Abstract
Signaling centers or organizers play a key role in axial patterning processes in animal embryogenesis. The function of most vertebrate organizers involves the activity of secreted antagonists of bone morphogenetic proteins (BMPs) such as Chordin or Noggin. Although BMP homologs have been isolated from many phyla, the evolutionary origin of the antagonistic BMP/Chordin system in organizer signaling is presently unknown. Here we describe a Chordin-like molecule (HyChdl) from Hydra that inhibits BMP activity in zebrafish embryos and acts in Hydra axis formation when new head organizers are formed during budding and regeneration. hychdl transcripts are also up-regulated in the head regeneration-deficient mutant strain reg-16. Accordingly, HyChdl has a function in organizer formation, but not in head differentiation. Our data indicate that the BMP/Chordin antagonism is a basic property of metazoan signaling centers that was invented in early metazoan evolution to set up axial polarity.
Keywords: axis formation, bone morphogenetic protein/Chordin, Cnidaria, regeneration, signaling
Localized signaling centers or organizers are widely used for the patterning of embryos or tissues during animal development. In general, organizers are able to generate polarity in surrounding tissue and to induce specific cell fates and cell behavior (1). One of the best-characterized examples is the amphibian embryonic organizer known as the Spemann–Mangold organizer (2). This organizer, localized at the dorsal blastopore lip, is able to induce the formation of a second body axis upon transplantation to the ventral side of a host embryo. The inductive capacity of the organizer is demonstrated by its ability to recruit host tissue into the second axis (3, 4). Functionally equivalent embryonic organizers are found in most vertebrates, including the node in amniotes and the embryonic shield in teleosts (5). The evolutionary origin of the embryonic organizer is unclear at present, but it was proposed that it is a vertebrate-specific invention (6).
There is evidence that the embryonic organizer is much older than commonly assumed. In the freshwater polyp Hydra, a member of the >500-million-year-old phylum Cnidaria, Browne (7) demonstrated the existence of an organizer-like activity already in 1909: Transplantation of a hypostome into the body column of a labeled host led to the formation of a second body axis including host-derived tissue (7, 8). The hypostome is the oral end of a cnidarian body axis and corresponds to the blastopore of a gastrula. However, whether the head organizer of Hydra represents the evolutionary origin of the vertebrate embryonic organizer or whether they arose independently is currently not known.
On the molecular level, Wnt/β-catenin signaling and the inhibition of bone morphogenetic protein (BMP) signaling by secreted antagonist such as Chordin and Noggin play a pivotal role in the establishment of the Spemann–Mangold organizer in Xenopus and the shield in zebrafish (4, 9, 10) as well as in the establishment of axial polarity in the mouse and sea urchin embryo (11–15). Several of these conserved genes have been described in cnidarians, i.e., members of the Wnt/β-catenin signaling pathway (16), and their expression pattern is consistent with a function in the Hydra head organizer. The role of BMP signaling is less clear, but analysis of a Hydra BMP homolog, hyBMP5-8b, indicated a role for BMP signaling in the specification of the aboral region of the polyp (17).
Here we report the isolation of a Chordin-like putative BMP antagonist (HyChdl). HyChdl is able to antagonize BMP signaling upon injection into zebrafish embryos. Strikingly, expression of hychdl is strongly up-regulated during budding and head regeneration, two processes in which new head organizers are formed. Our results reveal that the Hydra head organizer shares key molecular properties with the vertebrate embryonic organizer, suggesting that they have a common evolutionary origin.
Results
Hydra Has a Chordin-Like Cysteine-Rich Domain (CR Domain) Gene.
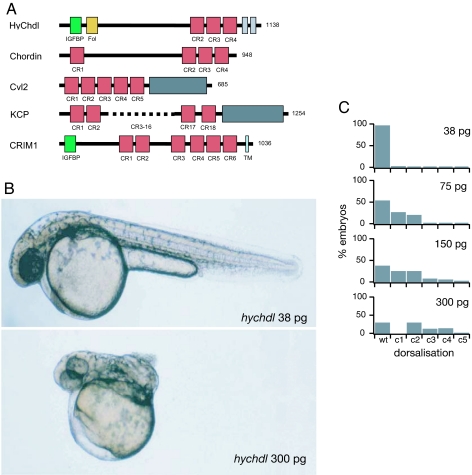
We isolated a 3.76-kb cDNA that contains a predicted ORF of 3,414 bp, encoding 1,138 aa. A functional signal peptide (amino acids 1–18; SignalP3.0, yeast signal peptide secretion assay) is followed by two CR domains (amino acids 33–85 and 107–174) in the N-terminal part of the deduced protein [see Fig. 1A and supporting information (SI) Fig. 5 and SI Methods]. These motifs are separated by a linker region (655 aa) from three Chordin-like CR domains (also called von Willebrand factor type C domain) in the C-terminal region (amino acids 822–877, 881–938, and 950–1009), and two additional, shorter CR domains (amino acids 1015–1047 and 1081–1116). Chordin orthologs from other species typically contain four BMP-binding CR domains, one at the N terminus and three in the C-terminal part (Fig. 1A). Although the overall length of the molecule (1,138 aa) and the presence of three CR domains in the C-terminal region reveal striking similarities to Chordin, the N-terminal CR domains differ from Chordin-type CR domains. BLAST and SMART analyses show that the first domain is most closely related to insulin-like growth factor binding protein (IGFBP) domains, but it also displays similarity to the BMP binding domain of Twisted gastrulation, another conserved regulator of BMP activity (SI Fig. 6A). The second domain is highly similar to follistatin (SI Fig. 6B), which consists of three so-called follistatin domains known to bind and inhibit BMPs as well as other TGF-β superfamily ligands. The three C-terminal CR domains of HyChdl all contain the two highly characteristic sequence motifs of the Chordin domain type: CX2CXC and CCX2C (SI Fig. 6C). Two additional, shorter CR domains that cannot be classified unambiguously follow the CR domains. However, the spacing of their cysteine residues corresponds very well to the first half of CR domains (SI Fig. 6C).
Fig. 1.
Structure and function of Hydra Chordin-like. (A) Comparison of the domain composition of HyChdl with CR domain proteins Chordin (58), Crossveinless 2 (59), CRIM-1 (60), and KCP (61). Red boxes indicate CR domains, gray boxes indicate von Willebrand factor type HC domain, green boxes indicate IGFBP domains, and yellow boxes indicates follistatin domain. Length of the proteins in amino acids is indicated on the right. (B and C) Microinjection of hychdl mRNA dorsalizes the zebrafish embryo. Shown are lateral views of live zebrafish embryos at 32 h after fertilization injected with 38 pg (B) and 300 pg (C) of hychdl mRNA, respectively. Embryos injected with low amounts of mRNA are phenotypically wild-type, whereas injection of high amounts of hychdl mRNA results in strong dorsalization (c4), indicated by curling up of the tail. (C) Embryos injected with increasing quantities of hychdl mRNA reveal a dosage-dependent dorsalization (c1–c4).
Taken together, the identified molecule contains one follistatin-type BMP binding domain in its N terminus and three Chordin-type BMP binding domains in its C terminus. Based on this domain architecture and the ability to inhibit BMP activity (see below), we termed the described molecule Hydra Chordin-like (HyChdl).
HyChdl Antagonizes BMP Activity.
Sequence analysis of hychdl suggested that it might act as a regulator of BMP activity. To support this idea experimentally we used the well established role of BMP signaling during early zebrafish development (Fig. 1 B and C). In this system, BMP activity is essential for the development of ventral structures, and hence overexpression of BMP antagonists leads to dose-dependent dorsalization of the embryos (18). The degree of dorsalization can be categorized by phenotypes ranging from loss of ventral tail fin tissue (c1, mild dorsalization) to complete curling up of the tail [c4, strong dorsalization (19)]. Injection of in vitro synthesized hychdl-mRNA into one- to two-cell zebrafish embryos led to dose-dependent dorsalization, similar to the described effects of zebrafish chordin mRNA (Fig. 1 B and C) (20), although the BMP antagonizing activity of hychdl mRNA was ≈20-fold weaker than that of zebrafish chordin mRNA (data not shown).
The N-terminal Chordin domain has been proposed to contribute significantly to the BMP inhibitory activity of vertebrate Chordin (21), however, in HyChdl this domain is replaced by two divergent CR domains. To analyze how these two domains affect the anti-BMP activity of HyChdl we used three constructs: one lacking only the IGFBP domain (hychdlΔIGFBP), one lacking the follistatin domain (hychdlΔFol), and one lacking both the IGFBP and follistatin domains (hychdlΔN). Whereas 150 pg of full-length hychdl mRNA per embryo led to dorsalization of 71.8% of the injected embryos (Table 1), only 54.6% of the embryos were dorsalized by the same amount of mRNA encoding HyChdlΔIGFBP and 52.2% by injection of hychdlΔFol mRNA (Table 1). Injection of 150 pg of hychdlΔN mRNA per embryo dorsalized 35.7% of the embryos (Table 1). These results show that HyChdl can act as a BMP antagonist in this heterologous assay and indicate that both the IGFBP and follistatin domains contribute to this activity.
Table 1.
Deletion of N-terminal domains reduces dorsalizing activity of HyChdl in zebrafish embryos
| mRNA | Picograms per embryo | n | Wild type, % | c1, % | c2, % | c3, % | c4, % |
|---|---|---|---|---|---|---|---|
| hychdl | 150 | 85 | 28.2 | 38.8 | 20 | 11.8 | 1.2 |
| hychdlΔIGFBP | 150 | 196 | 45.5 | 38.3 | 13.3 | 3.1 | 0 |
| hychdlΔFol | 150 | 134 | 47.8 | 32.8 | 16.4 | 3 | 0 |
| hychdlΔN | 150 | 221 | 64.3 | 28.9 | 6.8 | 0 | 0 |
Embryos injected with hychdl mRNA exhibit strong dorsalization (c1–c4), whereas injection with N-terminally truncated hychdl constructs lacking the IGFBP (hychdlΔIGFBP), the follistatin (hychdlΔFol), or both domains (hychdlΔN) reveal reduced dorsalized or wild-type phenotypes.
HyChdl Is Up-Regulated During Budding.
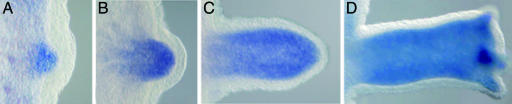
To study whether hychdl might be involved in patterning processes in adult polyps, we investigated its expression pattern by whole-mount in situ hybridization. First we analyzed hychdl expression during budding, Hydra's predominant way of reproduction. During this process, a new head organizer becomes established at the tip of the outgrowing bud (17, 22, 23). We observed a strong up-regulation of hychdl transcription commencing as soon as tissue evagination becomes visible (Fig. 2A). This up-regulation is maintained in the whole bud endoderm during the outgrowth of the bud (Fig. 2 B–D) and in young polyps (SI Fig. 7A). The high expression levels cease in steady-state adult polyps, with the pan-endodermal expression remaining higher in the head region than in the body column (SI Fig. 7 B and C).
Fig. 2.
hychdl expression during bud formation. (A) hychdl expression starts at bud stage 1–2 in the endoderm when the tissue layers begin to evaginate. hychdl expression is restricted to the endoderm at all following bud stages (B and C), and elevated expression levels along the entire bud continue until the bud has dropped off after stage 10–11 (D) (see SI Fig. 7A for comparison).
Up-Regulation of hychdl During Regeneration and de Novo Pattern Formation.
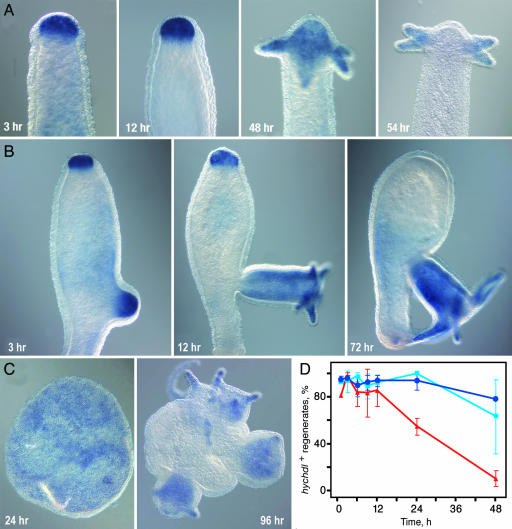
The regenerative capacity of Hydra allows the experimental induction of organizing centers. Upon bisection, wound healing occurs within 1 h and a new head organizer is established at the apical tip of the regenerate after 2–8 h (8, 24, 25). After 30–36 h tentacle buds and a new hypostome start to emerge, and after 48 h a complete head has been regenerated. Genes that are involved in the formation of the head organizer are expected to be up-regulated in the regenerating tip and indeed, we observed a strong up-regulation of hychdl transcription within 1 h after bisection at 80% body length (BL). This up-regulation is not a response to injury signals because RT-PCR experiments reveal that cutting alone never stimulates hychdl expression (SI Fig. 8). The enhanced expression was maintained for ≈48 h and was included in the endoderm of the newly formed tentacles (Fig. 3A). Shortly after completion of head regeneration (60 h), the expression level in the hypostome returned to basal levels, reestablishing the expression pattern of the adult polyp.
Fig. 3.
Expression of hychdl during regeneration and reaggregation. hychdl expression during regeneration in wild-type animals (A) and in the regeneration-deficient mutant strain reg-16 (B). Note the strong up-regulation of hychdl expression in apical tissue despite failure of head regeneration. (C) Expression of hychdl in reaggregates. Initially uniform endodermal staining becomes restricted to newly formed heads. (D) Kinetics of up-regulation of hychdl during regeneration. Percentage of hychdl-positive regenerates at different time points after bisection was analyzed in three independent experiments with >11 regenerates. No significant difference can be seen during head regeneration after bisection at 50% BL (light blue) and 80% BL (dark blue); red, foot regeneration (50% BL).
Tissue along the body column has a graded capacity to regenerate a head after decapitation with decreasing levels from head to foot (24–29). Therefore, the closer to the head polyps are bisected, the faster the regenerating tip acquires head organizer activity and the faster it regenerates a new head. Accordingly, up-regulation of genes that have an instructive function in head organizer formation should occur earlier after bisection close to the head than after bisection at more aboral levels, i.e., hywnt3a, which shows a temporal difference of ≈4 h (SI Fig. 9). When we compared the kinetics of up-regulation of hychdl after bisection at 80% and 50% BL, respectively, we could not detect a significant difference (Fig. 3D and SI Fig. 10). In both cases, up-regulation was evident in >90% of regenerates within 1 h and was maintained for 48 h. These results show that the up-regulation of hychdl does not strictly correlate with the acquisition of head activation potential of the regenerating tip.
To prove this notion, we analyzed the expression dynamics of hychdl in the regeneration-deficient Hydra strain reg-16. Animals of this mutant strain can form buds but fail to regenerate a head after decapitation or, in rare cases, regenerate a head with strong delay (30, 31). Despite the failure to regenerate a head, all reg-16 animals displayed a strong up-regulation of hychdl 6 h after decapitation (Fig. 3B). The elevated expression was maintained for 72–96 h if animals did not regenerate a head and ceased thereafter. In animals with delayed regeneration hychdl transcription returned to basal levels after completion of head formation regardless of the time point at which regeneration occurred. These data indicate that hychdl has no function in terminal head differentiation. Instead, the strong up-regulation in regenerates suggests that it acts in setting up the organizer, which is required for head differentiation. Further data on foot regeneration support this finding and show that hychdl was also transiently up-regulated in the endoderm of foot regenerates within 1 h after bisection and remained for 24–48 h (Fig. 3D and SI Fig. 11).
Finally, we analyzed hychdl expression in reaggregation experiments. Hydra reaggregates provide a unique de novo patterning assay (32). In this assay, body column tissue of several hundred polyps is dissociated into a single-cell suspension, and this suspension is then carefully centrifuged into small aggregates. In contrast to budding and regeneration, establishment of head organizers in this experiment starts from a situation in which all positional information has been lost. We found that after endoderm-ectoderm separation hychdl was strongly and uniformly expressed in the endodermal layer (Fig. 3C). After the emergence of new heads, expression of hychdl became restricted to the outgrowing body of the polyps. Thus, hychdl expression precedes the expression of hywnt3a and hybra1 that arise in discrete spots indicating the sites where new heads arise (16, 32).
Taken together, these results show that an up-regulation of hychdl during head regeneration occurs irrespective of whether a head is actually formed or not. It is therefore unlikely that hychdl has an instructive function in head differentiation. Instead, hychdl is a factor that is required in organizer formation itself.
Discussion
In this study we isolated a BMP antagonist belonging to the Chordin protein family, and we show that it acts during Hydra development in the establishment of organizing centers.
An Ancient Chordin Ortholog.
The overall domain architecture of this protein is similar to that of Chordin: three Chordin-like domains in the C-terminal and a cysteine-rich region in the N-terminal part, separated by ≈650 aa. This structure determines the function of Chordin proteins. The cysteine pattern and additional conserved residues clearly classify the three C-terminal cysteine-rich motifs as Chordin-type CR domains (SI Fig. 5C) that constitute BMP binding modules (21). Because the N-terminal cysteine-rich region of this protein contains an IGFBP and a follistatin domain (SI Fig. 5B) we named it Chordin-like (HyChdl). The functional relevance of the N-terminal Chordin motifs is yet not fully understood (21, 33). It is therefore of particular interest to see that both the IGFBP and the follistatin domain of the divergent N-terminal region of HyChdl contribute to its anti-BMP activity in the zebrafish microinjection experiments (Fig. 1D). In vertebrates, follistatin binds to BMPs and activin, a different TGF-β superfamily ligand (34–36). A direct binding of IGFBP domains to BMPs has not been reported, but IGFBP domains share similarities with the N-terminal CR domain of Twisted gastrulation (Tsg), which is required for binding of Tsg to BMPs (38, 39). Thus, the IGFBP domain of HyChdl could also contribute to its binding to BMPs. An effect of the IGFBP domain via an IGF-RTK-MAPK-Smad pathway, as described in vertebrates (40–42), is unlikely to occur in Hydra, because HySmad1 lacks the consensus phosphorylation sites required for this mode of regulation.
The finding that the Chordin ortholog from the sea anemone Nematostella exhibits the characteristic domain structure with four CR domains (43, 44) raises the question of whether the divergent N terminus of HyChdl represents a derived or ancestral feature. In the absence of data from a eumetazoan outgroup, e.g., sponges, this question remains unresolved. However, the Nematostella and the Hydra genomes do not contain any additional chordin-like genes, and our microinjection experiments suggest that HyChdl is indeed a Chordin ortholog.
Inhibition of BMP Signaling During Organizer Formation and Regeneration.
Our data reveal that hychdl has its primary function in organizer formation during budding and regeneration. During budding, a spot-like expression of hywnt3a marks the site of bud outgrowth (16). At a very similar time point, hychdl becomes up-regulated and a recently described BMP ortholog from Hydra, hybmp5-8b (17), becomes down-regulated. This leads to inversely related expression domains of hychdl and hybmp5-8b. Grafting experiments show that these changes in expression precede the acquisition of head activation capacity in the tip of the outgrowing tissue so that hyBMP5-8b expression is inversely correlated with organizer activity (17). However, the strong expression of hychdl throughout the developing bud suggests a function in addition to organizer formation: Because bud tissue displays an elevated level of neurogenesis (45) hychdl might play a role in this process. This would be in agreement with the neurogenic function described for chordin in vertebrates and invertebrates (46).
During head regeneration, regenerating stumps acquire head organizer properties within 2–8 h, depending on the axial level of bisection (8, 24–26, 29). After decapitation, hychdl is rapidly up-regulated within 1 h (Fig. 2A), similar to hyβ-catenin and hytcf in the Wnt pathway (16). However, the expression kinetics of hychdl during regeneration differs significantly from that of hywnt3a. The kinetics of hywnt3a up-regulation follows closely that of head regeneration (SI Fig. 9). The kinetics of hychdl up-regulation is also faster than that of hywnt3a and position-independent (Fig. 3D). This regeneration-specific up-regulation of hychdl occurs even in the absence of successful head regeneration in the mutant strain reg-16 (Fig. 3B) or during foot regeneration (SI Fig. 11). The transient up-regulation during foot regeneration might reflect a local release of inhibition observed after cutting, which results in a transient increase in head activation potential (24, 26, 27, 47, 48). This idea is supported by a similar transient up-regulation of several head-specific genes like hybra1, hytcf, and hyβcat during foot regeneration (49) (T.W.H. and B.H., unpublished observations). However, this activation is not a response to injury signals per se, as it has been shown for the up-regulation of hydkk1/2/4 after cutting (50). We therefore conclude that hychdl is required in the early establishment of organizer tissue, irrespective of the positional value the tissue will later adopt. This is further supported by the absence of significantly elevated expression levels of hychdl in the hypostome of adult polyps, suggesting that once the organizer is established hychdl is no longer required for its maintenance.
During head regeneration, hychdl and hybmp5-8b become simultaneously expressed at the site of regeneration (17). This is consistent with previous experimental and theoretical analyses showing a rapid up-regulation of tentacle markers (such as hybmp5-8b) before the expression of head markers in regenerates (32, 51). During budding, tentacle markers only appear when the organizer has formed. Because HyChdl can suppress BMP signaling by antagonizing BMP proteins, the transcriptional activation of hymbp5-8 during regeneration should not result in signaling activity.
An inhibition of BMP signaling has been recently found in other regenerative processes, e.g., in lens regeneration of newts. Grogg et al. (52) tested members of the BMP pathway for their ability to induce lens regeneration and found that inhibition of the BMP pathway by Chordin resulted in the induction of a lens from ventral explants, which normally cannot regenerate (52). Thus, their data reveal that lens regeneration can be achieved in noncompetent adult tissues after BMP inhibition. It will be therefore interesting to learn whether BMP inhibition is a more general mechanism in the recruitment of competent cells in other regenerative processes.
A Chordin-mediated inhibition of BMP signaling is also necessary during Xenopus development to establish the two dorsal signaling centers of the embryo. In cells located in the dorsal animal cap and marginal zone of the blastula (BCNE center), the action of β-catenin induces expression of chordin. In BMP gain of function experiments, both the BCNE and Spemann–Mangold organizers are completely suppressed (2, 4), and in antisense morpholino experiments it was shown that Chordin is absolutely required for the inductive activity of the Spemann–Mangold organizer (37).
In summary, we propose that BMP inhibition by HyChdl has a function during organizer formation to make cells competent, whereas canonical β-catenin Wnt signaling provides an instructive signal in this process.
Evolutionary Considerations.
Our results indicate that the BMP-Chordin antagonism is very old. The finding that HyChdl has its main function in setting up the Hydra organizer also suggests that it is primarily involved in the patterning of Hydra's oral–aboral body axis. This is also supported by recent data showing that Chordin also acts in Nematostella gastrulation. The Nematostella chordin ortholog nvchd is strongly expressed at the site of the blastopore, which transforms into the hypostome after gastrulation and corresponds to the head organizer in Hydra (43, 44). Similar to hychdl in Hydra, nvchd also displays a strong differential expression along the oral–aboral axis at later stages despite the fact that it additionally exhibits asymmetric expression along the directive axis (43, 44).
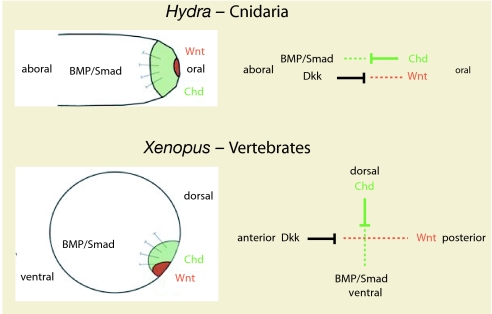
The other major signaling system that acts in cnidarian gastrulation and organizer formation is Wnt signaling (16, 54). We propose that both signaling systems, i.e., BMP/Chordin and Wnt/Dkk, represent main components of an ancient blastoporal signaling center that constitutes an archetypical organizer in metazoan evolution. This organizer is similar to the Spemann–Mangold organizer in vertebrates, where Wnt signaling also acts at the blastopore concomitantly with the BMP antagonist Chordin (Fig. 4).
Fig. 4.
Scheme illustrating the function of HyChdl in organizer formation. Cnidarians are characterized by two molecular vectors of antagonizing molecules, i.e., Wnt/Dkk and Chd/BMP, which act along the oral–aboral body axis of cnidarians; they give rise to the anterior–posterior and dorsoventral body axis of bilaterians.
Our results also have implications on the molecular mechanisms of axis evolution. There is general agreement that the BMP-Chordin antagonism patterns the dorsal-ventral axis in Bilaterians, whereas Wnt signaling and a cluster of hox genes patterns the anterior-posterior axis (53–55). Cnidaria are lacking an elaborated Hox system (55), and it is therefore striking that both axial patterning systems that are required for establishing the anterior–posterior and dorsoventral axes in bilaterians were already present in cnidarians, i.e., Wnt/Dkk and Chordin/BMP signaling. Here they control the formation of the main body axis, the oral–aboral axis. A clear assignment of these molecular systems to two perpendicular axes, however, occurs only in bilaterians. We therefore presume that, at the molecular level, the two axes of Bilateria did not evolve by “adding” an orthogonal axis to an already existing one, but rather by “splitting” one axis into two (Fig. 4).
Materials and Methods
Hydra Culture and Tissue Manipulation.
Hydra vulgaris strain Basel was used as wild-type strain and cultured as described previously (56). Additional regeneration experiments were carried out with the head regeneration-deficient strain Hydra magnipapillata reg-16 (30). For regeneration experiments, budless wild-type animals were bisected at 80% (close to the tentacles) and 50% (middle of the body column) BL, respectively, and transferred into fresh culture medium. Reg-16 polyps remain budless for a longer time than wild-type animals. To use animals at a defined developmental stage, reg-16 animals were bisected at bud stage 2 for regeneration experiments. For reaggregation experiments, heads and feet were removed from budless polyps and the body column tissue was mechanically dissociated in hypotonic dissociation medium. Aggregates were obtained by centrifugation of the cell suspension (32, 57).
Cloning of Hydra chordin-like.
An initial fragment containing the C-terminal part of hychdl was identified as a false positive in a yeast two-hybrid screen (16). Two rounds of 5′ RACE were performed to obtain the 5′ end. cDNA for 5′ RACE was synthesized with the SMART system (Clontech, Mountain View, CA) and gene-specific primers (CACACATGTAGTACATGGATCAGCTAGCC for 5′ RACE1; CAGGAAAGAATAATGTACCATTTACCCAC for 5′ RACE2). Antisense primers for 5′ RACE1 PCR were CTGAGAACTTGATATACAGCTG and CAAATTAGGTGCACATCTTGGC (nested). Antisense primers for 5′ RACE2 were GGCCGATGATAAATCTTCTTTGG and CTTTGGTTTCACATTCATTCATAGC. Obtained fragments were cloned into vector pGEM-T (Promega, Madison, WI) and sequenced on a ALF ExpressII automatic sequencer (Amersham Pharmacia, Little Chalfont, U.K.).
In Situ Hybridizations.
Whole-mount in situ hybridizations were done with a DIG-labeled hychdl-probe corresponding to the C-terminal part of the ORF (amino acids 779-1138) plus 3′ UTR, as described previously (16, 54). NBT/BCiP (Roche, Basel, Switzerland) was used as substrate.
Constructs for mRNA Injection, Preparation of mRNA, and Zebrafish Microinjection Experiments.
The complete ORF of hychdl was amplified from cDNA with Pfu DNA polymerase (Promega) and cloned into vector pCS2+. All constructs were linearized with NotI, and capped RNA was synthesized with the sp6 MessageMachine kit (Ambion, Austin, TX). Wild-type zebrafish embryos were injected at the one- to two-cell stage, and zebrafish culture was done under standard conditions (17).
Supplementary Material
Acknowledgments
We thank M. Hammerschmidt and T. Becker for support with zebrafish experiments, Kerstin Kuhn and Petra Snyder for help with the initial identification of the hychdl clone, and Herbert Steinbeisser for stimulating discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to B.H. and T.W.H.).
Abbreviations
- BL
body length
- BMP
bone morphogenetic protein
- IGFBP
insulin-like growth factor binding protein
- CR domain
cysteine-rich domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0604501104/DC1.
References
- 1.Gerhart J. Int J Dev Biol. 2001;45:133–153. [PubMed] [Google Scholar]
- 2.De Robertis EM. Nat Rev Mol Cell Biol. 2006;7:296–302. doi: 10.1038/nrm1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spemann H, Mangold H. Roux's Arch Entw Mech Org. 1924;100:599–638. [Google Scholar]
- 4.De Robertis EM, Kuroda H. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern CD. Gastrulation: From Cells to Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 6.Kourakis MJ, Smith WC. Trends Genet. 2005;21:506–510. doi: 10.1016/j.tig.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Browne EN. J Exp Zool. 1909;7:1–37. [Google Scholar]
- 8.Broun M, Bode HR. Development (Cambridge, UK) 2002;129:875–884. doi: 10.1242/dev.129.4.875. [DOI] [PubMed] [Google Scholar]
- 9.Niehrs C. Dev Cell. 2004;6:453–454. doi: 10.1016/s1534-5807(04)00102-9. [DOI] [PubMed] [Google Scholar]
- 10.Schier AF, Talbot WS. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 12.Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. J Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wikramanayake AH, Huang L, Klein WH. Proc Natl Acad Sci USA. 1998;95:9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Development (Cambridge, UK) 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 15.Kauffman JS, Raff RA. Dev Genes Evol. 2003;213:612–624. doi: 10.1007/s00427-003-0365-1. [DOI] [PubMed] [Google Scholar]
- 16.Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. Nature. 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 17.Reinhardt B, Broun M, Blitz IL, Bode HR. Dev Biol. 2004;267:43–59. doi: 10.1016/j.ydbio.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 18.Hammerschmidt M, Mullins MC. Results Probl Cell Differ. 2002;40:72–95. doi: 10.1007/978-3-540-46041-1_5. [DOI] [PubMed] [Google Scholar]
- 19.Kishimoto Y, Lee KH, Zon L, Hammerschmidt M, Schulte-Merker S. Development (Cambridge, UK) 1997;124:4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- 20.Miller-Bertoglio VE, Fisher S, Sanchez A, Mullins MC, Halpern ME. Dev Biol. 1997;192:537–550. doi: 10.1006/dbio.1997.8788. [DOI] [PubMed] [Google Scholar]
- 21.Larrain J, Bachiller D, Lu B, Agius E, Piccolo S, De Robertis EM. Development (Cambridge, UK) 2000;127:821–830. doi: 10.1242/dev.127.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broun M, Sokol S, Bode HR. Development (Cambridge, UK) 1999;126:5245–5254. doi: 10.1242/dev.126.23.5245. [DOI] [PubMed] [Google Scholar]
- 23.Li HP, Yao T. J Exp Biol. 1945;22:155–160. [Google Scholar]
- 24.MacWilliams HK. Dev Biol. 1983;96:239–257. doi: 10.1016/0012-1606(83)90325-1. [DOI] [PubMed] [Google Scholar]
- 25.Bode HR. Dev Dyn. 2003;226:225–236. doi: 10.1002/dvdy.10225. [DOI] [PubMed] [Google Scholar]
- 26.MacWilliams HK. Dev Biol. 1983;96:217–238. doi: 10.1016/0012-1606(83)90324-x. [DOI] [PubMed] [Google Scholar]
- 27.MacWilliams HK. J Theor Biol. 1982;99:681–703. doi: 10.1016/0022-5193(82)90194-1. [DOI] [PubMed] [Google Scholar]
- 28.Webster G, Wolpert L. J Embryol Exp Morphol. 1966;16:91–104. [PubMed] [Google Scholar]
- 29.Wilby OK, Webster G. J Embryol Exp Morphol. 1970;24:595–613. [PubMed] [Google Scholar]
- 30.Achermann J, Sugiyama T. Dev Biol. 1985;107:13–27. doi: 10.1016/0012-1606(85)90371-9. [DOI] [PubMed] [Google Scholar]
- 31.Kobatake E, Sugiyama T. Development (Cambridge, UK) 1989;105:521–528. doi: 10.1242/dev.105.3.521. [DOI] [PubMed] [Google Scholar]
- 32.Technau U, Cramer von Laue C, Rentzsch F, Luft S, Hobmayer B, Bode HR, Holstein TW. Proc Natl Acad Sci USA. 2000;97:12127–12131. doi: 10.1073/pnas.97.22.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu K, Kang KH, Heine P, Pyati U, Srinivasan S, Biehs B, Kimelman D, Bier E. Genetics. 2004;166:1323–1336. doi: 10.1534/genetics.166.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, Pillemer G, Steinbeisser H, Blum M. Mech Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- 35.Iemura S, Yamamoto TS, Takagi C, Uchiyama H, Natsume T, Shimasaki S, Sugino H, Ueno N. Proc Natl Acad Sci USA. 1998;95:9337–9342. doi: 10.1073/pnas.95.16.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura T, Takio K, Eto Y, Shibai H, Titani K, Sugino H. Science. 1990;247:836–838. doi: 10.1126/science.2106159. [DOI] [PubMed] [Google Scholar]
- 37.Oelgeschlager M, Kuroda H, Reversade B, De Robertis EM. Dev Cell. 2003;4:219–230. doi: 10.1016/s1534-5807(02)00404-5. [DOI] [PubMed] [Google Scholar]
- 38.Oelgeschlager M, Larrain J, Geissert D, De Robertis EM. Nature. 2000;405:757–763. doi: 10.1038/35015500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oelgeschlager M, Reversade B, Larrain J, Little S, Mullins MC, De Robertis EM. Development (Cambridge, UK) 2003;130:4047–4056. doi: 10.1242/dev.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pera EM, Wessely O, Li SY, De Robertis EM. Dev Cell. 2001;1:655–665. doi: 10.1016/s1534-5807(01)00069-7. [DOI] [PubMed] [Google Scholar]
- 41.Pera EM, Ikeda A, Eivers E, De Robertis EM. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eivers E, McCarthy K, Glynn C, Nolan CM, Byrnes L. Int J Dev Biol. 2004;48:1131–1140. doi: 10.1387/ijdb.041913ee. [DOI] [PubMed] [Google Scholar]
- 43.Rentzsch F, Anton R, Saina M, Hammerschmidt M, Holstein TW, Technau U. Dev Biol. 2006;296:375–387. doi: 10.1016/j.ydbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Matus DQ, Thomsen GH, Martindale MQ. Curr Biol. 2006;16:499–505. doi: 10.1016/j.cub.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 45.Berking S. J Embryol Exp Morphol. 1980;60:373–387. [PubMed] [Google Scholar]
- 46.Sasai Y. Int J Dev Biol. 2001;45:321–326. [PubMed] [Google Scholar]
- 47.Meinhardt H, Gierer A. J Theor Biol. 1980;85:429–450. doi: 10.1016/0022-5193(80)90318-5. [DOI] [PubMed] [Google Scholar]
- 48.Meinhardt H. BioEssays. 1994;16:627–632. doi: 10.1002/bies.950160906. [DOI] [PubMed] [Google Scholar]
- 49.Technau U, Bode HR. Development (Cambridge, UK) 1999;126:999–1010. doi: 10.1242/dev.126.5.999. [DOI] [PubMed] [Google Scholar]
- 50.Guder C, Pinho S, Nacak TG, Schmidt HA, Hobmayer B, Niehrs C, Holstein TW. Development (Cambridge, UK) 2006;133:901–911. doi: 10.1242/dev.02265. [DOI] [PubMed] [Google Scholar]
- 51.Bode PM, Awad TA, Koizumi O, Nakashima Y, Grimmelikhuijzen CJ, Bode HR. Development (Cambridge, UK) 1988;102:223–235. doi: 10.1242/dev.102.1.223. [DOI] [PubMed] [Google Scholar]
- 52.Grogg MW, Call MK, Okamoto M, Vergara MN, Del Rio-Tsonis K, Tsonis PA. Nature. 2005;438:858–862. doi: 10.1038/nature04175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Science. 2004;304:1335–1337. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- 54.Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 55.Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, Jensen MF, Zhu B, Steele R, Technau U. Nature. 2006;442:684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- 56.Technau U, Holstein TW. Dev Biol. 1996;177:599–615. doi: 10.1006/dbio.1996.0189. [DOI] [PubMed] [Google Scholar]
- 57.Gierer A, Berking S, Bode H, David CN, Flick K, Hansmann G, Schaller H, Trenkner E. Nat New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- 58.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coles E, Christiansen J, Economou A, Bronner-Fraser M, Wilkinson DG. Development (Cambridge, UK) 2004;131:5309–5317. doi: 10.1242/dev.01419. [DOI] [PubMed] [Google Scholar]
- 60.Coffinier C, Tran U, Larrain J, De Robertis EM. Mech Dev. 2001;100:119–122. doi: 10.1016/s0925-4773(00)00507-4. [DOI] [PubMed] [Google Scholar]
- 61.Matsui M, Mizuseki K, Nakatani J, Nakanishi S, Sasai Y. Proc Natl Acad Sci USA. 2000;97:5291–5296. doi: 10.1073/pnas.090020497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.