- 27.Tudzynski B, Holter K. Fungal Genet Biol. 1998;25:157–170. doi: 10.1006/fgbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 28.Toyomasu T, Nakaminami K, Toshima H, Mie T, Watanabe K, Ito H, Matsui H, Mitsuhashi W, Sassa T, Oikawa H. Biosci Biotechnol Biochem. 2004;68:146–152. doi: 10.1271/bbb.68.146. [DOI] [PubMed] [Google Scholar]
- 29.Hohn TM. In: Comprehensive Natural Products Chemistry. Cane D, editor. Vol 2. Oxford: Elsevier Science; 1999. pp. 201–215. [Google Scholar]
- 30.Sassa T, Kenmoku H, Nakayama K, Kato N. Biosci Biotechnol Biochem. 2004;68:1608–1610. doi: 10.1271/bbb.68.1608. [DOI] [PubMed] [Google Scholar]
- 31.Cultrone A, Scazzocchio C, Rochet M, Montero-Moran G, Drevet C, Fernandez-Martin R. Mol Microbiol. 2005;57:276–290. doi: 10.1111/j.1365-2958.2005.04686.x. [DOI] [PubMed] [Google Scholar]
- 32.Fujii I. In: Comprehensive Natural Products Chemistry. Sankawa U, editor. Vol 1. Oxford: Elsevier Science; 1999. pp. 409–441. [Google Scholar]
- 33.Priestle JP, Grutter MG, White JL, Vincent MG, Kania M, Wilson E, Jardetzky TS, Kirschner K, Jansonius JN. Proc Natl Acad Sci USA. 1987;84:5690–5694. doi: 10.1073/pnas.84.16.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukamoto Y, Wong H, Mattick JS, Wakil SJ. J Biol Chem. 1983;258:15312–15322. [PubMed] [Google Scholar]
- 35.Herrmann L, Bockau U, Tiedtke A, Hartmann MWW, Weide T. BMC Biotechnol. 2006;6:21. doi: 10.1186/1472-6750-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence MC, Iliades P, Fernley RT, Berglez J, Pilling PA, Macreadie IG. J Mol Biol. 2005;348:655–670. doi: 10.1016/j.jmb.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 37.Blume A, Benie AJ, Stolz F, Schmidt RR, Reutter W, Hinderlich S, Peters T. J Biol Chem. 2004;279:55715–55721. doi: 10.1074/jbc.M410238200. [DOI] [PubMed] [Google Scholar]
- 38.Chen SC, Chang YC, Lin CH, Lin CH, Liaw SH. J Biol Chem. 2006;281:7605–7613. doi: 10.1074/jbc.M510254200. [DOI] [PubMed] [Google Scholar]
- 39.Janowiak BE, Griffith OW. J Biol Chem. 2005;280:11829–11839. doi: 10.1074/jbc.M414326200. [DOI] [PubMed] [Google Scholar]
- 40.Mohamed HE, Vermaas WF. J Bacteriol. 2006;188:3337–3344. doi: 10.1128/JB.188.9.3337-3344.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabrielsen M, Bond CS, Hallyburton I, Hecht S, Bacher A, Eisenreich W, Rohdich F, Hunter WN. J Biol Chem. 2004;279:52753–52761. doi: 10.1074/jbc.M408895200. [DOI] [PubMed] [Google Scholar]
- 42.Peters RJ, Carter OA, Zhang Y, Matthews BW, Croteau RB. Biochemistry. 2003;42:2700–2707. doi: 10.1021/bi020492n. [DOI] [PubMed] [Google Scholar]
- 43.Kawaide H, Sassa T, Kamiya Y. J Biol Chem. 2000;275:2276–2280. doi: 10.1074/jbc.275.4.2276. [DOI] [PubMed] [Google Scholar]
- 44.Brodelius M, Lundgren A, Mercke P, Brodelius PE. Eur J Biochem. 2002;269:3570–3577. doi: 10.1046/j.1432-1033.2002.03044.x. [DOI] [PubMed] [Google Scholar]
- 45.Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higuchi R, Krummel B, Saiki RK. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. Plant J. 2004;39:886–893. doi: 10.1111/j.1365-313X.2004.02175.x. [DOI] [PubMed] [Google Scholar]
- 48.Fujii H, Koyama T, Ogura K. Biochim Biophys Acta. 1982;712:716–718. [PubMed] [Google Scholar]