Abstract
Germ-line transformation via transposable elements is a powerful tool to study gene function in Drosophila melanogaster. However, some inherent characteristics of transposon-mediated transgenesis limit its use for transgene analysis. Here, we circumvent these limitations by optimizing a φC31-based integration system. We generated a collection of lines with precisely mapped attP sites that allow the insertion of transgenes into many different predetermined intergenic locations throughout the fly genome. By using regulatory elements of the nanos and vasa genes, we established endogenous sources of the φC31 integrase, eliminating the difficulties of coinjecting integrase mRNA and raising the transformation efficiency. Moreover, to discriminate between specific and rare nonspecific integration events, a white gene-based reconstitution system was generated that enables visual selection for precise attP targeting. Finally, we demonstrate that our chromosomal attP sites can be modified in situ, extending their scope while retaining their properties as landing sites. The efficiency, ease-of-use, and versatility obtained here with the φC31-based integration system represents an important advance in transgenesis and opens up the possibility of systematic, high-throughput screening of large cDNA sets and regulatory elements.
Keywords: attP landing sites, germ-line transformation, site-specific integration
A major goal in the present era of genomics is to identify and functionally characterize all genes relevant to a specific pathway or biological process. With its powerful repertoire of genetic tools, the multicellular model organism Drosophila melanogaster has played an eminent role in this endeavor (1). One method to identify relevant genes is to perform chemical mutagenesis screens of various kinds. Another very fruitful approach in Drosophila has been the use of P-element-mediated germ-line transformation (2, 3), especially when combined with tools such as the Gal4/UAS expression system (4) or when it is used for insertional mutagenesis (5, 6). One characteristic of P-elements is their random integration behavior. Although this “randomness” is advantageous for generating mutations and deletions, it is generally not ideal for transgene analysis. The random integration of P-elements necessitates considerable effort to map insertions. Genomic position effects complicate the analysis of transgenes and render precise structure/function analyses nearly impossible. A further shortcoming of the P-element system is its relatively moderate transformation efficiency, a significant hurdle to any large-scale transgenesis effort.
Strategies have been developed to circumvent the problem of randomness by targeted integration systems in Drosophila, which are generally based on the FLP and Cre recombinases (7–9, 32). Such techniques permit precise targeting to genomic landing sites but are still handicapped by transformation rates that are, at best, moderately higher than those achieved with the P-element system (8). Furthermore, especially for FLP/FRT, these recombinase systems are often reserved for applications other than targeted integration, such as mosaic analysis (10), and are thus not suited to be used concomitantly for transgene integration.
Recently, another genome integration method has been developed, based on the site-specific φC31 integrase (11), and has subsequently been applied to Drosophila (12). The bacteriophage φC31 encodes a serine integrase that mediates sequence-directed recombination between a bacterial attachment site (attB) and a phage attachment site (attP) (13). Apart from the site-specificity, another feature of the φC31 system is that the integrase solely mediates integration (13), which distinguishes it from most other commonly used systems, such as Cre/loxP or FLP/FRT, where the recombinase can catalyze both the integration and the excision reactions.
Here we set out to bring the φC31 system to a level of efficiency, convenience, and expandability that renders it suitable for large-scale transgenesis approaches. In particular, we sought to make the system more robust by improving the delivery of the φC31 integrase and to create a library of well characterized, highly efficient landing sites throughout the four major chromosomes of the Drosophila genome. These landing sites were designed so as to not interfere with commonly used markers and transposon systems, and to be manipulatable in vivo by the Cre/loxP and attP/attB systems. Different “endogenous” φC31 integrase sources were generated and optimized to overcome the need of coinjecting capped, in vitro synthesized integrase mRNA. The combination of these tools allows for convenient and efficient site-specific germ-line transformation. Finally, we present the development of an integration system that utilizes an immediate visible readout for specific attP targeting and therefore should permit rapid selection for precise integration events.
Results
Design of a Versatile attP Landing Site.
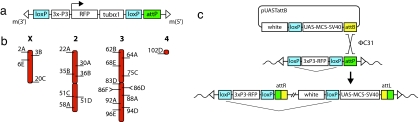
A landing site (referred to as M{3xP3-RFPattP}; Fig. 1a) was constructed with the following features. (i) It contains an attP site, which serves as the docking site for any incoming attB-containing plasmid. (ii) As a marker, we chose the red fluorescent protein (RFP) driven by the artificial 3xP3 promoter, leading to strong RFP expression in the eyes (14). RFP was chosen so as to not preclude the later use of the widespread markers yellow+ and white+. (iii) The 3xP3-RFP cassette is flanked by loxP sites, allowing for the elimination of this marker via Cre recombinase-mediated excision. (iv) The 3xP3-RFPattP construct is flanked by inverted repeats of the Mos1 mariner element to allow mariner-mediated transgenesis, making the landing site refractory to the P-element and piggyBac transposases, thus permitting unrestricted use of multipurpose transposon systems in conjunction with this landing site.
Fig. 1.
φC31-mediated integration into attP target sites. (a) Design of the pM{3xP3-RFPattP} landing site construct. This construct contains an attP docking site of 221 bp and a DsRFP marker gene attached to the tubulinα1 3′ UTR element. RFP expression is driven by the 3xP3 promoter, and the marker cassette is flanked by loxP sites. The landing site construct is bounded by mariner inverted repeats (inverted repeats, open triangles). (b) Diagram of the four major chromosomes indicating the cytological positions of the 25 ZH-attP landing sites that are located intergenically. Two lines exist at both positions 86D and 86F. (c) Integration mechanism of the pUASTattB vector into attP landing sites. The pUASTattB plasmid contains a 285-bp attB fragment, the white+ selectable marker, a UAS-MCS-SV40 cassette, and a single loxP site. The φC31 integrase mediates recombination between attB and attP sites, resulting in the integration of pUASTattB into the landing site, thereby creating the two hybrid sites attL and attR, which are refractory to the φC31 integrase. The final configuration at the landing site is directed by the orientation of the attB and attP elements. The loxP sites allow elimination of intervening sequences before or after integration of pUASTattB (indicated with flat arrowheads). In all of the injection experiments where we used the pUASTattB vector, it contained a lacZ reporter (not indicated). (The parallel diagonal lines indicate the presence of the plasmid backbone.) RFP, red fluorescence protein; UAS, upstream activating sequence; MCS, multiple cloning site.
A Large Collection of attP Lines.
Upon mariner-mediated germ-line transformation, 68 independent lines, designated as “ZH-attP” lines, were established (by screening the F1 generation for eye-specific RFP expression). Genomic DNA samples of these lines were subjected to an inverse PCR assay to determine the exact genomic location of their transgene. BLAST-based comparisons of the inverse PCR sequences with the D. melanogaster genome sequence (Ensembl v41 database) revealed an unambiguous location in 64 cases, while four lines yielded several matches, indicating that their transgene integrated into repetitive elements. Of the 64 precisely mapped lines, 25 lines were chosen for being pursued further because they fulfilled two criteria: these 25 landing sites are homozygous viable and are intergenically located [Fig. 1b and supporting information (SI) Table 4]. The accumulating genomics and proteomics data, however, may alter the assessment of the intergenic positioning and may therefore modify the number of these candidate lines.
Germ-Line-Specific Integrase Sources.
A few of the ZH-attP lines were then selected for germ-line transformation with a specifically generated pUASTattB plasmid (Fig. 1c) carrying a lacZ reporter and a white+ marker gene. This plasmid was coinjected with capped mRNA encoding the φC31 integrase. Adults obtained after injection were individually crossed with y w animals and their offspring were screened for white+ expression. We define the frequency of transgenesis (Ftrans) as the fraction of fertile crosses that gave at least one white+ offspring. The ZH-attP-86Fb showed the highest Ftrans (25%), which exceeded those of the previously described landing site attP2 (12), for which we have never reached values above 7% (data not shown). It also became clear, however, that the variable quality and stability of the capped, coinjected integrase mRNA poses a problem for reliably determining and comparing the Ftrans values of our attP lines and possibly also limit the efficiency of attB transgene integration.
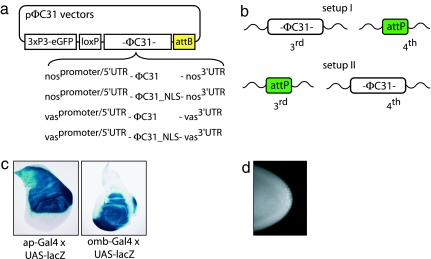
To circumvent this problem and to facilitate the injection process, we sought to generate an “endogenous” source of the φC31 integrase. To do this, we created a P-element that can express a φC31 integrase under the control of the nanos regulatory elements. This construct also attaches a nuclear localization sequence (NLS) to the C terminus of the integrase, because previous reports in mammalian Chinese hamster ovary cells suggest that the adding of a C-terminal NLS substantially enhances integration frequency (15). We obtained four independent P-element insertions and selected an insert on the X chromosome for further studies. Using this line, we were able to obtain substantially better integration frequencies than with the coinjected φC31 integrase mRNA used previously [integration rates were up to 30% for the landing site attP2 of Groth et al. (12)]. To optimize the integrase source further, we decided to modify various elements in the basic construct and determine how they affect integration frequencies. Two elements that were of particular interest to us were the promoters and the NLS. Therefore, constructs were assembled that express the φC31 integrase (+/− NLS) under the control of the regulatory elements of either the vasa or the nanos gene, including the promoter, 5′ UTR, and 3′ UTR sequences of these genes (Fig. 2a). Regulatory elements of these genes are known to cause germ cell-specific expression, either by specifically directing zygotic transcription in these cells (vasa) (16) or by spatially regulating translation and stability of maternal mRNA, which leads to expression at the posterior of the embryo where germ cells form (nanos) (17–23).
Fig. 2.
Establishment of germ-line-specific φC31 integrase lines. (a) nanos- and vasa-φC31 constructs used to generate transgenic integrase lines. The φC31 ORF is flanked by either nanos (nos) or vasa (vas) regulatory elements, including promoters, 5′ UTR, and 3′ UTR. NLS-tagged φC31 integrase versions were also tested. All constructs contain a 3xP3-EGFP marker cassette, a loxP site, and an attB site for site-specific integration into attP sites. (b) Schematic of the two different setups used to compare the four different integrase versions depicted in a. Set-up I contains the integrase constructs in an attP site on the third chromosome (3R 86F) in combination with a free attP site on the fourth (102D); set-up II represents the reverse situation. The animals used were homozygous for the depicted situation. (c) Detection of β-galactosidase activity in third-instar wing discs of the transgenic ZH-attP-86Fb-lacZ line (here, the pUAS-lacZattB plasmid was introduced via coinjection of φC31 integrase mRNA). The observed patterns correspond to the apterous (Left) and omb (Right) expression domains. (d) Antibody staining against φC31 integrase. An embryo at stage 4–5 shows enhanced staining in the posterior pole cells, indicating accumulation of φC31 integrase protein in these cells. The depicted embryo is homozygous for vas-φC31.
To compare these four constructs in a quantitative manner, it was imperative that they all be expressed from the same chromosomal context. We therefore made use of the attP/attB system. An attB site was introduced into the integrase constructs, allowing us to bring each of them to an identical position in the genome by using the same attP landing site for germ-line transformation. The constructs were tested in two locations: first at cytological position 86F of the third chromosome by employing line ZH-attP-86Fb (set-up I), and then at cytological position 102D of the fourth chromosome via ZH-attP-102D (set-up II) (Fig. 1b). For set-up I, the four integrants on the third chromosome were made homozygous for a free attP landing site on the fourth chromosome (from ZH-attP-102D), whereas for set-up II, the four integrase insertions on the fourth chromosome were made homozygous for a free attP site on the third chromosome (from ZH-attP-86Fb) (Fig. 2b). Antibody stainings were performed to confirm the specific expression of integrase in the posterior pole cells of lines transgenic for φC31 integrase (Fig. 2d and data not shown).
All resulting eight lines were then injected with pUAS-lacZattB and the frequencies of integration were scored (Table 1). The Ftrans values ranged from 32% to 43% in set-up I and from 16% to 55% in set-up II.
Table 1.
Germ-line transformation in flies transgenic for attP and various φC31 integrase constructs
| Line | Integrase construct | DNA injected pUAS-lacZattB, ng/μl | Giving w+ progeny |
|
|---|---|---|---|---|
| w+/total no. | % | |||
| Set-up I | ||||
| nos-φ-zh86Fb/attP-zh102D | nos-φC31-nos | 215 | 28/70 | 40 |
| nos-φNLS-zh86Fb/attP-zh102D | nos-φC31NLS-nos | 215 | 52/122 | 43 |
| vas-φ-zh86Fb/attP-zh102D | vas-φC31-vas | 215 | 29/69 | 42 |
| vas-φNLS-zh86Fb/attP-zh102D | vas-φC31NLS-vas | 215 | 25/77 | 32 |
| vas-φ-zh86Fb/attP-zh102D | vas-φC31-vas | 800 | 32/80 | 40 |
| Set-up II | ||||
| attP-zh86Fb/nos-φ-zh102D | nos-φC31-nos | 215 | 16/103 | 16 |
| attP-zh86Fb/nos-φNLS-zh102D | nos-φC31NLS-nos | 215 | 49/148 | 33 |
| attP-zh86Fb/vas-φ-zh102D | vas-φC31-vas | 215 | 47/93 | 51 |
| attP-zh86Fb/vas-φNLS-zh102D | vas-φC31NLS-vas | 215 | 16/49 | 33 |
| attP-zh86Fb/vas-φ-zh102D | vas-φC31-vas | 800 | 36/66 | 55 |
The lines injected were homozygous for both an attP site and a φ C31 integrase construct. G0 adults (male and females) obtained after injection were individually crossed with y w animals and progeny were screened for white+ expression. Line designations indicate the origin of the integrase constructs and the free attP site (e.g., nos-φ-zh86Fb/attP-zh102D:nos-φC31 construct is located in the attP site of line ZH-attP-86Fb, and the free attP site is derived from line ZH-attP-102D). Because single G0 females generally exhibited a low fertility and transgenesis rate compared with males, in all subsequent transformation experiments two injected females were used for one y w outcross. However, although we still tested females for transgenic frequencies, we present in the Tables 2 and 3 only the transgenic values obtained from G0 males. The percentages indicate the fraction of all the fertile G0 adults that gave white+ offspring.
A number of observations were made in these experiments. (i) The vasa-driven integrase transgenes were generally more efficient in mediating germ line transformation than the nanos constructs. (ii) Although the addition of the NLS did somewhat increase the efficiency of nanos-φC31, it had an adverse effect on the activity of the vasa-φC31 integrase. (iii) Set-up II led to higher integration rates than set-up I. (iv) The concentration of the injected plasmid did not critically affect the transgenic frequency (215 versus 800 ng/μl; Table 1). In summary, the above-described observations favor the vasa-φC31 construct for achieving highest transgenic frequencies.
To create an integrase source that could be quickly removed from a transgenic line, we inserted the vasa-φC31 construct into each of our four candidate attP sites on the X chromosome (Fig. 1b). The resulting lines were then tested by comparing their efficiency in catalyzing an insertion reaction at the attP site of ZH-attP-86Fb. The Ftrans values varied between 29% and 50% (Table 2; these values were derived from injections into embryos heterozygous for the attP landing site and hemizygous for the vasa-φC31 construct and therefore probably underestimate those expected from stocks homozygous for the attP site). These results indicate that vas-φC31-zh2A performs best.
Table 2.
Germ-line transformation with X-linked vas-φC31 integrase lines
| Integrase line | attP position | DNA injected pUAS-lacZattB, ng/μl | G0 males giving w+ progeny |
|
|---|---|---|---|---|
| w+/total no. | % | |||
| vas-φ-zh2A | X-2A | 215 | 72/144 | 50 |
| vas-φ-zh6E | X-6E | 215 | 23/56 | 41 |
| vas-φ-zh20C | X-20C | 215 | 27/67 | 40 |
| vas-φ-zh3B | X-3B | 215 | 15/51 | 29 |
Injected embryos were hemizygous for the vas-φC31 integrase and heterozygous for the attP site at 3R 86F (derived from ZH-attP-86Fb), where the integrase was provided through the parental females, and the attP site through the parental males. The numbers for the line vas-φ-zh2A are based on two injection experiments. The given transgenic frequencies are solely derived from the offspring of G0 males. X-linked integrase lines offer the advantage that the integrase can readily be eliminated by using only G0 males for the first outcross and then selecting white+ F1 males: these are always devoid of the integrase transgene.
We recently also generated a φC31 integrase construct in which the coding region was adapted to the D. melanogaster codon usage. This codon-optimized transgene, dφC31, differs in 172 nucleotides from the phage φC31 integrase ORF and further increased transformation rates (69% for G0 male outcrosses in a situation equivalent to set-up II; see above). This construct will be introduced into ZH-attP-2A to generate an X-linked vas-dφC31 line.
Germ Cell-Specific φC31 Integrase Has No Adverse Effects on Chromosomal Stability.
A possible concern about using constitutive expression of an integrase is the occurrence of chromosomal instability, especially in the light of a recent report that human fibroblasts stably transfected with φC31 integrase exhibit chromosomal aberrations (24). To investigate whether such φC31 integrase-mediated effects occur in Drosophila, we examined two lines, nos-φNLS-zh86Fb/attP-zh102D and attP-zh86Fb/vas-φ-zh102D; they were chosen because they represent the most efficient combinations of an autosomal nanos or vasa-driven integrase and a free attP site with respect to integration frequency (see Table 1). The concomitant presence of a free attP site in these lines not only resembles what one might use in practice, but might also increase the potential incidence of a chromosomal rearrangement. From each of the two lines, 10 randomly selected males were crossed with y w females, and the resulting larvae were subjected to a chromosomal analysis. For each cross, the salivary glands of six to eight F1 larvae were dissected and their polytene chromosomes were analyzed. In none of the 136 examined preparations did we observe any abnormal chromosomal configurations (SI Fig. 4 c and d). As a control we used a chromosomal deletion (of ≈500 kb) and various inversions that were readily detected in each case (SI Fig. 4 a and b). The two tested lines had been propagated for more than 7 months before this analysis. Together, these results indicate that, at least for the euchromatic portion of the genome, the mutagenic effects of the φC31 integrase are not a serious concern in Drosophila.
Accessibility, Transgene Expression, and Position Effects of Various attP Lines.
We next characterized 16 of the 25 candidate attP lines with respect to their integration frequencies and expression behavior. Unlike the set-ups shown in Table 1, where stocks were generated that are doubly homozygous for an attP site and an integrase construct, we directly crossed males homozygous or hemizygous for the attP site with females homozygous for the vasa-φC31 integrase located at 102D on the fourth chromosome. The resulting doubly heterozygous embryos were injected with pUAS-lacZattB. The transgenic frequencies ranged from 28% (ZH-attP-86Fa) to 60% (ZH-attP-51D) (Table 3). These numbers likely underestimate the Ftrans values expected for doubly homozygous set-ups. This can be concluded from line ZH-attP-86Fb, where the Ftrans values were determined for both heterozygotes (43% for G0 male outcrosses; see Table 3) and homozygotes (61% for G0 male outcrosses).
Table 3.
Comparison of targeting into various attP sites
| ZH-attP line | attP position | G0 males giving w+ progeny |
Eye color* | |
|---|---|---|---|---|
| w+/total no. | % | |||
| ZH-attP-2A | X-2A | 0/77 | 0 | Orange |
| ZH-attP-22A | 2L-22A | 22/56 | 39 | Light orange |
| ZH-attP-30A† | 2L-30A | 23/56 | 41 | Orange |
| ZH-attP-35B | 2L-35B | 21/53 | 40 | Light orange |
| ZH-attP-36B | 2L-36B | 26/46 | 57 | Orange |
| ZH-attP-51C | 2R-51C | 27/64 | 42 | Light orange |
| ZH-attP-51D | 2R-51D | 43/72 | 60 | Red |
| ZH-attP-58A | 2R-58A | 25/51 | 49 | Light orange |
| ZH-attP-62B | 3L-62B | 26/67 | 39 | Light orange |
| ZH-attP-64A | 3L-64A | 22/41 | 54 | Orange |
| ZH-attP-68E | 3L-68E | 32/85 | 37 | Light orange |
| ZH-attP-75C | 3L-75C | 19/52 | 37 | Light orange |
| ZH-attP-86Fa | 3R-86F | 13/46 | 28 | Red |
| ZH-attP-86Fb | 3R-86F | 40/92 | 43 | Orange |
| ZH-attP-96E | 3R-96E | 28/64 | 44 | Orange |
| ZH-attP-102D | 4–102D | 39/86 | 45 | Red |
Females homozygous for the vas-φ C31 construct, located at 102D on the fourth chromosome, were crossed to males homozygous (or hemizygous) for an indicated attP site, and the offspring were injected with pUAS-lacZattB (215 ng/μl). Because line ZH-attP-2A has the attP site on the X chromosome, G0 males are devoid of an attP site and therefore are not expected to give rise to transgenic offspring.
*Eye colors were determined 3 days after eclosion (heterozygous state). All flies transgenic for the injected plasmid reveal a reddish punctate color at the inner side of each of the three ocelli; this additional phenotype served as a convenient confirmation for transgenic flies in the cases of light orange eye color.
†Landing site ZH-attP-30A, though homozygous viable on its own, turned out to be homozygous lethal upon integration of UAS-lacZattB.
The transgenic lacZ flies from these injection experiments were then crossed to apterous (ap)- and omb-Gal4 driver lines. To detect β-galactosidase activity, third instar imaginal discs were subjected to X-Gal staining (SI Fig. 5). The observed wing disk patterns recapitulate the expression patterns expected from the drivers used. In none of the cases did we observe repression or ectopic expression of lacZ, indicating that they are not strongly influenced by nearby enhancers or repressors.
Unspecific Integrations Are Rare Events.
To estimate the extent of integrations occurring specifically at the provided attP site versus integrations occurring elsewhere in the genome, we relied on two assays. First, one of the 16 attP lines injected with pUAS-lacZattB carries the landing site on the X chromosome, namely ZH-attP-2A (Table 3). Because in these injections we always provided the attP chromosome through the males, the injected male G0 animals should not have a free attP site and therefore should not give rise to transgenic offspring, unless the integration occurred nonspecifically. None of the G0 males from the ZH-attP-2A yielded transgenic offspring (n = 77 fertile crosses), whereas G0 females did (Table 3 and data not shown), emphasizing a high degree of specificity. Second, we injected the pUAS-lacZattB plasmid at high concentration (800 ng/μl) into embryos that were homozygous for vas-φC31 on the fourth chromosome (102F) but were otherwise not equipped with a free attP landing site. Of the 53 fertile G0 outcrosses, only one gave transgenic progeny (two animals), which is an ≈27 times lower frequency than seen in the presence of an attP site at position 86F (Table 1). Subsequent inverse PCR on this transgenic line revealed an integration event at 2L 37F, into a site that shows no obvious homology to the 39-bp core attP sequence (25). The integration was accompanied by a microdeletion of 11 base pairs at the target site and both a 2-bp deletion and a single nucleotide exchange in the attB part of the hybrid attL site close to the crossover site. Taking these results together, it appears that unspecific integrations are rare events.
A “Split-white” Reconstitution System as an Immediate Readout for Specific attP Targeting.
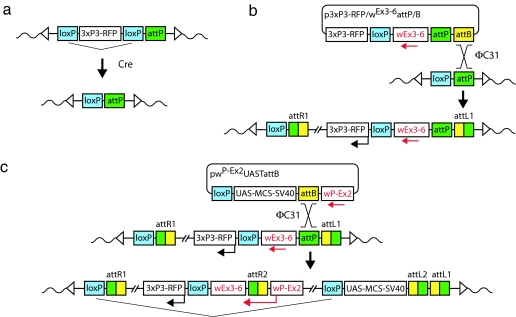
Although our analyses revealed a high degree of specificity with respect to attP targeting, an immediate readout confirming a specific integration event would nevertheless be convenient. In particular for structure/function analyses, which involve the comparison of subtly differing transgenes, an identical integration site is essential. When analyzing a large number of transgenes it would be convenient to eliminate PCR confirmation of each one. To establish such a system, a large part of the white gene (exons 3–6) was placed into the landing site. The remaining part (promoter and exons 1–2) is provided by the transformation vector pwP-Ex2UASTattB. Only if the incoming attB plasmid integrates into the donor attP site, located in the white intron between exons 2 and 3, will a functional white gene be reconstituted and result in the functional expression of white, indicating precise attP targeting. Instead of generating a new collection of specific split-white attP landing sites, we devised a strategy to permit the transformation of our existing attP sites into split-white attP sites. Line ZH-attP-86Fb was used to establish a proof-of-principle for this approach. First, the 3xP3-RFP cassette was eliminated by Cre-mediated excision, leaving one loxP site and the attP docking site at the genomic locus (Fig. 3a). We then introduced the split-white landing site construct into the attP site by φC31 integrase-mediated integration, re-marking the site with the 3xP3-RFP and providing a new attP site for subsequent φC31-mediated integration (Fig. 3b). Because the split-white landing site construct contains both an attP site and an attB site, intermolecular recombination between plasmids can occur, leading to the integration of more than one construct. Thus, transgenic lines resulting from this procedure were tested by PCR and sequencing (data not shown), and lines containing only a single split-white landing site construct were established. In addition to serving as an indicator for specificity, this split-white system reduces the size of the marker transgene and hence of the transformation vector, a property that should facilitate its handling and further increase the frequency of transgenesis.
Fig. 3.
Strategy for transforming an existing attP landing site into a split-white landing site. (a) Elimination of the 3xP3-RFP marker cassette. The attP line was crossed to a Cre-expressing line, leading to excision of the sequence between the loxP sites. (b) Placing of white (exons 3–6) into the modified attP site. Construct p3xP3-RFP/wEx3-6attP/B is introduced by φC31-mediated integration. To prevent intramolecular recombination between the two attachment sites, a shortened attP element of 54 bp was used and cloned immediately next to the attB element, which was trimmed at the 5′ region. (c) Germ-line transformation with the split-white vector pwP-Ex2UASTattB via φC31-mediated integration. Correct attP targeting establishes the white transcriptional unit, allowing expression of a functional white gene.
Discussion
The use of transgenes is essential for understanding the function of genes, their products, and their regulatory elements. The φC31 system described here should strongly facilitate such studies and should allow for the embarking of projects that until now have not been feasible with the available techniques. Below, we discuss how our strategy has advanced the recently described use of φC31 integrase in Drosophila (12).
We describe here the generation of a large collection of precisely mapped attP landing sites. These offer great flexibility regarding the choice of integration sites and the expression levels of transgenes. Predetermined integration sites effectively eliminate the time and effort needed to map transgene insertions, in contrast to those obtained by traditional transposon-mediated germ-line transformation. Defined attP sites allow precise in vivo structure/function analyses, as we have shown here in testing modified integrase constructs. Other advantages of using defined and precharacterized attP sites are that fewer lines for any given transgene will need to be generated, analyzed, and maintained. The need for long-term storage can be avoided altogether because the transgenic lines can be exactly reproduced as long as the original attP line(s) and constructs are available. The vast size of our landing site collection will also facilitate the simultaneous use of multiple transgenes, an increasing need in sophisticated fly genetics. The design of our landing site, intentionally lacking commonly used markers and withstanding commonly used transposases, offers further flexibility in this respect.
A significant improvement of our approach is the establishment of germ-line-specific φC31 integrases. The presence of an “endogenous” source of a transformation-mediating enzyme distinguishes this system from most other commonly used germ-line transformation methods for Drosophila. The use of a transgenic source of φC31 integrase eliminates the time and costs required for mRNA production and significantly reduces the complications associated with the injection process, such as the variability in efficiency caused by the quality and stability of the capped φC31 integrase mRNA. Importantly, our transgenic integrase sources also considerably enhance the integration rates.
Another attractive feature of our system is its amenability to additional modifications. These may include the design of the vectors as well as the existing landing sites; establishing a split-white system is just one example of how an attP site can be tailored to suit changing needs. Other modifications may include the pre-placing of promoters and/or reporters into the landing site. A further example for expandability of this system is the ability to make iterative in vivo assemblies (26), which will permit the analysis of large DNA units at a given locus. The elimination of vector sequences from integrating constructs, an advantage usually claimed by recombinase-mediated cassette exchange (RMCE) approaches (8, 9, 27), can also be achieved with our system by Cre-mediated excision, leaving essentially only one loxP site and the transgene with its regulatory elements at the genomic locus (see Figs. 1c and 3c).
Finally, a promising future application of this system is the handling of large DNA sets, as for instance cDNA libraries or collections of RNAi constructs. With the transformation system reported here, collections of constructs can be site-specifically and efficiently integrated into the genome, thus eliminating the efforts of mapping and testing. “UAS-ORF in vivo” libraries, for example, could be used for systematic mis- or overexpression screens that so far have been conducted mainly with EP-lines (28), an approach that suffered from not achieving saturation and from requiring complex analyses for the unambiguous identification of the phenotype-causing gene.
In summary, the system and resources presented in this study will simplify and accelerate the process of germ-line transformation in Drosophila and facilitate large-scale projects that so far have not been feasible with available techniques.
Materials and Methods
Plasmid Construction.
Details on plasmid construction are available in SI Text.
GenBank accession numbers are as follows: M{3xP3-RFPattP}, EF362407; M{3xP3-RFPattP′}, EF362408; pUASTattB, EF362409.
Germ-Line Transformation and Cre-Mediated Excision.
The mariner-mediated integration of the landing site construct pM{3xP3-RFPattP} into flies with y− w− background was performed according to standard germ-line transformation procedures by using the helper plasmid pKhsp82MOS (29). The generated attP lines were balanced by using standard fly stocks and procedures. All used integrase constructs (with the exception of the nos-φC31NLS construct on the X) and the split-white landing site construct p3xP3-RFP/wEx3-6attP/B were coinjected with φC31 integrase capped mRNA into the mentioned attP sites to generate stable lines. The φC31 integrase mRNAs were either transcribed from the plasmid pET11phiC31poly(A) (12) or from the plasmid pcDNA3.1-φC31, according to the protocol of the mMESSAGE mMACHINE kit (Ambion). Coinjections with mRNA were mainly done as described in ref. 12. DNAs were diluted in either TE buffer or water, mRNAs were diluted in nuclease-free water (Ambion). The 3xP3-RFP cassette was eliminated from line ZH-attP-86Fb via crossing to a hsp70-Cre line; excision was confirmed by standard PCR and sequencing.
Inverse PCR and Sequence Analysis.
Inverse PCR was performed as described on the web site of the Berkeley Drosophila Genome Project (www.fruitfly.org). The restriction enzymes used were MspI, Sau3AI, and XhoI (the latter for the unspecific integration event at 2L 37F). Primers used were designed according to the attP landing site construct. For the 25 “candidate” lines in SI Table 4, the 3′ flanking sequences were again PCR-amplified with more distant primers, followed by sequencing of the two independent PCR products. Inverse PCR sequences were blasted against the sequence data provided on the Ensembl fruit fly site (www.ensembl.org). Searches were done with Ensembl release 41, and the search tool SSAHA2 was used.
X-Gal Staining.
To detect β-galactosidase activity, third-instar larval discs were fixed and subjected to a standard X-Gal color reaction for 20 min at room temperature.
Antibody Staining.
The antibody staining was done as described in ref. 30. The primary antibody was diluted at 1/500. As a secondary antibody, we used a goat anti-rabbit conjugated to DTAF from Jackson ImmunoResearch at a dilution of 1:400.
Chromosome Squashes.
Preparation of salvary gland polytene chromosomes was performed as described in ref. 31. Chromosomes were analyzed by phase-contrast microscopy at a magnification of ×100.
Supplementary Material
Acknowledgments
We thank P. Brugger for alerting us to the φC31 system; M. P. Calos, E. R. Gavis, A. Nakamura, and E. A. Wimmer for plasmids; W. R. Brown and D. Egli for the φC31 integrase antibody; P. Gallant and D. Steiger for suggestions and information about their experience with the described system; B. Dickson and H. Stocker for suggestions regarding the split-white system; T. Laverty for exchange of experience; M. C. M. Smith for valuable information on the φC31 system; N. Kampik and members of the laboratory for help with fly work; and P. Gallant, A. Smith, and G. Hausmann for comments on the manuscript. This work was fully supported by Frontiers in Genetics, National Center of Competence in Research.
Abbreviations
- NLS
nuclear localization sequence
- RFP
red fluorescent protein.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF362407–EF362409).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611511104/DC1.
References
- 1.St Johnston D. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 2.Rubin GM, Spradling AC. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 3.Spradling AC, Rubin GM. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 4.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 5.Cooley L, Kelley R, Spradling A. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- 6.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn C, Handler AM. Proc Natl Acad Sci USA. 2005;102:12483–12488. doi: 10.1073/pnas.0504305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberstein A, Pare A, Kaplan L, Small S. Nat Methods. 2005;2:583–585. doi: 10.1038/nmeth775. [DOI] [PubMed] [Google Scholar]
- 10.Blair SS. Development (Cambridge, UK) 2003;130:5065–5072. doi: 10.1242/dev.00774. [DOI] [PubMed] [Google Scholar]
- 11.Thorpe HM, Smith MC. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groth AC, Fish M, Nusse R, Calos MP. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorpe HM, Wilson SE, Smith MC. Mol Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 14.Horn C, Jaunich B, Wimmer EA. Dev Genes Evol. 2000;210:623–629. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- 15.Andreas S, Schwenk F, Kuter-Luks B, Faust N, Kuhn R. Nucleic Acids Res. 2002;30:2299–2306. doi: 10.1093/nar/30.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sano H, Nakamura A, Kobayashi S. Mech Dev. 2002;112:129–139. doi: 10.1016/s0925-4773(01)00654-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Lehmann R. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- 18.Gavis ER, Lehmann R. Nature. 1994;369:315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 19.Dahanukar A, Wharton RP. Genes Dev. 1996;10:2610–2620. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- 20.Smibert CA, Wilson JE, Kerr K, Macdonald PM. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 21.Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. EMBO J. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahanukar A, Walker JA, Wharton RP. Mol Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- 23.Johnstone O, Lasko P. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Jeppesen I, Nielsen K, Jensen TG. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 25.Groth AC, Olivares EC, Thyagarajan B, Calos MP. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dafhnis-Calas F, Xu Z, Haines S, Malla SK, Smith MC, Brown WR. Nucleic Acids Res. 2005;33:e189. doi: 10.1093/nar/gni192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bateman JR, Lee AM, Wu CT. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rorth P. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coates CJ, Turney CL, Frommer M, O'Brochta DA, Atkinson PW. Mol Gen Genet. 1997;253:728–733. doi: 10.1007/s004380050377. [DOI] [PubMed] [Google Scholar]
- 30.Karch F, Bender W, Weiffenbach B. Genes Dev. 1990;4:1573–1587. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- 31.Pardue M. In: Drosophila: A Practical Approach. Roberts D, editor. Oxford: IRL; 1986. pp. 111–137. [Google Scholar]
- 32.Siegal ML, Hartl DL. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.