Abstract
Protein-binding DNA sequence elements encode a variety of regulated functions of genomes. Information about such elements is currently in a state of rapid growth, but improved methods are required to characterize the sequence specificity of DNA-binding proteins. We have established an in vitro method for specific and sensitive solution-phase analysis of interactions between proteins and nucleic acids in nuclear extracts, based on the proximity ligation assay. The reagent consumption is very low, and the excellent sensitivity of the assay enables analysis of as few as 1–10 cells. We show that our results are highly reproducible, quantitative, and in good agreement with both EMSA and predictions obtained by using a motif finding software. This assay can be a valuable tool to characterize in-depth the sequence specificity of DNA-binding proteins and to evaluate effects of polymorphisms in known transcription factor binding sites.
Keywords: ChIP, EMSA, proximity ligation assay
Sequence-specific DNA binding by proteins controls processes such as replication, recombination, and transcription. Distinct DNA-binding proteins are also associated with centromers, telomers, and other specialized regions in the genome, where they regulate chromosome condensation, cohesion, and other aspects of genome maintenance. Activation of genes by DNA–protein interactions is a fundamental regulatory mechanism involving the recruitment of chromatin-modifying complexes and transcription complexes to initiate RNA synthesis (1). Of all human genes, 6% are estimated to code for DNA-binding transcription factors (2). Alterations of DNA-binding proteins are frequently involved in human disease, particularly in cancer, but information about the sequence specificity of DNA-binding proteins remains limited.
Genome-wide location maps have been generated for binding sites for most transcription factors in yeast by combining ChIP (3) with DNA microarray analysis (ChIP-chip) (4–6). Other studies have focused on chromatin remodeling factors, such as components of the RCS complex (7) and the DNA replication machinery, e.g., ORC and MCMs (8). To identify human transcription factor binding sites, promoter or CpG island microarrays have been used (9, 10). Moreover, high-resolution tiling path arrays have been used for transcription factor binding studies (11). In this regard, the ENCODE (Encyclopedia of DNA Elements) project was launched in 2003 to identify all functional elements in the human genome (12), assisted by the development of PCR-product and oligonucleotide tiling path arrays initially covering 1% of the genome. Despite its power and utility, ChIP-chip analysis has to be considered a screening technology of moderate resolution.
Several other array-based techniques have been developed to identify genomic binding sites of transcriptional activators or other DNA-binding proteins (13–15), but in all cases results must be confirmed by independent reference methods. EMSA (16) is frequently used to verify binding specificity in vitro. The supershift assay is an improvement of the technique (17) that allows antibody-based identification of DNA-binding proteins in the gel-retardation assays. EMSA allows resolution of complexes of different stoichiometry or conformation, and it can identify DNA-binding proteins and their binding sequences within a gene's regulatory region. EMSA may also be used for quantitative analysis of thermodynamic and kinetic parameters. However, the method is associated with several drawbacks because it is time-consuming, requires large amounts of input material, and involves the use of both acrylamide and radioactivity. Furthermore, EMSA is difficult to adapt to high-throughput analysis.
Here, we present a procedure for measuring DNA–protein interactions that is based on the proximity ligation assay (PLA) for ultrasensitive protein analysis (18, 19). In this technique, specific DNA representations of detected proteins are created and amplified in place of the proteins themselves, analogously to the detection of mRNA as cDNAs via reverse-transcription reactions. Briefly, oligonucleotides are attached to specific protein-binding reagents, typically mono- or polyclonal antibodies. When two proximity probes recognize and bind the same target molecule or a complex of two interacting target molecules, the ends of their conjugated oligonucleotides come sufficiently close to allow them to be joined by enzymatic ligation, assisted by the addition of a connector oligonucleotide. The detected protein molecules thus promote the ligation reactions by ensuring sufficient proximity between the ends of the proximity probes' oligonucleotide extensions. As two independent recognition events are required for detection of the target protein to create an amplifiable DNA molecule, there is a conceptual similarity to PCR, ensuring highly specific detection. The ligation products can then be replicated, detected, and quantified by real-time PCR, whereas unreacted probes remain silent. In this fashion, complicated and insensitive detection of protein molecules is replaced by a straightforward, sensitive, and specific detection reaction for nucleic acids. The conversion of target molecule identities to DNA reporter sequences offers the additional advantage that large sets of proteins could be analyzed in parallel, e.g., by including distinct tag sequences in the amplified segments and subsequent readout by hybridization to oligonucleotide arrays (20). To use PLA for sensitive and specific detection of interactions between proteins and nucleic acids, one of the proximity probes is a partly double-stranded oligonucleotide with a single-stranded 3′ extension. The other probe is an antibody directed against the DNA-binding protein, and it has an attached DNA strand with a free 5′ end (Fig. 1). We demonstrate the benefits of this new assay by characterizing the DNA sequence specificity of the proteins p53, HNF-4α, and USF1.
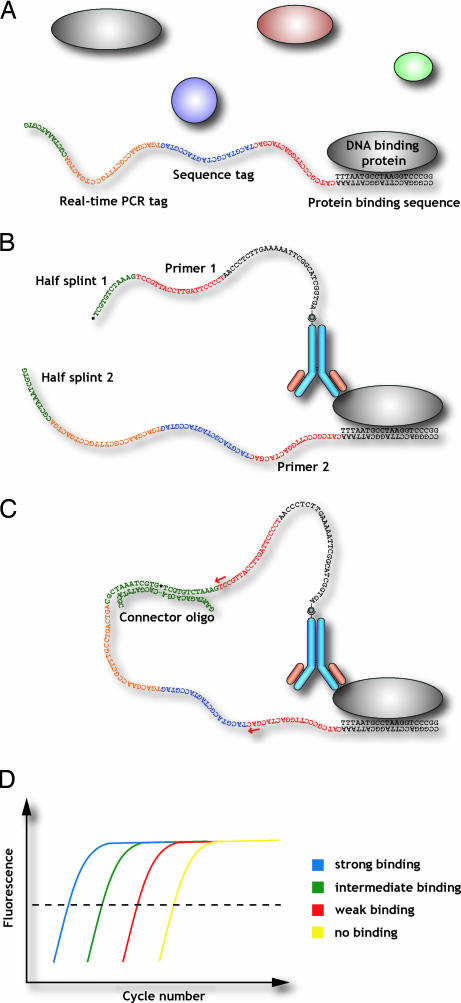
Fig. 1.
Analysis of DNA–protein interactions by proximity ligation. A DNA-binding protein can be investigated by using two affinity probes, each carrying an oligonucleotide extension. One of the probes would be an antibody directed against a particular DNA-binding protein, whereas the other would consist of a partially double-stranded DNA sequence potentially recognized by the same DNA-binding protein. If the protein were simultaneously bound by both affinity probes (A and B), the ends of their appended oligonucleotides would be brought sufficiently close so that they could hybridize together to a subsequently added connector oligonucleotide, allowing them to be joined by enzymatic ligation (C). The ligated DNA sequence, which would serve as a specific DNA representation of the binding event between the protein and the investigated recognition sequence, would be subsequently amplified and detected by real-time PCR (D).
Results
We chose to analyze the transcription factors p53, HNF-4α, and USF1 because they have been intensely studied and because they have all been associated with human disease.
p53.
The tumor suppressor p53 can mediate several different functions by activating or repressing a large number of target genes. TP53 is one of the most commonly mutated genes in human cancer, and mutations generally affect amino acids within the DNA-binding domain (21). In addition to its role in transcriptional regulation, p53 is also involved in transcription-independent regulation of apoptosis, genome integrity, DNA repair, and DNA recombination (22).
As a proof of principle, proximity ligation was used to analyze nuclear extracts from the breast carcinoma cell line MCF-7 for evidence of interactions between the DNA-binding protein p53, recognized by a p53 antibody-DNA conjugate, and partially double-stranded DNA probes including putative recognition sequences for the p53 protein. Three previously described p53-binding DNA sequences were analyzed together with specificity controls (Fig. 2A). The following sequences were shown to be positive by PLA: (i) a consensus P53-binding sequence (23), (ii) a focal adhesion kinase promoter region shown to interact with p53 (24), and (iii) a polymorphic microsatellite, (TGYCC)15, that mediates induction of p53-inducible gene 3 by p53 (25). Additionally, we included the following specificity controls: (i) a modified p53 consensus sequence in which four nucleotides within the consensus binding site were altered, (ii) the above p53 consensus sequence but now analyzed in the presence of a 1,000-fold excess of consensus probe without the ligatable sequence extension, (iii) a negative control sequence shown to bind HNF-4α, and (iv) a control for which no nuclear lysate was added to the reaction. The nuclear extract was diluted and analyzed with the consensus probe and the mutated consensus probe to determine the limit of detection, which was shown to be 2.5 ng of the MCF-7 nuclear extract (Fig. 2B). The experiments were additionally performed with recombinant P53 protein (Active Motif, Rixensart, Belgium) instead of the MCF-7 nuclear extract (results not shown).
Fig. 2.
Analysis of the DNA binding specificity of p53. (A) Three sequences previously reported to bind p53 were analyzed by PLA along with specificity controls in nuclear lysates prepared from MCF-7 cells. Positive controls: Pos I was a p53 consensus sequence published by Kastan et al. (23), Pos II (24) was a focal adhesion kinase promoter region shown to interact with p53, and Pos III was a polymorphic microsatellite that mediates induction of p53-inducible gene 3 by p53 (25). Negative controls: Pos I mut was a probe in which four nucleotides within the consensus binding site had been altered, and Pos I inhib control, a 1,000-fold excess of the consensus p53 DNA probe without the proximity probe extension, was added to the reaction together with the full-length consensus probe. The negative control was a probe positive for HNF-4α binding but with no known affinity for p53, and in the no lysate control, no nuclear lysate was added to the reaction. The S/N is shown on the y axis. (B) Dilutions of MCF-7 nuclear lysate were analyzed with the Pos I (filled bar) and the Pos I Mut probe (open bar) to determine the limit of detection of p53 in the extract.
HNF-4α.
Hepatocyte differentiation and metabolism are controlled both by ubiquitous and liver-specific transcription factors. HNF-4α belongs to the nuclear receptor family and is considered the major regulator of the hepatocyte phenotype (26). Mutations in HNF-4α resulting in an early stop codon or amino acid substitutions have been associated with an autosomal dominant form of diabetes, MODY1 (27), and polymorphisms in the HNF-4α promoter have been linked to the common form of type 2 diabetes (28).
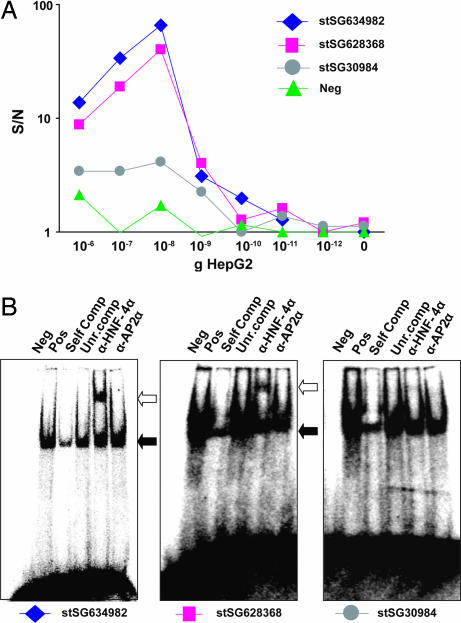
Three sequences identified by ChIP-chip analysis in HepG2 cells (29) and containing tentative HNF-4α binding sites predicted by the BioProspector software (30) were analyzed by PLA in HepG2 nuclear extracts, along with a probe containing the consensus sequence for p53-binding, serving as a negative control. The nuclear extract was diluted to find the amount that resulted in the highest signal-to-noise ratio (S/N) and to identify the highest detectable dilution (Fig. 3A). The optimal total protein mass in the nuclear extract for a maximal S/N was 20 ng, and the limit of detection was ≈1 ng, corresponding to extracts from 1 to 10 cells. Larger amounts of protein resulted in a decrease of signal, most probably because of a “high-dose hook effect,” known to occur when the amount of analyte exceeds the amount of affinity probes in an assay (31). By analyzing each consensus sequence with two or three different lysate dilutions, accurate measurements were obtained.
Fig. 3.
PLA analysis and EMSA of tentative HNF-4α-binding sequences. (A) Three sequences identified by ChIP-chip analysis were analyzed by PLA for interactions with HNF-4α in HepG2 nuclear extracts together with a negative control (a p53 consensus oligonucleotide). The nuclear extract was diluted as indicated along the x axis to investigate the lowest dilution with detectable amounts of binding protein. S/N values are shown on the y axis. (B) EMSAs of sequences identified by ChIP-chip (Pos) along with negative control sequences (Neg), competitor oligonucleotides having the same (Self Comp) or an unrelated sequence (Unr.comp), and supershift reactions with HNF-4α antibodies (α-HNF-4α and with irrelevant AP2α antibodies (α-AP2α.
The PLA measurements were verified by supershift EMSA analysis of the three HNF-4α-positive sequences (Fig. 3B), showing good agreement between the two methods. The signal generated by the DNA fragment stSG30984 was barely positive in EMSA (with ≈4 μg of nuclear lysate per lane), but a signal significantly higher than background was observed with the PLA, demonstrating that this assay is more sensitive than EMSA and has a wider dynamic range.
An additional 50 tentative HNF-4α binding sequences were randomly selected from the same ChIP-chip study and analyzed by PLA in HepG2 nuclear extracts. The S/N for the proximity ligation analysis of each of the probes was compared with the previously calculated prediction score that indicates the estimated likelihood that HNF-4α would bind to the actual sequence motifs in each of the probes [supporting information (SI) Fig. 6]. The correlation was good in that probes with the highest prediction score gave rise to high S/N, whereas probes with lower prediction score, from 4 to 7, with few exceptions demonstrated specific binding to the protein but at lower S/N.
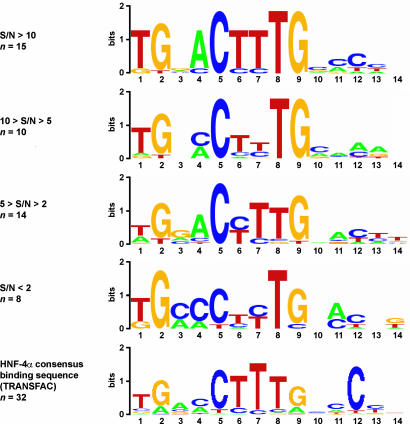
The different probe sequences were divided in groups based on their S/N values. The S/N defining the respective groups were selected so that a similar and sufficiently high number of sequences was assigned to each of them. Subsequently, each of the groups of sequences was analyzed with the motif-finding program BioProspector (30). The consensus motif found for each of the groups together with the previously known binding consensus motif for HNF-4α from the TRANSFAC database (32) are presented (Fig. 4) as a consensus logo created with WebLogo (33). The sequences that gave rise to the highest S/N were also those most similar to the TRANSFAC database consensus motif. As the S/N decreases, the similarity to the TRANSFAC motif decreases as well, but the most important nucleotide positions within the binding site are conserved (positions 1, 2, 5, 8, and 9).
Fig. 4.
HNF-4α consensus motives as identified by PLA and TRANSFAC. The 50 sequences shown in Fig. 4 were divided in groups based on their S/N values. The values defining each group were selected so that similar numbers of sequences were assigned to each of them. Subsequently, each of the group of sequences was analyzed by using the motif-finding program BioProspector (30), which identifies the most common motifs found among such sequences. The consensus motif found for each of the groups is illustrated with the program WebLogo (33). The consensus for each of the sequence groups represents the overall results obtained when BioProspector was run several times for each of the group, and very similar results were obtained in all cases. The previously known binding consensus sequence for HNF-4α as found in TRANSFAC (32) is indicated at the bottom.
These data indicate that the binding site predictions as previously calculated (29) are very accurate and of higher quality than ones previously reported (46–60), where in vitro methods such as EMSA and DNA footprinting were used to map the binding sites. The 50 sequences analyzed in this study contain sequences that were commonly identified as tentative HNF-4α binding sites by Rada-Iglesias et al. (29) and in other previous studies (46–50) and that generally performed well in the PLA (red sequences in SI Table 1, e.g., APOA4/C3 intergenic and F10 promoter). The present study also includes sequences for which there is a discrepancy between previous reports (47, 50) and the predictions by Rada-Iglesias et al. (29) but for which the PLA results support the Rada-Iglesias et al. prediction calculations (green and blue sequences, SI Table 1). The APOA1 promoter includes three previously identified binding sequences (50) not identified by Rada-Iglesias et al. (29). A putative binding site in the promoter was identified by Rada-Iglesias et al. and performed better in PLA than did the three previously reported sequences (50). Similarly, the APOA4 promoter includes sequences for which there is a discrepancy between binding predictions by Rada-Iglesias et al. and sites and previously reports (47). Again, the PLA supported these findings (SI Table 1).
USF1.
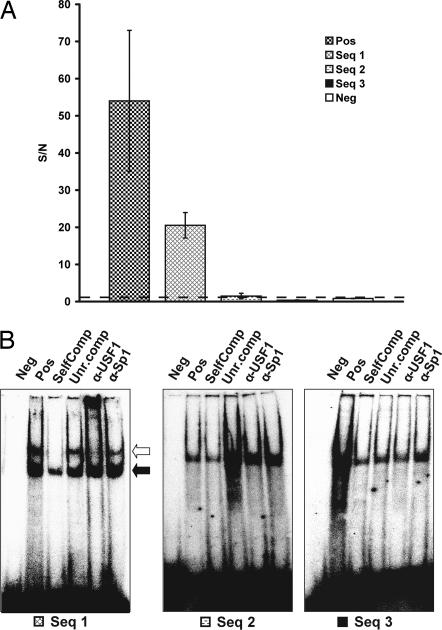
The ubiquitous transcription factor USF1 was recently implicated in familial combined hyperlipidemia, characterized by elevated levels of total serum cholesterol or triglycerides or both (34), type 2 diabetes, and elevated risk of cardiovascular disease. This was done by identification of SNP haplotypes associated with this form of hyperlipidemia. Genomic binding sites for USF1 and derived consensus binding sequences have been previously described (29). Four potential USF1 binding sequences identified by ChIP-chip analysis, as well as a HNF-4α consensus sequence used as a negative control, were analyzed by PLA for interactions with USF1 in 20 ng HepG2 nuclear extracts (Fig. 5). The positive control sequence and sequence 1 (both show high similarity with USF1 consensus E-box sequence) yielded high S/N by PLA, whereas sequences 2 and 3 and the negative control failed to generate signals significantly different from background. These results were confirmed by EMSAs (with ≈4 μg nuclear lysate per lane) along with specificity controls, introduction of competitive probes, competitive probes with an unrelated binding sequence, and supershift reactions with USF1 and Sp1 antibodies for sequences 1, 2, and 3. There is a good agreement between the two methods; only sequence 1 was observed to undergo bandshift in EMSA (Fig. 5).
Fig. 5.
DNA sequence specificity of USF1. (A) Four potential USF1 binding sequences identified by ChIP-chip analysis (Pos, positive control; Seq 1, sequence 1; Seq 2, sequence 2; and Seq 3, sequence 3), as well as a negative control (Neg; an HNF-4α consensus oligonucleotide) were analyzed with PLA for interactions with USF1 in 10-ng HepG2 nuclear extracts. S/N values are shown on the y axis. (B) EMSAs of three of the positive sequences as identified by ChIP-chip along with negative controls, competitive probes with the same (SelfComp) or an unrelated (Unr.comp) binding sequence, and supershift reactions with USF1 (α-USF1) and Sp1 (α-Sp1) antibodies.
Discussion
Much remains to be learned about the interplay between specific DNA-binding proteins and their target sequences to functionally annotate the genome. Parallel projects are underway to describe common genetic variation caused by SNPs and copy number variation and other mechanisms. Most protein-coding genes have now been identified, but many alternative promoters have recently been detected, and it is anticipated that more remain to be identified. Conserved regions in the human genome that are not known to be protein-coding (1.5%) may nonetheless be undetected coding sequences, or they may have structural importance or be involved in gene regulation. High-throughput techniques for analysis of DNA–protein interactions are therefore important for the functional annotation of the genome. Technologies for analysis of DNA–protein interactions like ChIP-chip or with sequence readout [ChIP-PET (pair end-tag sequencing) or ChIP-STAGE (sequence tag analysis of genomic enrichment)] (51–54) are powerful means to screen genomes for sites of DNA–protein interaction in vivo. The resolution of these techniques is in the hundreds of base pairs, i.e., the size of standard enhancers or promoters. The same is true for other array-based methods, such as DIP-chip (15) or DamID (14). Verification of binding is typically done by real-time PCR, which has a similar window size. However, individual proteins typically bind sequence elements of 10 bp or less, so other techniques are needed to more precisely identify sites for DNA–protein interaction at base-pair resolution.
A considerable number of methods exist for the investigation of DNA–protein interactions, addressing single pairs of interacting biomolecules at a time. Traditional methods for DNA–protein interaction analysis include Southwestern blotting (35), gel-retardation techniques, nitrocellulose-binding assays (36), and reporter constructs in yeast (37). Biochemical assays, such as DNase I footprinting, methylation protection, ethylation interference, or hydroxylradical footprinting, can be valuable tools to locate DNA-binding sites of proteins. They do not, however, generate information concerning the strength of the interactions. Biophysical approaches, such as fluorescence anisotropy (38), fluorescence correlation spectroscopy (39), and single-molecule force spectroscopy (40), can be powerful means to analyze DNA–protein interactions, but they require expert knowledge as well as nonstandard equipment and are therefore less applicable for routine laboratory use. Other drawbacks of many of the above mentioned techniques are high sample consumption and the need for protein purification before analysis. Additionally, these methods are poorly suited for multiplexing.
Protein-binding microarrays (13, 41) allow analysis of transcription factor binding patterns in vitro. The DNA-binding protein to be analyzed is expressed with an epitope tag and bound to double-stranded DNA microarrays. After removing nonbound proteins by washes, the array is labeled with a fluorophore-conjugated antibody specific for the epitope tag on the expressed DNA-binding protein. The method offers high-throughput characterization of DNA–protein binding with a very high resolution, but tagged proteins may exhibit altered binding properties compared with the endogenous proteins, and exact quantification of the interactions is difficult.
In vitro selection has been successfully used to generate highly accurate descriptions of protein binding sites (42). However, SELEX will, by design of the assay, only yield “optimal” binding sequences, which might be different from those found in vivo. In that regard, PLA offers the advantage of allowing comparison of protein affinities for any known or natural binding sites.
The PLA technique has important advantages over existing methods for in vitro analysis of protein–DNA interactions. This assay is nonradioactive and has a very low reagent and sample consumption rate and extremely high sensitivity through the use of nucleic acid reporters that enable analysis of a few nanograms of nuclear extracts, corresponding to between 1 and 10 cells. Accordingly, the method is several orders of magnitude more sensitive than techniques like EMSA for investigating DNA binding of proteins, and it yields quantitative results, allowing affinities to be calculated. The assay procedure takes less than 4 h, including the PCR step. The PLA assay can be performed in a homogenous format as shown herein or on a solid support with the protein-binding DNA probe immobilized to a surface prior to interaction analysis, allowing nonspecifically bound reagents to be removed by washes (data not shown). The homogeneous assay format requires no washes or phase separations and the hands-on time is very limited, with only three additions of reagents, which makes it suitable for automation.
The required instrumentation, including the real-time PCR instrument, is standard laboratory equipment. Reagents for these assays are inexpensive and/or easy to prepare. The technique should also be suitable for multiplexing, i.e., allowing the simultaneous analysis of many different proteins and/or different DNA probe molecules. For that purpose, unique tag sequences are included in the DNA or antibody probes (Fig. 1), permitting individual amplification products to be identified on DNA tag arrays (20). Libraries of potential binding sites can be identified either by using bioinformatic approaches or by mining results from high-throughput techniques, as shown in this manuscript. As prices for oligonucleotide synthesis continue to fall, costs for synthesis of such libraries will be comparable to those charged for commercial microarray probe collections.
Proximity ligation of transiently interacting molecules may permit the analysis of interactions with affinities lower than those detectable by physical capture, and the mechanism has furthermore been shown to allow detection of complexes involving more than two components (43). The proximity ligation mechanism can also be adapted to allow in situ analysis of DNA–protein interactions for localized detection (O. Söderberg, personal communication), rendering the technique promising for studies of tissue heterogeneity in terms of promoter occupancy.
Cantor and coworkers (55) have developed an alternative approach for in vitro analysis of protein–DNA interactions. Double-stranded oligonucleotides bound by transcription factors were physically isolated and amplified, and the identities and amounts of bound sequences were identified by mass spectrometric quantification of DNA mass tags. The authors report successful multiplexing, which is limited only by the resolution defined by the length of the mass tags. We conclude that DNA molecules bound by specific proteins can be captured either by physical isolation or proximity-dependent ligation, and amplified representations of captured sequences can be identified via real-time PCR or mass spectrophotometric tags, and read-out on tag arrays could allow even greater numbers of sequences to be evaluated.
In summary, the PLA-based technique described here is suitable for highly sensitive, quantitative analyses of interactions between transcription factors and regulatory sequence elements with single-base-pair resolution. It should thus be useful for characterization of transcription factor levels across different tissues and cell types in various normal and disease states.
Materials and Methods
Antibody Biotinylation and Conjugation.
Biotinylated affinity-purified polyclonal antibody against p53 was purchased from R&D Systems (Abingdon, U.K.). The HNF-4α antibodies were purchased from Active Motif and from Santa Cruz Biotechnology (Santa Cruz, CA) (catalog no. C19). The polyclonal antibody against USF1 (H86) was purchased from Santa Cruz Biotechnology.
HNF-4α and USF1 antibodies were biotinylated with d-biotin-N-hydroxysuccinimide (Nordic Biosite, Taby, Sweden) according to the recommendations of the manufacturer.
Streptavidin–oligonucleotide conjugates were prepared by coupling a 5′ free thiol-modified oligonucleotide to maleimide-derivatized streptavidin, as previously described (19). The sequence of the 5′STV is 5′-P-TCGTGTCTAAAGTCCGTTACCTTGATTCCC CTAACCCTCT TGAAAAATTCGGCATCGGTGA-3′. The biotinylated antibodies were coupled with the STV-oligonucleotide conjugates as follows. The biotinylated antibodies were diluted in PBS with 1% (wt/vol) BSA (Sigma, St Louis, MO) to a final concentration of 30 nM. The antibodies were then combined with the streptavidin–oligonucleotide conjugations in a 1:1 ratio in a volume of 5 μl at room temperature for 1 h. Thereafter, the antibody-oligonucleotide probes were further diluted to a concentration of 1.2 nM in a probe-dilution buffer [1× PBS/1% (wt/vol) BSA/16 μg/ml sheared polyA bulk nucleic acid (Sigma–Aldrich, Stockholm, Sweden)/1 mM d-biotin (Molecular Probes, Eugene, OR)] and stored at 4°C until use.
DNA Probes.
All DNA probes for this study were designed by using the software framework ProbeMaker (44). HPLC-purified oligonucleotide probes were purchased from Biomers.net (Ulm, Germany). The common sequence, i.e., primer 2, the sequence tag, and the real-time PCR tag, as well as 12 nucleotides complementary to one half of the connector oligonucleotide (Fig. 1), is CATCGCCCTTGGACTACGACTGACGAACCGCTTTGCCTGAC-TGATCGCT AAATCGTG.
The variable sequences contain the predicted binding sites (10–12 bp) that have been extended by 5 bp on both 5′ and 3′ ends.
The p53 variable sequences were as follows: p53 consensus oligonucleotide (23), TAGAGAACATGTCTAAGCATGCTGGGGACT; p53 consensus oligonucleotide (mutated binding site), TACAGAATCGCTCTAAGCATGCTGGGGACT; “FAK-promoter region”, AGGCACCTCCGCAAGCCCCACGCAGC; and polymorphic microsatellite, (TGYCC)15 (where Y is C/T).
The HNF-4α variable sequences were as follows: stSG634982, AACTCAGGGCAAAGTTCAGCTGCTG; stSG628368, CCTCTCTGAACCTTGGATTCCTCAC; and stSG30984, CTGGAAGGCCAAAGACCCTAGTCAA.
The 50 additional HNF-4α binding sequences are shown in SI Table 1.
The USF1 variable sequences were as follows: positive control (stSG627950), CCTGCCCACGTGACCCGGCCT; sequence 1 (stSG627950), TGCACGGTCACGTGCTCGAGC; sequence 2 (stSG609339), TGGATAGTCACTTAATGCTTA; sequence 3 (stSG628088), CATACGGTTGCGTGGATGCTC; and negative control (HNF-4α consensus), AACTCAGGGCAAAGTTCAGCTGCTG.
The probes were made partially double-stranded by hybridizing an equimolar amount of a 20-mer oligonucleotide complementary to the variable part of the probe in 0.3× standard saline citrate (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7).
Nuclear Lysates and EMSA.
Nuclear lysate from peroxide-treated MCF-7 cells was purchased from Active Motif. HepG2 nuclear extracts were prepared as previously described (45). EMSAs were performed as described by Rada-Iglesias et al. (29).
Proximity Ligation Reaction.
Incubation of samples with proximity probes p53 and HNF-4α/USF1.
For p53, DNA probes (25 pM) were incubated with 1 μl of the nuclear extract in 10 mM Tris buffer (pH 7.5) with 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 5% (vol/vol) glycerol, and 1 μg of poly(dI-dC) in optical PCR tubes (Applied Biosystems, Foster City, CA) in a total volume of 3 μl for 30 min at room temperature. Two microliters of 5′ free anti-p53-DNA conjugate was added to the mixture at a concentration of 20 pM, and the incubation was continued for 2 h at room temperature. The reaction conditions were adapted from a protocol provided by Santa Cruz Biotechnology.
For HNF-4α/USF1, DNA probes (25 pM) were incubated with 1 μl of the nuclear extract in 10 mM Hepes-KOH buffer (pH 9.7), 10% (vol/vol) glycerol, 50 mM KCl, 5 mM MgCl2, and 0.6 mM DTT in optical PCR tubes (Applied Biosystems) in a total volume of 3 μl for 60 min on ice. Two microliters of the 5′ free anti-HNF-4α/USF1 conjugate was added to the mixture in a concentration of 20 pM, and the incubation was continued for at least 4 h at 4°C. Reaction conditions were adapted from the EMSA protocol published by Rada-Iglesias et al. (29).
Ligation and quantitative real-time PCR.
After the initial incubations, 45 μl of a combined mixture for ligation and amplification was added to the 5-μl incubations [final concentrations: 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 3.15 mM MgCl2, 0.4 Weiss units of T4 DNA ligase (Fermentas, St. Leon-Rot, Germany), 400 nM connector oligonucleotide (TACTTAGACACGACACGATTTAGTTT; biomers.net), 80 μM ATP, 200 μM dNTP mixed with dUTP, 100 nM primers (forward, CATCGCCCTTGGACTACGA; reverse, GGGAATCAAGGTAACGGACTTTAG; biomers.net), 100 nM TaqMan probe (FAM-TGACGAACCGCTTTGCCTGA-MGBNFQ; Applied Biosystems), and 1.5 units of Platinum TaqDNA polymerase (Invitrogen)]. After the addition of the combined ligation and amplification mix, the tubes were sealed with optical PCR lids (Applied Biosystems) and transferred to a real-time PCR instrument (Mx3000P quantitative PCR system; Stratagene, Amsterdam, The Netherlands) or a PRISM 7000 sequence detection system (Applied Biosystems). Thermal cycling conditions comprised an initial activation step of 2 min at 95°C, then 45 cycles of 15-s denaturation at 95°C, and 60 s annealing/extension at 60°C.
Analysis of the results.
The samples were analyzed in duplicates or triplicates. The results presented in the figures are mean values of the S/N, where the number of ligations of proximity probe pairs in the sample are divided by the number of ligations in a negative control.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Councils for Medicine and Natural and Engineering Sciences, the Wallenberg Consortium North for Functional Genomics, the Graduate Research School in Genomics and Bioinformatics, Uppsala Bio-X, and the EU-FP6 integrated project MolTools. J.S. was supported by a Marie Curie Action Grant.
Abbreviations
- PLA
proximity ligation assay
- S/N
signal-to-noise ratio.
Footnotes
Conflict of interest statement: U.L. is the inventor of patents describing the proximity ligation technology. He is one of the founders of the company Olink AB, which exploits the proximity ligation technology.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611229104/DC1.
References
- 1.Ptashne M, Gann A. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Orlando V, Paro R. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 4.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 5.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 6.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 7.Ng HH, Robert F, Young RA, Struhl K. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 9.Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. Proc Natl Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 13.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Nat Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 14.van Steensel B, Delrow J, Henikoff S. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Noll DM, Lieb JD, Clarke ND. Genome Res. 2005;15:421–427. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fried M, Crothers DM. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristie TM, Roizman B. Proc Natl Acad Sci USA. 1986;83:3218–3222. doi: 10.1073/pnas.83.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 19.Gullberg M, Gustafsdottir SM, Schallmeiner E, Jarvius J, Bjarnegard M, Betsholtz C, Landegren U, Fredriksson S. Proc Natl Acad Sci USA. 2004;101:8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 21.Hollstein M, Sidransky D, Vogelstein B, Harris CC. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 22.Sengupta S, Harris CC. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 23.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 24.Golubovskaya V, Kaur A, Cance W. Biochim Biophys Acta. 2004;1678:111–125. doi: 10.1016/j.bbaexp.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 26.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 27.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 28.Silander K, Mohlke KL, Scott LJ, Peck EC, Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G, et al. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 29.Rada-Iglesias A, Wallerman O, Koch C, Ameur A, Enroth S, Clelland G, Wester K, Wilcox S, Dovey OM, Ellis PD, et al. Hum Mol Genet. 2005;14:3435–3447. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Brutlag DL, Liu JS. Pac Symp Biocomput. 2001;2001:127–138. [PubMed] [Google Scholar]
- 31.Wolf BA, Garrett NC, Nahm MH. N Engl J Med. 1989;320:1755–1756. doi: 10.1056/NEJM198906293202614. [DOI] [PubMed] [Google Scholar]
- 32.Wingender E. Nucleic Acids Res. 1988;16:1879–1902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooks GE, Hon G, Chandonia JM, Brenner SE. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, et al. Nat Genet. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- 35.Bowen B, Steinberg J, Laemmli UK, Weintraub H. Nucleic Acids Res. 1980;8:1–20. doi: 10.1093/nar/8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodbury CP, Jr, von Hippel PH. Biochemistry. 1983;22:4730–4737. doi: 10.1021/bi00289a018. [DOI] [PubMed] [Google Scholar]
- 37.Hanes SD, Brent R. Science. 1991;251:426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi M, Sakumi K, Sekiguchi M. Biochemistry. 1990;29:3431–3436. doi: 10.1021/bi00466a002. [DOI] [PubMed] [Google Scholar]
- 39.Heyduk T, Lee JC. Proc Natl Acad Sci USA. 1990;87:1744–1748. doi: 10.1073/pnas.87.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch SJ, Shundrovsky A, Jantzen BC, Wang MD. Biophys J. 2002;83:1098–1105. doi: 10.1016/S0006-3495(02)75233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P. Nat Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 43.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 44.Stenberg J, Nilsson M, Landegren U. BMC Bioinformatics. 2005;6:229. doi: 10.1186/1471-2105-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrews NC, Faller DV. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vergnes L, Taniguchi T, Omori K, Zakin MM, Ochoa A. Biochim Biophys Acta. 1997;1348:299–310. doi: 10.1016/s0005-2760(97)00071-4. [DOI] [PubMed] [Google Scholar]
- 47.Carriere V, Vidal R, Lazou K, Lacasa M, Delers F, Ribeiro A, Rousset M, Chambaz J, Lacorte JM. J Biol Chem. 2005;280:5406–5413. doi: 10.1074/jbc.M408002200. [DOI] [PubMed] [Google Scholar]
- 48.Ktistaki E, Lacorte JM, Katrakili N, Zannis VI, Talianidis I. Nucleic Acids Res. 1994;22:4689–4696. doi: 10.1093/nar/22.22.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung HL, High KA. J Biol Chem. 1996;271:2323–2331. doi: 10.1074/jbc.271.4.2323. [DOI] [PubMed] [Google Scholar]
- 50.Tzameli I, Zannis VI. J Biol Chem. 1996;271:8402–8415. doi: 10.1074/jbc.271.14.8402. [DOI] [PubMed] [Google Scholar]
- 51.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 52.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 53.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, Bhinge AA, Morgan XC, Iyer VR. Nat Methods. 2005;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L, Kasif S, Cantor CR. Proc Natl Acad Sci USA. 2007;104:3061–3066. doi: 10.1073/pnas.0611075104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.