Abstract
Thioredoxins belong to a large family of enzymatic proteins that function as general protein disulfide reductases, therefore participating in several cellular processes via redox-mediated reactions. So far, none of the 18 members of this family has been involved in human pathology. Here we identified TXNDC3, which encodes a thioredoxin–nucleoside diphosphate kinase, as a gene implicated in primary ciliary dyskinesia (PCD), a genetic condition characterized by chronic respiratory tract infections, left–right asymmetry randomization, and male infertility. We show that the disease, which segregates as a recessive trait, results from the unusual combination of the following two transallelic defects: a nonsense mutation and a common intronic variant found in 1% of control chromosomes. This variant affects the ratio of two physiological TXNDC3 transcripts: the full-length isoform and a novel isoform, TXNDC3d7, carrying an in-frame deletion of exon 7. In vivo and in vitro expression data unveiled the physiological importance of TXNDC3d7 (whose expression was reduced in the patient) and the corresponding protein that was shown to bind microtubules. PCD is known to result from defects of the axoneme, an organelle common to respiratory cilia, embryonic nodal cilia, and sperm flagella, containing dynein arms, with, to date, the implication of genes encoding dynein proteins. Our findings, which identify a another class of molecules involved in PCD, disclose the key role of TXNDC3 in ciliary function; they also point to an unusual mechanism underlying a Mendelian disorder, which is an SNP-induced modification of the ratio of two physiological isoforms generated by alternative splicing.
Keywords: cilia, situs inversus, splice
Primary ciliary dyskinesia (PCD) is a rare genetic condition (MIM 242650) characterized by respiratory tract infections, male infertility, and, in approximately half of the patients, abnormal left–right asymmetry, thereby defining Kartagener's syndrome (MIM 244400) (1, 2). This complex disease phenotype, which is transmitted in a Mendelian fashion, results from abnormal functioning of respiratory and embryonic nodal cilia, as well as sperm flagella, three different cellular organelles that share a common structure called axoneme (3, 4). The main components of this highly organized structure include nine peripheral microtubule doublets surrounding a central pair of two microtubules (“9+2” structure), which are tied up with nexin links and radial spokes. Attached to the peripheral doublets, the outer and inner dynein arms are multiprotein complexes that are essential for normal ciliary movements. The respiratory cilia of patients with PCD have been shown to carry various axonemal abnormalities; although the main defects involve the outer and/or the inner arms, such ultrastructural variability, together with the fact that dynein arms are multiprotein complexes encoded by numerous genes (5), disclose the important genetic heterogeneity underlying PCD. This has greatly hampered the identification of causal genes. To date, despite substantial efforts made for several years by different teams including ours (6–18) and the recent availability of huge sequence information, only four genes have been involved in PCD: after a candidate gene approach based on data from the unicellular algae Chlamydomonas reinhardtii and the evolutionary conservation of genes encoding axonemal proteins, we have isolated a human dynein sequence named DNAI1, in which mutations were subsequently identified (19–21). Afterward, thanks to a genetic linkage analysis performed in one large consanguineous family, mutations have been identified in a second gene encoding a dynein chain, DNAH5 (22, 23). Recently, mutations in the X-linked RPGR gene have been found occasionally in males with a complex phenotype associating PCD and retinitis pigmentosa (24). Last, mutations in the DNAH11 gene, which encodes a dynein heavy chain, have been identified in two patients with respiratory tract infections and situs inversus (13). The molecular basis of PCD is, therefore, just beginning to be elucidated; and although DNAI1 and DNAH5 mutations underlie PCD in nearly half of patients with outer dynein arm defects, the cause remains unknown in the other patients (25, 26). We therefore searched for candidate genes that may account for the disease in those patients.
Results and Discussion
TXNDC3 Is Expressed in Testis and Respiratory Epithelial Cells.
TXNDC3, a gene also known as SPTRX-2, NM23-H8, or NME8, was first identified as a sequence encoding a protein containing thioredoxin and nucleoside diphosphate kinase (NDK) domains (27). It eventually turned out that TXNDC3 represents the human ortholog of the sea urchin IC1 gene that encodes a component of sperm outer dynein arms (27–29), an observation that prompted us to test its involvement in PCD. So far, IC1 orthologs have also been described in other species like Ciona intestinalis, rats, mice, and humans (27, 29, 30). The corresponding proteins were identified as the first members of a novel family of proteins with an N-terminal region homologous to the sequence of thioredoxin, followed by two or three NDK domains (28). NDKs are a family of proteins, also known as NME proteins, which catalyze nonsubstrate-specific conversions of nucleoside diphosphates to nucleoside triphosphates; these proteins, present in all organisms from bacteria to mammals, participate in many biological functions such as proliferation, differentiation, or development (31). In humans, this protein family comprises at least nine members that can be divided into two sets according to their sequence: while group I is composed of proteins that share high homology with each other and have the classic enzymatic activity of an NDK (i.e., NME1 to NME4), members of group II (i.e., NME5 to NME7, TXNDC3, and TXNDC6) are less conserved, and enzymatic activity has been demonstrated only for NME6. Of particular interest, members of group II share high homology with the NDK domains of the sea urchin IC1 chain, and, except for NME6, they are mainly expressed in testis (31, 32). Within the NDK family, TXNDC3 and TXNDC6 are the only two proteins containing an NDK domain and a thioredoxin domain. Thioredoxins are enzymes that participate in redox reactions via the reversible oxidation of a conserved active site (Cys-Gly-Pro-Cys) with two cystein residues that switch from a dithiol to a disulfide form. In humans, eight genes encoding thioredoxins and containing this site have been identified (33). Among them, TXNDC3 was found to be expressed specifically in testis (27), more precisely in the sperm fibrous sheath in rats (30), whereas TXNDC6 was found to be expressed at very low levels in a variety of adult tissues with highest levels essentially in testis and lung, along the microtubules of the spermatid manchette and the flagellar axoneme, as well as those of the ciliary axoneme (32).
Here we considered TXNDC3 as a candidate gene for PCD because of the participation of IC1 in sperm outer dynein arms. We therefore first tested its expression in human trachea and respiratory epithelial cells; this was done by means of RT-PCR, because it was previously detected in testis only, and at very low levels in that tissue (27, 30). We indeed detected TXNDC3 transcripts through amplification of overlapping fragments encompassing the coding region (data not shown and see below).
Identification of a Nonsense Mutation (p.Leu426X) and a Common Intronic Variant (c.271–27C>T) in the TXNDC3 Gene of a Patient with PCD.
The finding that TXNDC3 is expressed in the respiratory tract encouraged us to further test the hypothesis that patients with a PCD phenotype characterized by structural or functional defects of their outer dynein arms may carry TXNDC3 mutations. For each patient of our PCD population, the ultrastructural anomaly of respiratory cilia was precisely determined by means of transmission electron microscopy, and the ciliary motility was assessed through standard procedures (34). We assumed that TXNDC3 defects could underlie the PCD phenotype of patients with abnormal outer dynein arm structure (33 patients), or of those with typical clinical symptoms of Kartagener's syndrome and cilia that are structurally normal but immotile (eight patients). Given the testis expression of TXNDC3, we also included in our study six independent male patients with a sterility resulting from an absence of outer dynein arms in their flagellar axonemes. We therefore screened for mutations the eighteen TXNDC3 exons (Fig. 1A) and their intronic flanking regions in those 47 patients. In one girl (D50X1), we identified a heterozygous T>A transversion in exon 15, causing a nonsense mutation at codon 426 (c.1277T>A, p.Leu426X). This gene defect, which is inherited from the mother [supporting information (SI) Fig. 5], predicts a truncated protein devoid of the putative active site of the first NDK domain and the entire second NDK domain (Fig. 1). This 13-year-old patient, born to healthy parents (family D50; SI Fig. 5), is affected by chronic respiratory infections with bronchiectasis, chronic sinusitis, and serous otitis; she also has a situs ambiguous with the heart and the liver located centrally. The ciliary beat frequency appeared normal, and transmission electron microscopy revealed that 66% of her respiratory cilia have shortened or absent outer dynein arms (Fig. 2). Because the patient's mother has no respiratory symptom, we hypothesized that the patient is a compound heterozygote, despite the fact that she was born to related parents. We therefore screened her TXNDC3 gene for a second mutation and identified a heterozygous C>T transition in intron 6 (c.271–27C>T) inherited from her father (Fig. 1A and SI Fig. 5), whereas, in keeping with a recessive transmission of the disease phenotype, her two healthy brothers were found to be only heterozygous carriers: one (D50S1) carries the c.271–27C>T substitution inherited from his father, whereas the other (D50S2) carries the nonsense mutation inherited from his mother (SI Fig. 5). The c.271–27C>T sequence variation was subsequently found in the heterozygous state in another PCD patient (D65X1), whereas all of the other patients were found to be homozygous for a cytosine at this position. The same c.271–27C>T substitution was also identified in the heterozygous state in 2 of 98 control subjects, raising its allele frequency to 1% in a control population. As for patient D65X1, the disease phenotype differed from that of patient D50X1, in that it was characterized by chronic respiratory infections with bronchiectasis, chronic sinusitis, and serous otitis associated with a situs inversus totalis; transmission electron microscopy examination of his respiratory cilia revealed no ultrastructural defect, and ≈40% of the cilia were immotile, while the remaining cilia displayed a normal beating. As patient D65X1 carried the c.271–27C>T change in the heterozygous state and no other molecular defect was identified in his TXNDC3 genomic sequence, we searched for a splicing defect that would have escaped the genomic analysis, by amplifying overlapping fragments encompassing the entire coding sequence; this analysis revealed no abnormal splice product (data not shown). Taken together, these data therefore strongly suggest that the c.271–27C>T variant alone does not contribute to the PCD phenotype of D65X1.
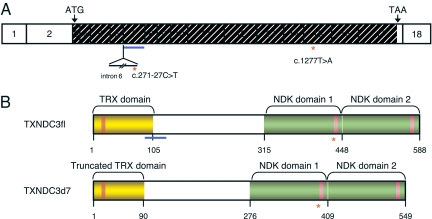
Fig. 1.
The human TXNDC3 gene and related products. (A) TXNDC3 cDNA structure showing the location of the exons drawn to scale. The translation start and stop codons are labeled with ATG and TAA, respectively. The translated region is hashed. Exon 7 is underlined in blue, and intron 6 is shown as a thin line below exons 6 and 7. The red asterisks mark the locations of the c.271–27C>T and c.1277T>A nucleotide variations, located in intron 6 and exon 15, respectively. (B) Structure of the TXNDC3 isoforms. The TXNDC3fl isoform (Upper), and the TXNDC3d7 isoform (Lower). The thioredoxin (TRX) domain and the two NDK domains are shown in yellow and green, respectively. Within the TRX domain, the active site (GCPC) is shown by an orange box, and, within the NDK domains, the putative NDP kinase active sites are shown by pink boxes. The location of the region encoded by exon 7 is underlined in blue. The location of the p.Leu426X mutation is shown by a red asterisk.
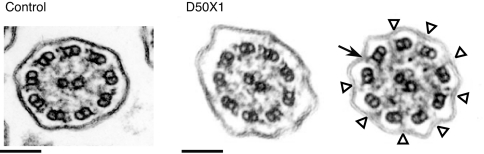
Fig. 2.
Electron micrograph of cross-sections of respiratory cilia. (Left) Normal ciliary ultrastructure. (Right) Two cross-sections of respiratory cilia from patient D50X1 showing a heterogeneous ultrastructure: whereas some cilia appear normal (Left), others (66%) display outer dynein arm defects (Right). The black arrow indicates a normal outer dynein arm, and white arrowheads indicate shorter or missing outer dynein arms. (Scale bars: 100 nm.)
The c.271–27C>T Intronic Variant Alters the Ratio of Two Physiological TXNDC3 Isoforms.
The c.271–27C>T variant occurs within a polypyrimidine tract in the vicinity of putative branch points of intron 6, a region believed to be important for splicing. Indeed, similar intronic substitutions located upstream of a splice-acceptor site have already been shown to affect splicing (35–37). To test whether the c.271–27C>T variant has an effect on splicing, we first amplified TXNDC3 cDNA fragments spanning exons 6–8 from RNA samples obtained from testis, trachea, and nasal cells. Interestingly, these experiments enabled us to detect two physiological TXNDC3 transcripts: a full-length isoform designated TXNDC3fl, and a second isoform carrying an in-frame deletion of the 117 bp of exon 7 and named TXNDC3d7 (Figs. 1B and 3A). This observation prompted us to examine whether the c.271–27C>T variant modulates this alternative splice event. To this end, we amplified the TXNDC3 cDNA region encompassing exon 7 from lymphoblastoid cell lines established in all members of family D50, as well as in D65X1 and in control samples. Two groups were subsequently considered, the first one referring to DNA samples with the control sequence (i.e., c.271–27C/C), and the second to DNA samples bearing the c.271–27C>T variant (i.e., c.271–27C/T) (Fig. 3A). In each case, we determined the respective amounts of TXNDC3fl and TXNDC3d7 transcripts (Fig. 3B). We were then able to determine the TXNDC3fl/(TXNDC3fl+TXNDC3d7) ratio (named the R ratio) for each individual. As shown in SI Fig. 6A, the R ratios of the second group differ dramatically from those of the first group with P = 9.96 × 10−8, 2.09 × 10−8, 1.12 × 10−5, 8.52 × 10−7 and 5.18 × 10−3 for D50X1, D50F, D50S1, D65X1, and control C15, respectively. Although these data were generated by means of conventional PCR, it is important to underline that it was obtained with the use of a single primer set bracketing the spliced exon so that the two isoforms (i.e., TXNDC3fl and TXNDC3d7) were co-amplified in the same tube. To confirm this result, we performed a similar experiment using a real-time quantitative PCR assay (SI Fig. 6B). As deduced from this analysis, the TXNDC3fl/[TXNDC3d7+TXNDC3fl] ratio (R′) was significantly higher in cells from subjects who carry the C>T SNP in the heterozygous state than in cells from individuals homozygous for the C allele (P = 3.90 × 10−3, 1.38 × 10−4, 5.84 × 10−5, 9.04 × 10−5, and 2.85 × 10−2 for D50X1, D50F, D50S1, D65X1, and C15, respectively) (SI Fig. 6B).
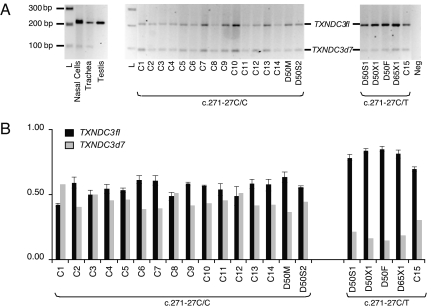
Fig. 3.
In vivo expression of TXNDC3fl and TXNDC3d7 isoforms. (A) Expression of the TXNDC3fl and TXNDC3d7 isoforms by means of conventional RT-PCR using primers F1 and R1 located in exons 6 and 8, respectively. RT-PCR amplifications were performed on RNAs extracted from human testis, trachea, and nasal cells, and from lymphoblastoid cell lines established for all members of family D50 (D50F, D50M, D50X1, D50S1, and D50S2), for patient D65X1 with a c.271–27C/T genotype, and for 15 controls, C1 to C15, with C1 to C14 having a c.271–27C/C genotype and C15 having a c.271–27C/T genotype. One representative experiment of three independent experiments is shown. L, 1-kb+ ladder; Neg, reaction without RNA. (B) Relative expression of TXNDC3fl and TXNDC3d7 transcripts generated by conventional PCR. The amounts of TXNDC3fl and TXNDC3d7 transcripts, calculated by the GeneTools program, are represented with black and gray bars, respectively. The sum of the two isoforms was arbitrarily set to 1. (Left) The results for individuals who carry the c.271–27C/C genotype, i.e., the patient's mother (D50M) and brother (D50S2) and controls (C1 to C14). (Right) The results for the five individuals bearing the c.271–27C>T variant, i.e., patient D50X1, her father (D50F), her brother (D50S1), patient D65X1, and control C15. Results are represented as means ± SEM of three independent experiments.
To test whether the c.271–27C>T substitution is directly involved in the regulation of TXNDC3 splicing, we compared the splicing patterns of TXNDC3 transcripts obtained after transfection of TXNDC3 minigenes containing the genomic sequence spanning intron 5 to intron 8 and carrying a cytosine or a thymine at position c.271–27 (Fig. 4). To this end, we generated the following eight TXNDC3 minigenes: four independent clones were obtained after subcloning of the PCR-amplified genomic DNA from patient D50X1, who is heterozygous for the c.271–27C>T substitution (i.e., pTXNDC3_C1 and pTXNDC3_C2, containing a C at position c.271–27, and pTXNDC3_T1 and pTXNDC3_T2, containing a T at the same position); these clones were subsequently subjected to site-directed mutagenesis so that the cytosine at position c.271–27 in clones pTXNDC3_C1 and pTXNDC3_C2 was replaced by a thymine (generating plasmids pTXNDC3_C1>T and pTXNDC3_C2>T) and, conversely, the thymine at position c.271–27 in clones pTXNDC3_T1 and pTXNDC3_T2 was replaced by a cytosine (generating plasmids pTXNDC3_T1>C and pTXNDC3_T2>C). As shown in Fig. 4, in all cases, two molecular species were generated, one corresponding to TXNDC3fl and the other to TXNDC3d7; however, the ratio of TXNDC3d7 to TXNDC3fl was always higher in cells transfected with minigenes carrying a cytosine in intron 6 than in cells transfected with minigenes carrying a thymine at that position. These results, therefore, not only confirm the data obtained on lymphoblastoid cell lines, but also provide a clear-cut demonstration of the direct involvement of this intronic SNP on the splicing of TXNDC3 transcripts.
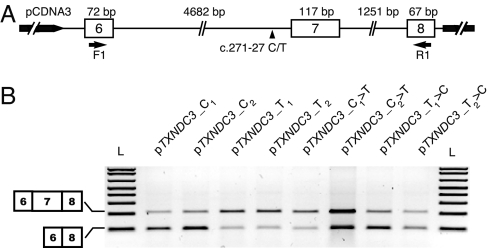
Fig. 4.
Impact of the c.271–27C>T variant on splicing of TXNDC3 transcripts. (A) Schematic representation of the different TXNDC3 constructs used in this experiment. Exons and introns are represented by open boxes and thin lines, respectively. The location of the c.271–27C/T SNP is indicated by a black arrowhead. The primers used in the RT-PCR experiments (F1 and R1) are represented by horizontal arrows. (B) RT-PCR amplification of TXNDC3 transcripts from HeLa cells transfected with minigenes carrying either a cytosine in intron 6 (i.e., pTXNDC3_C1, pTXNDC3_C2, pTXNDC3_T1>C, and pTXNDC3_T2>C) or a thymine at that position (i.e., pTXNDC3_T1, pTXNDC3_T2, pTXNDC3_C1>T, and pTXNDC3_C2>T) (see Materials and Methods). L, 1-kb+ ladder.
A similar combination of an unambiguous mutation with a functional polymorphism has been reported in severe poikilocytic anemia (36) and erythropoietic protoporphyria (38). In extremely rare cases, the identified polymorphism acts on splicing and modifies the ratio of two alternative transcripts; this is the case of two intronic polymorphisms of the CFTR gene, which affect splicing of exon 9 and contribute to atypical cystic fibrosis phenotypes (39) through a mechanism that is therefore reminiscent of the situation described here.
Presumed Functions of Human TXNDC3 Isoforms in Light of Their Paralogs and Orthologs.
The identification of the c.271–27C>T variant raises the larger question of the mechanism by which such low expression of the TXNDC3d7 isoform is detrimental to ciliary function. First, one should note that the thioredoxin domain of this isoform is most likely nonfunctional, because it lacks its last 15 residues (Fig. 1B). However, our current knowledge on TXNDC6, the closest paralog of TXNDC3, may shed light on the function of the truncated TXNDC3d7 isoform. As reported by Miranda-Vizuete et al. (33), TXNDC3 and TXNDC6, which could have arisen from a common ancestor, share an identical intron/exon organization of their 5′ extremity. Interestingly, we found that the sequence of TXNDC3 exon 7 matches perfectly that of TXNDC6 exon 5, with a residue conservation reaching 71% (SI Fig. 7B). Strikingly, TXNDC6 has been shown to be expressed at low levels in testis and lung, as two alternative transcripts: a full-length product and a variant lacking exon 5, named Δ5-TXNDC6 (32). The corresponding proteins were detected in the close vicinity of ciliary and flagellar axonemes. Most importantly, it was shown that the Δ5-TXNDC6 isoform was able to bind microtubules with high affinity, which is not the case of the full-length isoform (32). It is, therefore, tempting to speculate that the TXNDC3d7 isoform is also able to bind the microtubules of the respiratory cilia. To test this hypothesis, we assessed the ability of radiolabeled recombinant TXNDC3fl and TXNDC3d7 proteins to bind pure polymerized microtubules. In fact, the two TXNDC3 isoforms were found to be able to bind the microtubules. Although in both cases only a small fraction of TXNDC3 was indeed found to sediment with the microtubules, this result was highly reproducible; and most importantly, we always observed a slightly higher binding of TXNDC3d7 to microtubules, as compared with TXNDC3fl (SI Fig. 8). If this alternative splice event is indeed functionally important, we expect the TXNDC3 protein sequence to be highly conserved throughout evolution and especially the genomic organization around exon 7. To test this hypothesis, we looked for TXNDC3 orthologs and identified, in addition to the five known sequences, seven sequences that are likely to represent TXNDC3 orthologs in several species. Comparison of these 12 sequences, from C. intestinalis to the chimpanzee, revealed a high conservation degree with the human sequence, ranging from 34% to 97%, respectively (SI Fig. 7A). Careful determination or examination of the genomic structure of all but one TXNDC3 orthologs (i.e., the sea urchin gene whose genomic sequence is not yet available) showed that the reading frame of exon 7 is perfectly maintained throughout evolution, suggesting that the alternative splicing mechanism may have been conserved from C. intestinalis to humans. In addition, from C. intestinalis to the chimpanzee, the residue similarity of exon 7 with the human sequence is high, ranging from 54% to 100%, respectively (SI Fig. 7C).
The role of TXNDC3 in ciliary function is also supported by the following observations: thioredoxin-like proteins have been found in close association with outer dynein arms in a large number of species, from lower eukaryotes to mammals. For instance, the unicellular algae Chlamydomonas has two thioredoxin-like chains, LC3 and LC5, closely linked to the outer dynein arms (40) and believed to represent the homologs of TXNDC3 (5). Just recently, these two thioredoxins have been involved in the control of flagellar motility through their interactions with flagellar components (41). In addition, from Chlamydomonas to mammals, NDP kinases are also found in ciliary structures where they are supposed to play essential functions, particularly through their ability to produce GTP (42–44). Taken together, these observations strongly support the hypothesis that TXNDC3 is a bifunctional protein with biological properties that have been highly conserved throughout evolution, with the TXNDC3d7 isoform that would play a critical role in the outer dynein arms, at least thanks to its ability to bind microtubules. In this regard, the existence of two TXNDC3 isoforms expressed at different levels in ciliated tissues like trachea, and in testis that contains flagellar structures, raises the question of the function of each isoform, in the light of the known differences between the biology of cilia and flagella. The finding that testis expresses virtually only the TXNDC3fl isoform, together with the observation that the TXNDC3 protein is localized essentially to the fibrous sheath of sperm tails (30), which is a periaxonemal structure, suggests that another, unidentified, member of the thioredoxin system may be necessary for the sperm flagellar axoneme. Indeed, the fact that ciliary and flagellar axonemes share a similar morphological structure does not imply that their protein content is identical. This notion is supported by the report of male patients with PCD and dynein arm defects in their respiratory cilia, in the absence of any detectable axonemal abnormality in their spermatozoa; and reverse situations have been observed in infertile male patients (reviewed in ref. 45). Interestingly, as for TXNDC3, it has previously been suggested that during evolution, the TXNDC3 ancestor shifted from an axonemal to a periaxonemal localization and that the paralog TXNDC6 might substitute functionally for the TXNDC3 ancestor in the sperm flagellar axoneme (30). Given the ability of TXNDC3d7 to bind microtubules, it is now tempting to speculate that the Δ5-TXNDC6 isoform is involved in that function.
Thioredoxins in Human Pathology.
So far, there is no disease that has been shown to result from mutations in a member of the thioredoxin family. However, interestingly enough, the RP2 gene, which encodes a protein with one NDK domain, has been shown to be involved in X-linked retinitis pigmentosa (RP), an inherited affection of the retina due to photoreceptor degeneration (46) (MIM 312600). These cells contain a structure called connecting cilium, which bridges the outer and inner segments of the retinal photoreceptors; the structure of the connecting cilium is similar to that of nonmotile primary cilia with 9 peripheral microtubule doublets and no central doublets (“9+0” structure). Previous studies suggested that alteration of this structure may result from intraflagellar transport dysfunction and lead to retinal degeneration (47–50). Noteworthy, a link has been established between photoreceptor and respiratory cilia with the identification of mutations in RPGR, which encodes a GTPase regulator involved in X-linked RP, in families presenting with a complex disease phenotype combining RP with various defects of respiratory cilia and even PCD (24, 51, 52).
In addition, although no thioredoxin-like protein has been shown to be directly involved in RP, TXNL6 (also known as RdCVF), which is a truncated thioredoxin-like protein, seems to play an essential role in cone viability; indeed, in the rd1 mouse, a model for RP, TXNL6 has been shown to prevent cones from degenerating (53). Overall, such structural homologies between RP2/TXNL6 (two proteins expressed in retinal cells and containing either an NDK domain or a thioredoxin-like domain) and TXNDC3 (a protein expressed in other kinds of ciliated cells and containing these two domains) further strengthen the hypothesis that TXNDC3 plays an important role in ciliary function.
Conclusion.
This study identified a patient born to healthy parents and presenting with a PCD phenotype resulting from two transallelic defects in TXNDC3, a gene that encodes protein isoforms belonging to the large family of thioredoxins. A nonsense mutation predicting a truncated protein devoid of the NDK domains was indeed identified together with a common intronic sequence variation reducing the amount of one particular isoform. These data, which are therefore consistent with a disease phenotype segregating in a recessive manner, point to an unusual mechanism underlying a Mendelian disorder, which is an SNP-induced modification of the ratio of two physiological isoforms generated by alternative splicing. In addition, the ultrastructural abnormalities displayed by our patient are particular in that a partial lack and shortening of outer dynein arms are found in 66% of her respiratory cilia. The persistent beating of cilia may, therefore, reveal the activity of the remaining normal cilia; and we can assume that a similar dysfunction occurs in embryonic nodal cilia, thereby accounting for the situs ambiguous documented in the patient. This assumption is further strengthened by the very recent report of a family with patients presenting with a Kartagener's syndrome potentially linked to the 7p14.1 region known to contain the TXNDC3 gene (54). Finally, our data, which underline the key role played by thioredoxin-like and NDK proteins in ciliary structure and function, identify TXNDC3 as a new gene involved in PCD; they consequently disclose members of the thioredoxin family, like TXNDC6, as other excellent candidates for PCD.
Materials and Methods
Patients.
Forty-one patients with PCD were investigated. Electron microscopic examination of ciliary sections from nasal or bronchial biopsy showed abnormal outer dynein arms in 33 patients, whereas cilia were structurally normal but immotile in 8 patients with typical Kartagener's syndrome. Six independent male patients with sterility resulting from an absence of outer dynein arms in their flagella were also included in our study. A permanent lymphoblastoid cell line was obtained by EBV transformation of lymphocytes from all members of family D50 and from patient D65X1.
The PCD genetic study was approved by the ethical committee of our institution (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale-Henri Mondor, Créteil, France), and informed consent was given by patients and/or their parents.
Sequencing Reactions.
For each individual, genomic DNA was isolated from blood sample according to standard techniques. To screen the 18 exons of the TXNDC3 gene, we used 17 sets of primers (SI Table 1, including the sequence of all of the primers used in the study) in PCRs with genomic DNA extracted from blood samples according to standard techniques. PCR products were used as sequencing templates. The sequencing primers were the same as the amplification primers. Sequencing reactions were performed by using the Big Dye Terminator kit (Applied Biosystems, Foster City, CA), and an ABI-3100 DNA sequencer (Applied Biosystems) was used to obtain sequences.
Conventional RT-PCR.
A first series of RT-PCR experiments was performed under standard conditions (OneStep RT-PCR; Qiagen, Valencia, CA), with 40 cycles of amplification on RNA from human testis, trachea, and nasal cells, and from lymphoblastoid cell lines established for all members of family D50, patient D65X1, and 15 controls (C1 to C15). Sense (F1) and antisense (R1) primers located in exon 6 and exon 8, respectively, enabled the amplification of a cDNA fragment encompassing exon 7. The products were separated on agarose gel, and a quantitative evaluation of the products was done by using the GeneTools program (Syngene, Cambridge, U.K.). The normality of the distribution of the R ratios for D50M, D50S2, and C1 to C14 was positively tested by the Shapiro–Wilk test.
Constructs.
The genomic DNA from patient D50X1, who carries the c.271–27C>T sequence variation in the heterozygous state, was used as a template to amplify the sequence surrounding the alternatively spliced exon 7 of TXNDC3 with primers located in introns 5 and 8 (F4 and R4). The PCR products were cloned into the pCDNA3.1/V5-His TOPO TA expression vector (Invitrogen, Paisley, U.K.) according to standard procedures. Two independent clones containing a TXNDC3 minigene with a cytosine residue at position c.271–27, designated pTXNDC3_C1 and pTXNDC3_C2, were subsequently subjected to site-directed mutagenesis by the QuikChange kit (Stratagene, Cedar Creek, TX), with oligonucleotides (F5 and R5) designed to introduce the C>T substitution; the resulting plasmids were named pTXNDC3_C1>T and pTXNDC3_C2>T. Similarly, two other independent clones obtained after subcloning of the PCR-amplified genomic DNA from patient D50X1 and containing a T at position c.271–27 (designated pTXNDC3_T1 and pTXNDC3_T2) were used as templates to generate pTXNDC3_T1>C and pTXNDC3_T2>C (carrying a C at position c.271–27) using primers F6 and R6.
Transfections.
Transfections of HeLa cells obtained from American Type Culture Collection (Manassas, VA) were performed by using the Lipofectamine-Plus method (Invitrogen) in OptiMEM medium, according to the manufacturer's standard protocol. Approximately 24 h after transfection of each of the eight TXNDC3 minigenes, total RNA was isolated. RT-PCR was performed by using two primers (F1 and R1) located on exons 6 and 8, respectively.
Web Resources.
URLs for data presented herein are as follows: Online Mendelian Inheritance in Man, www.ncbi.nlm.nih.gov/Omim (for PCD and Kartagener syndrome); Ensembl, www.ensembl.org; NCBI, www.ncbi.nlm.nih.gov; and Softberry, www.softberry.com.
Supplementary Material
Acknowledgments
We are grateful to the patients and their family members, whose cooperation made this study possible. We thank Roberto Incitti for advice on statistical tests. Franck Perez and Odile Valiron are gratefully acknowledged for helpful discussions. This work was supported by grants from the Groupement d'Intérêt Scientifique-Institut des Maladies Rares (A03091), the Assistance Publique–Hôpitaux de Paris (CRC 96125), the Legs Poix from the Chancellerie des Universités, the Agence Nationale pour la Recherche (ANR-05-MRAR-022-01), and the Université Pierre et Marie Curie (Bonus Qualité Recherche).
Abbreviations
- PCD
primary ciliary dyskinesia
- NDK
nucleoside diphosphate kinase
- RP
retinitis pigmentosa.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611405104/DC1.
References
- 1.Afzelius BA. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 2.Kartagener M. Beitr Klin Tuberk. 1933;83:489–501. [Google Scholar]
- 3.Fliegauf M, Omran H. Trends Genet. 2006;22:241–245. doi: 10.1016/j.tig.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Badano JL, Mitsuma N, Beales PL, Katsanis N. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 5.Pazour GJ, Agrin N, Walker BL, Witman GB. J Med Genet. 2006;43:62–73. doi: 10.1136/jmg.2005.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witt M, Wang Y, Wang S, Sun C, Pawlik J, Rutkiewicz E, Zebrak J, Diehl SR. Am J Hum Genet. 1999;64:313–318. doi: 10.1086/302203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blouin JL, Meeks M, Radhakrishna U, Sainsbury A, Gehring C, Sail GD, Bartoloni L, Dombi V, O'Rawe A, Walne A, et al. Eur J Hum Genet. 2000;8:109–118. doi: 10.1038/sj.ejhg.5200429. [DOI] [PubMed] [Google Scholar]
- 8.Maiti AK, Bartoloni L, Mitchison HM, Meeks M, Chung E, Spiden S, Gehrig C, Rossier C, DeLozier-Blanchet CD, Blouin J, et al. Cytogenet Cell Genet. 2000;90:119–122. doi: 10.1159/000015645. [DOI] [PubMed] [Google Scholar]
- 9.Meeks M, Bush A. Pediatr Pulmonol. 2000;29:307–316. doi: 10.1002/(sici)1099-0496(200004)29:4<307::aid-ppul11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Pennarun G, Chapelin C, Escudier E, Bridoux AM, Dastot F, Cacheux V, Goossens M, Amselem S, Duriez B. Hum Genet. 2000;107:642–649. doi: 10.1007/s004390000427. [DOI] [PubMed] [Google Scholar]
- 11.Bartoloni L, Blouin JL, Maiti AK, Sainsbury A, Rossier C, Gehrig C, She JX, Marron MP, Lander ES, Meeks M, et al. Genomics. 2001;72:21–33. doi: 10.1006/geno.2000.6462. [DOI] [PubMed] [Google Scholar]
- 12.Pennarun G, Bridoux AM, Escudier E, Dastot-Le Moal F, Cacheux V, Amselem S, Duriez B. Am J Respir Cell Mol Biol. 2002;26:362–370. doi: 10.1165/ajrcmb.26.3.4738. [DOI] [PubMed] [Google Scholar]
- 13.Bartoloni L, Blouin JL, Pan Y, Gehrig C, Maiti AK, Scamuffa N, Rossier C, Jorissen M, Armengot M, Meeks M, et al. Proc Natl Acad Sci USA. 2002;99:10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neesen J, Drenckhahn JD, Tiede S, Burfeind P, Grzmil M, Konietzko J, Dixkens C, Kreutzberger J, Laccone F, Omran H. Cytogenet Genome Res. 2002;98:38–44. doi: 10.1159/000068545. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YJ, O'Neal WK, Randell SH, Blackburn K, Moyer MB, Boucher RC, Ostrowski LE. J Biol Chem. 2002;277:17906–17915. doi: 10.1074/jbc.M200348200. [DOI] [PubMed] [Google Scholar]
- 16.Jeganathan D, Chodhari R, Meeks M, Faeroe O, Smyth D, Nielsen K, Amirav I, Luder AS, Bisgaard H, Gardiner RM, et al. J Med Genet. 2004;41:233–240. doi: 10.1136/jmg.2003.014084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zariwala M, O'Neal WK, Noone PG, Leigh MW, Knowles MR, Ostrowski LE. Am J Respir Cell Mol Biol. 2004;30:428–434. doi: 10.1165/rcmb.2003-0338RC. [DOI] [PubMed] [Google Scholar]
- 18.Horvath J, Fliegauf M, Olbrich H, Kispert A, King SM, Mitchison H, Zariwala MA, Knowles MR, Sudbrak R, Fekete G, et al. Am J Respir Cell Mol Biol. 2005;33:41–47. doi: 10.1165/rcmb.2004-0335OC. [DOI] [PubMed] [Google Scholar]
- 19.Pennarun G, Escudier E, Chapelin C, Bridoux AM, Cacheux V, Roger G, Clement A, Goossens M, Amselem S, Duriez B. Am J Hum Genet. 1999;65:1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guichard C, Harricane MC, Lafitte JJ, Godard P, Zaegel M, Tack V, Lalau G, Bouvagnet P. Am J Hum Genet. 2001;68:1030–1035. doi: 10.1086/319511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zariwala M, Noone PG, Sannuti A, Minnix S, Zhou Z, Leigh MW, Hazucha M, Carson JL, Knowles MR. Am J Respir Cell Mol Biol. 2001;25:577–583. doi: 10.1165/ajrcmb.25.5.4619. [DOI] [PubMed] [Google Scholar]
- 22.Omran H, Haffner K, Volkel A, Kuehr J, Ketelsen UP, Ross UH, Konietzko N, Wienker T, Brandis M, Hildebrandt F. Am J Respir Cell Mol Biol. 2000;23:696–702. doi: 10.1165/ajrcmb.23.5.4257. [DOI] [PubMed] [Google Scholar]
- 23.Olbrich H, Haffner K, Kispert A, Volkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, et al. Nat Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 24.Moore A, Escudier E, Roger G, Tamalet A, Pelosse B, Marlin S, Clement A, Geremek M, Delaisi B, Bridoux AM, et al. J Med Genet. 2006;43:326–333. doi: 10.1136/jmg.2005.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornef N, Olbrich H, Horvath J, Zariwala MA, Fliegauf M, Loges NT, Wildhaber J, Noone PG, Kennedy M, Antonarakis SE, et al. Am J Respir Crit Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zariwala MA, Leigh MW, Ceppa F, Kennedy MP, Noone PG, Carson JL, Hazucha MJ, Lori A, Horvath J, Olbrich H, et al. Am J Respir Crit Care Med. 2006;174:858–866. doi: 10.1164/rccm.200603-370OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadek CM, Damdimopoulos AE, Pelto-Huikko M, Gustafsson JA, Spyrou G, Miranda-Vizuete A. Genes Cells. 2001;6:1077–1090. doi: 10.1046/j.1365-2443.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, Inaba K, Mohri H. Mol Biol Cell. 1996;7:1895–1907. doi: 10.1091/mbc.7.12.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padma P, Hozumi A, Ogawa K, Inaba K. Gene. 2001;275:177–183. doi: 10.1016/s0378-1119(01)00661-8. [DOI] [PubMed] [Google Scholar]
- 30.Miranda-Vizuete A, Tsang K, Yu Y, Jimenez A, Pelto-Huikko M, Flickinger CJ, Sutovsky P, Oko R. J Biol Chem. 2003;278:44874–44885. doi: 10.1074/jbc.M305475200. [DOI] [PubMed] [Google Scholar]
- 31.Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. J Bionenerg Biomembr. 2000;32:247–258. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- 32.Sadek CM, Jimenez A, Damdimopoulos AE, Kieselbach T, Nord M, Gustafsson JA, Spyrou G, Davis EC, Oko R, van der Hoorn FA, Miranda-Vizuete A. J Biol Chem. 2003;278:13133–13142. doi: 10.1074/jbc.M300369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda-Vizuete A, Sadek CM, Jimenez A, Krause WJ, Sutovsky P, Oko R. Antioxid Redox Signal. 2004;6:25–40. doi: 10.1089/152308604771978327. [DOI] [PubMed] [Google Scholar]
- 34.Tamalet A, Clement A, Roudot-Thoraval F, Desmarquest P, Roger G, Boule M, Millepied MC, Baculard TA, Escudier E. Pediatrics. 2001;108:E86. doi: 10.1542/peds.108.5.e86. [DOI] [PubMed] [Google Scholar]
- 35.Wichterle H, Hanspal M, Palek J, Jarolim P. J Clin Invest. 1996;98:2300–2307. doi: 10.1172/JCI119041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fournier CM, Nicolas G, Gallagher PG, Dhermy D, Grandchamp B, Lecomte MC. Blood. 1997;89:4584–4590. [PubMed] [Google Scholar]
- 37.Hong SH, Rhyne J, Miller M. Circ Res. 2003;93:1006–1012. doi: 10.1161/01.RES.0000102957.84247.8F. [DOI] [PubMed] [Google Scholar]
- 38.Gouya L, Martin-Schmitt C, Robreau AM, Austerlitz F, Da Silva V, Brun P, Simonin S, Lyoumi S, Grandchamp B, Beaumont C, et al. Am J Hum Genet. 2006;78:2–14. doi: 10.1086/498620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, Jorissen M, Droogmans G, Reynaert I, Goossens M, et al. J Clin Invest. 1998;101:487–496. doi: 10.1172/JCI639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison A, Sakato M, Tedford HW, Benashski SE, Patel-King RS, King SM. Cell Motil Cytoskeleton. 2002;52:131–143. doi: 10.1002/cm.10044. [DOI] [PubMed] [Google Scholar]
- 41.Wakabayashi K, King SM. J Cell Biol. 2006;173:743–754. doi: 10.1083/jcb.200603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel-King RS, Benashski SE, King SM. J Biol Chem. 2002;277:34271–34279. doi: 10.1074/jbc.M204137200. [DOI] [PubMed] [Google Scholar]
- 43.Patel-King RS, Gorbatyuk O, Takebe S, King SM. Mol Biol Cell. 2004;15:3891–3902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell KA, Gallagher BC, Szabo G, Otero Ade S. Cell Motil Cytoskeleton. 2004;59:62–73. doi: 10.1002/cm.20025. [DOI] [PubMed] [Google Scholar]
- 45.Afzelius BA. J Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, et al. Nat Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- 47.Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. J Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong DH, Pawlyk B, Sokolov M, Strissel KJ, Yang J, Tulloch B, Wright AF, Arshavsky VY, Li T. Invest Ophthalmol Vis Sci. 2003;44:2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 49.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, et al. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 50.Eley L, Yates LM, Goodship JA. Curr Opin Genet Dev. 2005;15:308–314. doi: 10.1016/j.gde.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 51.van Dorp DB, Wright AF, Carothers AD, Bleeker-Wagemakers EM. Hum Genet. 1992;88:331–334. doi: 10.1007/BF00197269. [DOI] [PubMed] [Google Scholar]
- 52.Zito I, Downes SM, Patel RJ, Cheetham ME, Ebenezer ND, Jenkins SA, Bhattacharya SS, Webster AR, Holder GE, Bird AC, et al. J Med Genet. 2003;40:609–615. doi: 10.1136/jmg.40.8.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leveillard T, Mohand-Said S, Lorentz O, Hicks D, Fintz AC, Clerin E, Simonutti M, Forster V, Cavusoglu N, Chalmel F, et al. Nat Genet. 2004;36:755–759. doi: 10.1038/ng1386. [DOI] [PubMed] [Google Scholar]
- 54.Gutierrez-Roelens I, Sluysmans T, Jorissen M, Amyere M, Vikkula M. Eur J Hum Genet. 2006;14:809–815. doi: 10.1038/sj.ejhg.5201631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.