Abstract
Networks of interacting genes are responsible for generating life's complexity and for mediating how organisms respond to their environment. Thus, a basic understanding of genetic variation in gene networks in natural populations is important for elucidating how changes at the genetic level map to higher levels of biological organization. Here, using the well-characterized phototransduction network in Drosophila, we analyze variation in gene expression within and between two closely related species, Drosophila melanogaster and Drosophila simulans, under different environmental conditions. Gene expression levels in the pathway are largely conserved between these two sibling species. For most genes in the network, differences in level of gene expression between species are correlated with degree of polymorphism within species. However, one gene encoding the light-induced ion channel TRPL (transient receptor potential-like) shows an excess of expression divergence relative to polymorphism, suggesting a possible role for natural selection in shaping this expression difference between species. Finally, this difference in TRPL expression likely has significant functional consequences, because it is known that a high level of rhabdomeral TRPL leads to increased sensitivity to dim background light and an increased response to a wider range of light intensities. These results provide a preliminary quantification of variation and divergence of gene expression between species in a known gene network and provide a foundation for a system-level understanding of functional and evolutionary change.
Keywords: natural selection, gene expression, network
Genes act together in networks to generate important organismal phenotypes. Understanding the general properties of gene networks and how they adapt to environmental changes is of fundamental importance to evolutionary biology. Elucidating the structure of gene networks represents a major goal of systems biology and ecological genomics in the postgenomic era. Recent approaches have primarily exploited genomic and computational methods to focus on distantly related organisms (1). However, very little is known about the basic microevolutionary properties of known genetic networks, information that is crucial for understanding network function, regulation, and evolution, because key adaptations first emerge at the population level. For example, very little is known about natural variation within gene expression networks in different populations, different species, and under different environmental conditions. How much expression variation is present in a gene network within a population? Do genes that show low variation within one species also show low variation in closely related species? To attempt to answer these fundamental questions, we analyzed variation in gene expression in the phototransduction network under two different environmental conditions within and between species of Drosophila melanogaster and its sibling species Drosophila simulans, which diverged ≈3 Mya (2).
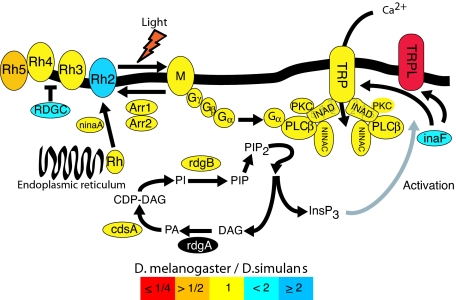
The Drosophila phototransduction network is one of the best characterized genetic networks to date at the biochemical and cellular level. The network is composed of ≈25 genes that encode structural proteins and signal transducers responsible for transforming light energy entering the eye into a nerve impulse (3). The topology of this network has been extensively studied, and the molecular functions of the majority of the molecules in the network are known. The cascade of transduction signals is triggered by as little as one photon of light inducing the activation of rhodopsin into metarhodopsin, which in turn activates downstream targets, subsequently leading to the activation of the calcium channels, TRPL and TRP, and ultimately neural activation (details in Fig. 1).
Fig. 1.
Major proteins of the phototransduction network. Colors indicate the observed relative levels of gene expression between D. melanogaster and D. simulans. Color coding indicates expression difference between species as a ratio (D. melanogaster/D. simulans). Red is ≤1/4; orange is >1/2; yellow is 1/1; light blue is <2/1; dark blue is ≥2/1; black, no data. The Drosophila phototransduction network is a paradigm for the study of signal transduction (3, 14, 23, 31, 32). The end point of the cascade is the depolarization of the photoreceptor cell, resulting from the opening of Na+- and Ca2+-permeable channels, TRP and TRPL. Light initially activates rhodopsin into active metarhodopsin, which catalyzes the activation of a heterotrimeric G protein. The Gα subunit of the activated heterotrimer then activates an effector enzyme, phosphoinositide-specific phospholipase C (PLCβ, encoded by the gene norpA), which leads to the production of two signaling molecules, DAG (diacylglycerol) and InsP3 (inositol triphosphate) from PIP2 (phosphatidylinositol-4,5-biphosphate). This ultimately leads to the activation of the light-sensitive channels TRP, TRPL, and TRPγ (the latter is not studied here). The gray arrow indicates a mechanism of action that is not totally elucidated. TRPL and TRPγ form heteromultimers with TRP and with each other. Modulation of the ratio of TRP/TRPL subunits in the rabdhomeres through the light-sensitive translocation of TRPL has been suggested as a physiological mechanism increasing sensitivity to dim background light and allowing response to a wider dynamic range of light intensities (13). A sharp response is provided by the inactivation reactions that take place soon after activation. Two important players in the inactivation are the arrestins, which bind to and inactivate rhodopsin. Activity of both the GTP-bound Gqα subunit and PLC is terminated by the GTPase activity of the G protein and other components. Light-sensitive channels are inactivated by Ca2+ through the action of calmodulin. RDGC is a rhodopsin phosphatase that is regulated by calmodulin. Its function in deactivation of the response, by means of the deactivation of rhodopsin-mediated signaling, is mostly inferred from mutant phenotypes. Many of the molecules are assembled in a signalplex whose core component is INAD, a scaffold protein with five PDZ protein-interaction modules. Other proteins interact with INAD, but their localization in the rabdhomeres does not depend on INAD. InaF serves as a regulator of the calcium-channel activity, but its mode of action is not completely elucidated. NinaA is responsible for transporting rhodopsin from the endoplasmic reticulum to the cell membrane. PKC is a protein kinase encoded by inaC, whose role is not fully understood, but mutant phenotypes suggest a role in adaptation to light and termination response.
Light perception is central to many ecological and sexual traits from foraging behavior to mate recognition. Although the number and type of genes involved in phototransduction exhibits extensive conservation across animal evolution (3), recent investigations of specific genes, such as rhodopsin, have revealed functional regulatory variation associated with ecological adaptations in vertebrates (4). Genetic variation within and among Drosophila species in characters related to vision is abundant (5–7). Although few physiological and behavioral differences of ecological importance are known to differentiate D. melanogaster and D. simulans (8, 9), traits related to vision stand out as being important. For instance, it has been reported that D. melanogaster preferably courts in the dark and that females of D. simulans are more responsive to the visual aspects of male courtship than are females of D. melanogaster (10). Locomotion in response to light also differs between these species. A study of phototaxis along a gradient of light intensity by Parsons (11) demonstrated that Drosophila simulans shows greater positive phototaxis than Drosophila melanogaster, and it is more tolerant of high light intensity (10). Furthermore, the genetic architecture of phototactic behavior is different between D. simulans and D. melanogaster (12).
Here, we quantify absolute levels of mRNA abundance for 20 genes in the phototransduction network for 11 different strains each in D. melanogaster and D. simulans, using real-time quantitative PCR (rt-qPCR). Between species, transcript abundance of orthologous nodes (genes) in the network was highly correlated, which implies conservation of the gene network over a short evolutionary time span. We find that levels of expression polymorphism and divergence at nodes in the gene network are generally correlated. However, a single gene, encoding the light-induced ion channel TRPL, stands out from the others in showing substantially greater between-species divergence relative to within-species polymorphism. This expression difference is likely to have important functional consequences for phototransduction between the two sibling species, because rhabdomeral TRPL abundance plays a role in the fine tuning of light perception in D. melanogaster (13).
Results and Discussion
Transcript abundance for 20 genes in the visual network was quantified in 11 geographically diverse strains of D. melanogaster and 11 strains of D. simulans. Individuals were collected midway through the light portion of a 12-h:12-h light:dark cycle. Additionally, individuals from seven D. melanogaster strains and five D. simulans strains, chosen randomly, were collected after a 1-h exposure to total darkness. In total, there were 34 strain-treatment combinations (22 light, 12 dark) × 20 genes × 2 rt-qPCR replicates each, for a total of 1,360 rt-qPCRs plus several controls. The experimental design that was used pooled cDNA reactions to reduce experimental error and biological noise, which entailed 68 separate mRNA extractions and two rounds of cDNA synthesis per extraction for a total of 136 reverse-transcriptase reactions. Using cloned DNA fragments of each of the 20 genes as controls, we estimated the absolute number of transcripts for each gene (see Methods). Naturally occurring transcript abundance was found to range across three orders of magnitude from 5,600 to 1.5 million transcripts per gene. The reproducibility between replicate rt-qPCR experiments was excellent (Pearson correlation, r = 0.99, P < 2 × 10−16, n = 660). The expression level of all genes in each experiment and the significance of particular factors on expression were determined by using a mixed-model analysis of variance [supporting information (SI) Table 1].
Transcript Abundance in the Network Is Similar Between Species.
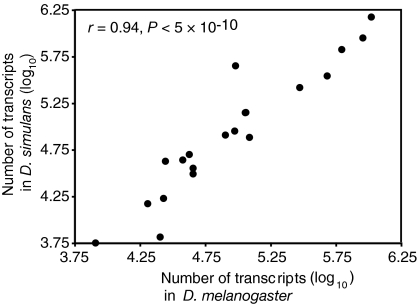
Examination of 20 genes in the phototransduction networks of D. melanogaster and D. simulans showed that mean transcript abundance was highly correlated between species (r = 0.94, P = 4.6 × 10−10) (Fig. 2). The general constancy in expression level across the network, maintained between species over the last 3 MY, suggests that the topology of the network has likely not changed substantially between D. melanogaster and D. simulans. Likewise, there was no coordinated directional change in expression across the network between species, because roughly half the genes (11/20) showed higher average expression in D. melanogaster relative to D. simulans (binomial test, n = 20, P = 0.82). This comparison eliminates the possibility that morphological differences between species, such as eye size, are responsible for differences in transcript abundance between species.
Fig. 2.
Relative transcript abundance. Expression level of the genes of the phototransduction pathway in D. melanogaster and D. simulans are largely conserved between the two sibling species (Pearson correlation, r = 0.94, P = 4.6 × 10−10, n = 20).
Up-Regulation of Entire Network Under Dark Conditions.
We found no significant species × environment effect, which suggests that the two species are affected similarly by the short treatment in the dark. Five genes showed significant up-regulation in the dark treatment (one hour of total darkness) compared with the light treatment (ANOVA, P < 0.05). The most significant change (1.26-fold, P = 0.0023) is observed for the gene encoding the first signal transducer in the cascade, Gγ30A, which acts downstream of metarhodopsin (Fig. 1). As might be expected of genes encoding interacting proteins, Gβ76C, which encodes the next protein in the cascade, is also significantly induced (1.23-fold, ANOVA, P = 0.028). These two proteins interact with each other and form a heterotrimer with the product of Gα49. We saw no significant increase in expression of the gene for this last signaling molecule (ANOVA, P = 0.87). However, any increase in expression might be obscured by the fact that Gα49 is the only G protein of the three whose expression appears not to be limited to the Drosophila eye, whereas most other genes we studied have their expression localized in the photoreceptor cells (www.flybase.org) (14). Because expression in the eye is likely to be only a small fraction of expression in the body, a slight increase in abundance would be more difficult to detect for Gα49. Our results also support this interpretation, because the gene is expressed at much a higher level than that of the two other genes whose products form the heterotrimer (SI Table 1). Interestingly, the gene for two calcium-channel proteins (TRP and TRPL), which act as heterodimers, were also significantly induced in the dark (1.27- and 1.21-fold, P < 0.05). One surprising result is that all 20 genes in the phototransduction network were qualitatively up-regulated in the dark compared with the light. This coordinated directional change in gene expression is highly unlikely by chance alone (binomial test, P = 1.9 × 10−6). This finding strongly suggests that all of the genes respond to dark conditions via up-regulation of transcription. The up-regulation of the entire network conflicts with models in which the modification of gene network outputs under different conditions is achieved by the manipulation of particular key nodes to increase sensitivity, for example, the up-regulation of activators such as the rhodopsins and down-regulation of repressors such as the arrestins. Here, in the dark treatment compared with the light treatment, all components of the network, including activators, repressors, structural genes, and genes involved in signal transduction, are up-regulated, albeit not necessarily stoichiometrically. This transcriptional response represents a unique mode of network regulation, one where the entire network acts as a single module. A large-scale gene-expression study in Drosophila also recently reported changes in transcript abundance in response to light conditions for several of the genes in the phototransduction pathway with a longer dark treatment (15). Our findings also contrast with the classical view that transcription has little to do with short-term light adaptation, because it is a slow process, and rather support Hardie's view (16) that modulation of transcription may be an important part of the eye's overall physiological strategy for light adaptation.
Polymorphism and Divergence.
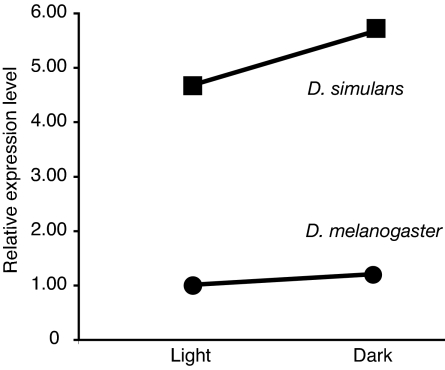
Our data also address the extent to which the level of genetic variation at specific nodes of a network in a species can be used to predict how much variation would be found in a closely related species. We found that polymorphism for expression levels in D. melanogaster, as estimated by the mean sum of squares (MS) for lines, is significantly correlated with that found in D. simulans (Spearman correlation, r = 0.47, P = 0.038), suggesting that genetic variation tends to be found at the same nodes of the network in these two closely related species. This variation among strains does not appear to depend on the total expression level, because the polymorphism did not correlate with absolute expression level (Spearman correlation, rmel = −0.14, P = 0.54, rsim = −0.08, P = 0.73). Five genes showed a significant difference in average expression between the two species (Fig. 1, P < 0.05). For instance, inaF shows a 1.6-fold relative up-regulation in D. melanogaster (P = 0.0099). The INAF protein serves as an activator of calcium-channel activity, but its mode of action is not completely elucidated (17). Another transcript, rdgc, whose product is involved in response termination, also shows relative up-regulation in D. melanogaster (1.4-fold, P = 0.0089). The largest and most significant change in expression was observed in the gene encoding TRPL, which showed a 4.7-fold up-regulation in expression in D. simulans relative to D. melanogaster (P < 0.0001) (Fig. 3). TRPL is a major structural component in a calcium channel located in the rhabdomere membrane, whose opening is the final step of light activation (see Functional Significance of TRPL and Fig. 1).
Fig. 3.
Expression level of trpl in D. melanogaster and D. simulans in light and after a 1-h exposure to complete darkness. Expression level is shown relative to the lowest expression level, which is arbitrarily set to one. ANOVA: Psp < 0.0001; Penv = 0.03; Pint = 0.88.
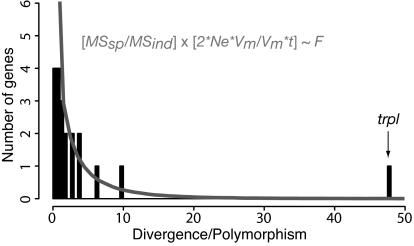
To assess which evolutionary forces may be acting on the expression of the genes in the visual network, we compared the variance in expression level among strains within each species to the average difference in expression level between species. Under stabilizing selection, the average value of a phenotype is expected to be conserved between species, and most variation should be found within species, because the variance in the trait will be maintained by slightly deleterious mutations (18, 19). Thus, we tested whether differences in transcript abundance observed between species (divergence) were correlated with differences within species (polymorphism). We found that polymorphism within species was generally correlated with divergence observed between species (Spearman correlation, r = 0.65, P = 0.0025). However, one gene, encoding TRPL, showed a large excess of expression divergence compared with polymorphism (47-fold). This excess of divergence relative to polymorphism suggests that diversification or directional selection has acted to change the expression of TRPL between the two species. The probability of observing different levels of divergence relative to polymorphism can be estimated by using neutral models of quantitative trait evolution (20). Following the procedures described in refs. 21 and 22, the ratio of the variation between species to variation within species (scaled by their expectations) should follow an F distribution:
where Ne is the effective population size, Vm is the mutational variance, and t is the number of generations separating the two species. If we assume that these two species diverged for 32 × 106 generations and that their effective population size is ≈3 × 106, the expected distribution of F ratios should be 2.7 times the standard F1,10 distribution. Because F1,10,0.025 = 6.94, a ratio of polymorphism to divergence exceeding 21 is indicative of evolution by diversifying selection (Fig. 4). The high level of divergence observed for trpl and the low amount of polymorphism observed for this gene suggest that the action of directional selection has shaped the difference of TRPL expression between the species.
Fig. 4.
Distribution of divergence (MSspecies) to polymorphism (MSstrain) ratio across the phototransduction pathway. The solid lines represent the theoretical expectation; i.e., 2.7 times the F1,10 distribution. The gene at the far right of the distribution is trpl.
Functional Significance of TRPL.
TRPL is a member of the TRP cation channels, which are key to animal physiology and sensory perception and thus play fundamental roles in ecological adaptations. In Drosophila, they form a superfamily of 13 proteins with roles in phototransduction, taste, thermosensation, osmosensation, touch, and hearing (23). TRP is the main cation channel in Drosophila, and the importance of TRPL has been underappreciated because of the lack of apparent phenotype of a trpl-null mutant in standard assays (24). TRPL is unique among the three-phototransduction ion channels, because under dark conditions, it is translocated from the main cell body to the rhabdomere, where phototransduction takes place. High levels of rhabdomeral TRPL enable flies to be more sensitive to dim or background light and allow them to respond to a wider range of light intensities (13). Increasing rhabdomeral TRPL has been suggested to be a mechanism to fine tune the visual response (16). TRPL expression in D. melanogaster peaks at the end of the day, and it has recently been proposed that the accumulation of trpl transcript at night would facilitate efficient accumulation of TRPL protein in the rhabdomeres at dusk (15). This suggests that up-regulation of trpl in D. simulans would result in an increased sensitivity to light relative to D. melanogaster.
Our study provides an estimate of genetic variation and divergence in gene expression of the well characterized phototransduction gene network. Analysis of these data reveals that the network appears globally conserved between D. melanogaster and D. simulans, because few genes show significant differences in expression level between species. However, the expression of a single gene, trpl, has dramatically diverged between D. simulans and D. melanogaster, while at the same time showing very little variation in expression within each species from a world-wide population sample. This result suggests that the change in trpl expression in the network was driven by directional selection with functional consequences. The nature of the selective forces acting on the expression level of trpl remains to be investigated. Given the importance of visual cues in mate choice in D. simulans (10), one can speculate that sexual selection may have played a role. Another possibility is that the association of D. melanogaster with human activities may have been associated with new visual requirements that resulted in selection for different expression phenotypes in this species, as suggested by David et al. (9). Functional analyses of the consequence of high TRPL expression difference in D. simulans and how this expression relates to the species' enhanced phototaxic behavior (11) and its reported tolerance to higher light intensity (10) will help to refine further these hypotheses. Finally, investigation of visual physiological activity and sensitivity coupled with the analysis of variation in network components within species will help to elucidate the evolutionary and ecological mechanisms that may drive selection for differences in gene expression between species in the phototransduction network.
Materials and Methods
Fly Species and Strains.
D. melanogaster and D. simulans strains were obtained from stock centers and individual collections. D. simulans strains: Yun Tao kindly provided S5 (South Africa), S9 (Southern France), S13 (Tunisia), S17 (Congo), S25 (Australia), S30 (Japan), S34 (Polynesia), and Japan/S2 (Japan); Turrumurra (Australia) was obtained from the Tucson Drosophila Stock Center (Tucson, AZ); Sim1 (Chapel Hill, NC) was obtained from D.L.H. D. melanogaster strains: Harwich (England), Oregon-R (Oregon), and Hikone-R (Japan) were obtained from D.L.H.; ICA (Peru), Toonda (Australia), and Taiwan (Taiwan) were obtained from the Tucson Drosophila Stock Center; BS1 (Spain), CA1 (South Africa), M2 (Australia), PYR2 (Spain), and Reids3 (Portugal) were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). BS1, CA1, Harwich, M2, PYR2, Taiwan, Toonda, S25, S30, S34, Sim1, and Japan were subjected to the dark treatment.
Both species were reared in glass vials on standard Drosophila medium at 25°C for several generations until 30–60 flies were available for RNA extraction. Flies were then transferred to a controlled 12-h:12-h light:dark-cycle environment at 25°C for one generation in a light chamber with UVA/UVB full-spectrum bulbs, with vials positioned randomly. Recently eclosed (<5 h old) males were then collected and transferred to fresh vials and again arranged randomly in the light chamber. After 3 days, male flies of the same strain were transferred to empty plastic 15-ml transparent tubes (BD Biosciences, Carlsbad, CA), with 10–15 flies per tube. Tubes with strains marked for dark exposure were covered completely with aluminum foil. Both sets of tubes were returned to the light chamber for 1 h for CO2 anesthesia recovery and then flash-frozen in situ by rapid submersion in liquid nitrogen halfway through the day light cycle. To obtain at least 15 flies per strain per mRNA extraction, collections were made on two separate days and combined. Flash-frozen flies were stored at −80°C. All fly handling was done by using CO2.
Expression Analysis.
Two RNA extractions were performed for each strain-treatment for a total of 68 extractions. Pools of 12–15 frozen flies were crushed on dry ice, and RNA was extracted by using the SV RNA extraction system (Promega, Madison, WI) as described in ref. 25 (DNAfree; Ambion, Austin, TX). Because the majority of the proteins known to function in Drosophila phototransduction are either highly enriched or expressed solely in photoreceptor cells (14), we extracted RNA from whole flies. RNA quality and concentration were estimated first by using a spectrophotometer and second by using RiboGreen (Molecular Probes, Eugene, OR) in quadruplicate, according to manufacturer protocols. One microgram of total RNA per extraction was used for each of two reverse transcription cDNA syntheses (SuperScript II; Invitrogen, Carlsbad, CA), using both poly(dT) and random hexamers, for a total of four reverse transcription reactions per strain-treatment combinations (136 in total). cDNA products from the four reactions were pooled for each strain treatment and diluted 1:10. Ten microliters of this 1:10 dilution of cDNA was used for each rt-qPCR. cDNA for all treatments was stored at −80°C. From each cDNA reaction, 0.5 μl was used to verify the success of each reaction by means of PCR amplification and gel imaging of one of the target genes (Arr2).
We set out to study all of the known genes in the pathway, excluding genes that were obviously expressed ubiquitously, such as calmodulin. We created PCR primers for 22 genes. To create perfect-match primers that could be used for amplification in both species, we aligned the longest coding sequence of each D. melanogaster gene (26) with the unassembled draft sequence of the D. simulans genomes (Genome Sequencing Center, Washington University, St. Louis, MO; July 2004), using BLASTN (27). Primers were designed only for conserved regions between the two species by using Primer 3 software (http://fokker.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and then checked for amplification performance in silico with Amplify (28). Primers were then compared with the D. melanogaster and D. simulans genome sequences by using BLASTN to ensure that each primer had no significant homology to other regions of the genome. Primers were synthesized by Qiagen (Valencia, CA). Amplicons were between 125 and 318 bp long. Primer sequences are available in SI Table 2. While completing the manuscript, we again verified our primers with more advanced genome drafts of the D. simulans genome. We aligned the coding regions of the genes against the most completed genome of the strain w501 available at http://biology574.dhcp.unc.edu/∼cdjones/blastsim.html (accessed on April 25, 2006, and September 20, 2006). We found that all the primers used had perfect matches except Rh6 and Rdga, which had mismatches. Because rt-qPCR artifact seemed most likely to account for the apparent strong difference in expression of these genes between the species (data not shown), we excluded these genes in the study.
DNA controls for use in creating standard curves to determine absolute transcript abundances via rt-qPCR for each of the 20 genes were constructed as follows: PCR products for each gene were individually cloned into plasmids and transformed into Escherichia coli by using the pGEM-T Vector System (Promega). Transformed colonies of E. coli were transferred to LB media and allowed to grow for 24 h at 30°C in an orbital shaker, after which they were purified by using PCR purification columns from Qiagen. Incorporation of each amplicon was verified by restriction digestion and gel visualization. Purified plasmids were then linearized by using the restriction enzyme XmnI, which targets a single site in the vector. Next, the DNA concentration of linearized plasmid (nanograms per microliter) was determined by light absorbance, using a low-volume spectrophotometer with two to four replicate measurements per gene (NanoDrop, Wilmington, DE). The absolute number of plasmid copies per microliter was then calculated by determining the mass of each plasmid + amplicon for each gene, using the forumla massamplicon = total number of base pairs × nucleotide mass (660 g/mol of nucleotides). Dilutions (1:10) of linearized amplicon-incorporated plasmids were then used to construct standards for each gene. The mean range of standards across all genes was 1,270–12.7 billion molecules per 10 μl used in each rt-qPCR.
Sample and control cDNA for each gene was analyzed in 96-well plate format: one plate was used for each gene and included 78 sample wells (two replicates per sample, arranged randomly), two no-template control wells, and eight wells comprising the plasmid DNA standards. Sample template cDNA and rt-qPCR mix plus primer for 14 targets of the genes were aliquoted from 96-well plates by using a Beckman BioMek FX liquid handling robot (Beckman Coulter, Fullerton, CA). Eight genes were prepared manually. All DNA standards were aliquoted into 96-well plates by hand. rt-qPCRs were run on an MX 3000p rt-qPCR instrument (Stratagene, La Jolla, CA) by using the following cycling protocol: 15 min at 94°C, and 15 s at 94°C, 30 s at 55°C, and 30 s at 72°C for 45 cycles. The gene Rh4 showed extremely low cDNA abundance according to its amplification curve and was run de novo for 66 cycles. The threshold cycle for all genes was manually set at the cycle where log-linear amplification began. Melting-curve analysis was performed after completion of each rt-qPCR, to confirm the absence of primer dimers or the amplification of nonspecific DNA sequences. No primer dimers or nonspecific amplification was observed.
trpl Expression Difference in Heads.
During the course of the experiment, it was reported that TRPL was expressed not only in Drosophila eyes but also in the Malpighian tubules (29). We therefore tested whether the highly significant difference in TRPL expression was maintained if RNA from heads only was measured. We therefore raised flies as described in Fly Species and Strains for one D. melanogaster (Hikone R) and one D. simulans strain (S17). Heads were detached from the body by plunging tubes with flies into liquid nitrogen for 5 s. This process was repeated five times. The heads and body were then placed on a plastic dish floating in a liquid nitrogen bath. Heads were then handpicked by using a sterile pipette tip. All of the subsequent steps were the same as those performed for whole-flies extraction, except that RNA concentration was estimated by using a low-volume spectrophotometer. Consistent with whole-body results, trpl showed a significant up-regulation in D. simulans relative to D. melanogaster (SI Fig. 5).
The differential expression of trpl could result from a sequencing error in the D. melanogaster reference strain that would result in a mismatch in the region covered by the PCR primers. To verify that this was not the case, we sequenced the region covered by the primers in six strains of D. melanogaster and two strains of D. simulans and extracted the region from two additional strains from the D. simulans genome project. Genomic DNA was amplified by using the primers seqTRPL-F (5′-GCATCTTCGGGACGACCAGG-3′) and seqTRPL-R (5′-TAATGACGCTGTACGAGCCG-3′), and both strands of the purified PCR products were sequenced. We found that our PCR primers perfectly matched both species (SI Fig. 6).
Statistical Analysis.
Individual rt-qPCRs for a given treatment that failed were excluded, as were both replicate reactions if significant disagreement between replicates was observed; together, these comprised ≈1% of 1,360 reactions. All statistical calculations were carried out by using R (www.r-project.org) (30) with the exception of the mixed model ANOVA, which was carried out by using SAS, Version 9 (SAS Institute, Cary, NC). The log-transformed data were subjected to a linear model: expression = species + environment + species × environment, where replicates are nested within strains. We used the SAS procedure PROC MIXED to estimate the effect on gene expression of species, of environment, and potential species × environment interactions and their significance levels. The results are presented in SI Table 1. Polymorphism and divergence data were obtained by analyzing data that were gathered in the light with a standard ANOVA by extracting the mean sum of squares for lines and species in R (30).
Supplementary Material
Acknowledgments
We thank Pierre Fontanillas, Scott Rifkin, and Nadia Aubin-Horth for constructive comments and discussions; Pierre Capy for helpful comments on the manuscript; Hung-Tat Leung and William L. Pak for undertaking related experimental work; Rima Izem for help with SAS; Christian Daly for training on the robots and quantitative PCR equipment; and the Faculty of Arts and Sciences Center for Systems Biology for the use of their facility. C.R.L. is a Frank Knox Memorial Fellow and was supported by the Natural Sciences and Engineering Research Council, le Fonds Québécois de la Recheche sur la Nature et les Technologies, and the Québec Fondation Desjardins. This work was supported by National Institutes of Health Grant GM068465 (to D.L.H.).
Abbreviation
- rt-qPCR
real-time quantitative PCR.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611402104/DC1.
References
- 1.Bergmann S, Ihmels J, Barkai N. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell JR. Progress and Prospects in Evolutionary Biology: the Drosophila Model. New York: Oxford Univ Press; 1997. [Google Scholar]
- 3.Hardie RC, Raghu P. Nature. 2001;413:186–193. doi: 10.1038/35093002. [DOI] [PubMed] [Google Scholar]
- 4.Spady TC, Seehausen O, Loew ER, Jordan RC, Kocher TD, Carleton KL. Mol Biol Evol. 2005;22:1412–1422. doi: 10.1093/molbev/msi137. [DOI] [PubMed] [Google Scholar]
- 5.Dobzhansky T, Pavan C, Burla H. Ecology. 1950;31:36–43. [Google Scholar]
- 6.Hadler N. Genetics. 1964;50:1269–1277. doi: 10.1093/genetics/50.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markow T. Am Nat. 1979;114:884–892. [Google Scholar]
- 8.Capy P, Gibert P. Genetica. 2004;120:5–16. doi: 10.1023/b:gene.0000017626.41548.97. [DOI] [PubMed] [Google Scholar]
- 9.David JR, Allemand R, Capy P, Chakir M, Gibert P, Petavy G, Moreteau B. Genetica. 2004;120:151–163. doi: 10.1023/b:gene.0000017638.02813.5a. [DOI] [PubMed] [Google Scholar]
- 10.Parsons P. Q Rev Biol. 1975;50:151–169. doi: 10.1086/408437. [DOI] [PubMed] [Google Scholar]
- 11.Parsons P. Behav Genet. 1975;5:17–25. doi: 10.1007/BF01067578. [DOI] [PubMed] [Google Scholar]
- 12.Markow T, Smith W. Genetics. 1977;85:273–278. doi: 10.1093/genetics/85.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahner M, Frechter S, Da Silva N, Minke B, Paulsen R, Huber A. Neuron. 2002;34:83–93. doi: 10.1016/s0896-6273(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 14.Montell C. Annu Rev Cell Dev Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- 15.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie R. Neuron. 2002;34:3–5. doi: 10.1016/s0896-6273(02)00650-5. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Geng C, Leung HT, Hong YS, Strong LL, Schneuwly S, Pak WL. Proc Natl Acad Sci USA. 1999;96:13474–13479. doi: 10.1073/pnas.96.23.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilad Y, Oshlack A, Rifkin SA. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XS, Wang J, Hill WG. Genetics. 2004;166:597–610. doi: 10.1534/genetics.166.1.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M, Hill W. Evolution (Lawrence, Kans) 1986;40:915–935. doi: 10.1111/j.1558-5646.1986.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 21.Rifkin SA, Kim J, White KP. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh WP, Chu TM, Wolfinger RD, Gibson G. Genetics. 2003;165:747–757. doi: 10.1093/genetics/165.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montell C. J Physiol. 2005;567:45–51. doi: 10.1113/jphysiol.2005.092551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niemeyer BA, Suzuki E, Scott K, Jalink K, Zuker CS. Cell. 1996;85:651–659. doi: 10.1016/s0092-8674(00)81232-5. [DOI] [PubMed] [Google Scholar]
- 25.Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engels WR. Trends Biochem Sci. 1993;18:448–450. doi: 10.1016/0968-0004(93)90148-g. [DOI] [PubMed] [Google Scholar]
- 29.MacPherson MR, Pollock VP, Kean L, Southall TD, Giannakou ME, Broderick KE, Dow JA, Hardie RC, Davies SA. Genetics. 2005;169:1541–1552. doi: 10.1534/genetics.104.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ihaka R, Gentleman R. J Comput Graphic Stat. 1996;5:299–314. [Google Scholar]
- 31.Hardie RC. J Exp Biol. 2001;204:3403–3409. doi: 10.1242/jeb.204.20.3403. [DOI] [PubMed] [Google Scholar]
- 32.Lee SJ, Montell C. Neuron. 2001;32:1097–1106. doi: 10.1016/s0896-6273(01)00538-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.