Abstract
During pregnancy the uterine decidua is populated by large numbers of natural killer (NK) cells with a phenotype CD56superbrightCD16−CD9+KIR+ distinct from both subsets of peripheral blood NK cells. Culture of highly purified CD16+CD9− peripheral blood NK cells in medium containing TGFβ1 resulted in a transition to CD16−CD9+ NK cells resembling decidual NK cells. Decidual stromal cells, when isolated and cultured in vitro, were found to produce TGFβ1. Incubation of peripheral blood NK cells with conditioned medium from decidual stromal cells mirrored the effects of TGFβ1. Similar changes may occur upon NK cell entry into the decidua or other tissues expressing substantial TGFβ. In addition, Lin−CD34+CD45+ hematopoietic stem/progenitor cells could be isolated from decidual tissue. These progenitors also produced NK cells when cultured in conditioned medium from decidual stromal cells supplemented with IL-15 and stem cell factor.
Keywords: pregnancy
During the late luteal phase and continuing during pregnancy, the uterine endometrial lining undergoes a process termed decidualization under the control of progesterone and estradiol. One of the most distinctive features of this process is the appearance of a large population of decidual natural killer (dNK) cells. Approximately 70% of human decidual lymphocytes and ≈30% of all cells in the decidual compartment are CD56+CD3− natural killer (NK) cells, a much larger fraction than is present in peripheral blood (1). Decidualization is also characterized by a morphological and biochemical differentiation of fibroblast-like endometrial stromal cells into decidual stromal cells (2). dNK cells reside in close cell-to-cell contact with decidual stromal cells (3). Interestingly, decidualized stromal cells have been reported to secrete a number of characteristic factors including prolactin, insulin-like growth factor binding protein 1, tissue factor, VEGF, IL-11, and IL-15 (2). Because of this close interaction and secretory potential, it has been proposed that decidual stromal cells may profoundly affect dNK cells.
Human peripheral blood NK (pNK) cells are divided into two main populations. Approximately 95% of pNK cells are CD56dimCD16+ (hereinafter termed CD16+ pNK). The minor population of pNK cells (≈5% of the total) is CD56brightCD16− (hereinafter termed CD16− pNK). CD16+ pNK cells have high quantities of cytolytic granules and are cytotoxic (4). The majority of these CD16+ pNK cells express members of the killer Ig-like receptor (KIR) family. In contrast, most CD16− pNK cells do not express KIR, have few cytotoxic granules, and have low cytotoxic effector function (4, 5). Conversely, they express higher levels of CD94/NKG2A than the CD16+ pNK cell subset (4). The two subsets also differ in expression of IL-2 receptor α, certain chemokine receptors, and adhesion molecules, which may effect their recruitment into different tissues (4). Interestingly, the minor CD16− pNK cell subset appears to contain the main cytokine-producing pNK cells and may be important in directing immune responses (4).
Comparison of dNK cells with the two subsets of pNK cells indicates that an appropriate nomenclature for these cells may be CD56superbrightCD16− dNK cells because they express even higher levels of CD56 than CD16− pNK cells. Like CD16− pNK cells, dNK cells lack CD16, express high levels of CD94/NKG2A receptors (6), and show low cytotoxicity against NK cell targets (7). However, many dNK cells possess KIR and large quantities of cytolytic granules, in this way resembling CD16+ pNK cells (1, 8). dNK cells are transcriptionally unique and differentially express a substantial number of genes when compared with pNK cells (9). The relationship of pNK cells to decidual or uterine NK cells is controversial. The CD16− phenotype of dNK cells has prompted the hypothesis that dNK cells originate from CD16− pNK cells that migrate to the uterus and undergo further tissue-specific differentiation (1). Mechanisms involving differential CXCR4, CCR5, and CXCR3 expression by CD16− pNK cells (10), CXCL12 expression by extravillous trophoblast (11), MIP-1β expression by endometrial cells (12), or induction of CXC chemokine ligands 10 and 11 in human endometrium by estradiol and progesterone (13) may induce recruitment of pNK cells. The possibility that dNK cells could originate from CD16+ pNK cells has received minimal attention.
An alternative hypothesis that does not invoke pNK cells also exists. Adult tissues contain resident stem cells for self renewal and repair in supportive niche environments (14). Hematopoietic progenitor cells (HPC) have the potential to differentiate into NK cells when stimulated with IL-15 with or without stem cell factor (SCF) or Flt-3 ligand (15). It is possible that dNK cells may originate from precursor cells which reside in uterine tissues or migrate there from blood.
Results
Conditioned Medium (CM) from Decidual Stromal Cells Causes Peripheral NK Cells to Acquire Some Characteristics of dNK Cells.
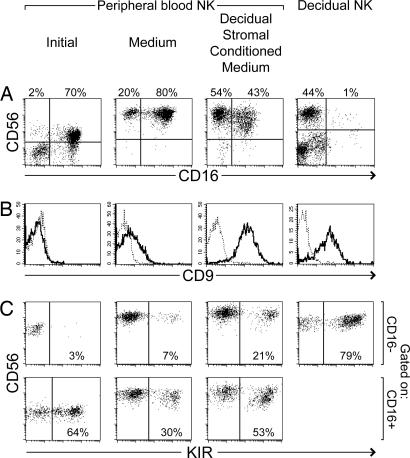
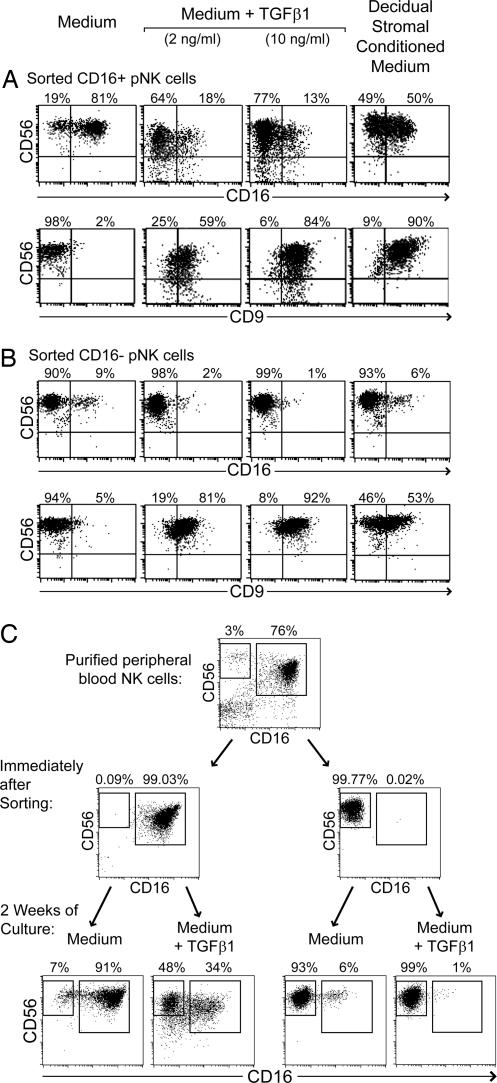
dNK cells have a distinct phenotype, CD56superbrightCD16−CD9+, with a substantial proportion expressing KIR (Fig. 1) (1, 9) Precursors of dNK cells may include one or both subsets of pNK cells, which may migrate into the decidua and differentiate in the local decidual environment. To investigate the potential influences of decidual stromal cells, stromal cells were isolated from human decidua, cultured, and used to prepare CM. Isolated pNK cells (containing a mixture of the CD16+ and CD16− pNK cell subsets) were then incubated with or without this decidual stromal CM. All cultures in this study were supplemented with IL-15 and SCF to promote the survival of NK cells. After 2 weeks of culture with normal medium, the proportion of CD16− cells among pNK cells was increased to some extent. However, in the presence of decidual stromal CM this fraction increased much more [Fig. 1A and supporting information (SI) Table 1]. Increases of the CD56+CD16− subset after culture with stromal CM, compared with medium alone, were observed in 12 of 16 independent experiments (P = 0.002, paired t test) (see SI Table 1). Both culture conditions appeared to increase the intensity of CD56 staining (Fig. 1A). Previous transcriptional profiling and flow cytometry studies have identified CD9 as a marker characteristic of dNK cells but not pNK cells (9) (Fig. 1B). In the presence of stromal CM, CD9 was consistently expressed at relatively high levels on essentially all of the pNK cells (Fig. 1B and SI Table 1). pNK cells cultured in normal medium remained CD9 negative or low. Whereas the proportion of KIR+ cells decreased in pNK cells grown in normal medium, cells in CM retained a larger percentage of KIR+ cells (SI Table 1). Thus, incubation with decidual stromal CM consistently resulted in greater percentages of cells expressing KIR compared with cells in normal medium (Fig. 1C and SI Table 1). This was apparent within both the CD16+ and CD16− gated NK cell populations in all 15 experiments performed. Most strikingly, cultures with decidual stromal CM accumulated substantial numbers of CD16− NK cells that also expressed KIR (Fig. 1C and SI Table 1). Therefore, culture of pNK cells in the presence of decidual stromal CM appeared to favor a transition toward a phenotype showing some dNK cell characteristics, increasing the proportion of CD56+CD16− cells, inducing CD9 expression, and displaying a high percentage of KIR+ cells.
Fig. 1.
Culture of peripheral NK cells with CM from decidual stromal cells causes accumulation of cells showing some characteristics of dNK cells. NK cells purified from peripheral blood (and containing both CD16+ and CD16− NK cells) were cultured in normal medium or CM harvested from decidual stromal cells. Both conditioned and normal media were supplemented with IL-15 and SCF. After 2 weeks, expression of CD56, CD16, CD9, and KIR were analyzed by flow cytometry. KIR were detected with pooled HP3-E4, CH-L, and DX9 mAb (recognizing KIR 2DL1, 2DS1, 2DL2, 2DS2, 2DL3, and 3DL1). Staining for these markers at the start of the experiments is shown in the first column. For comparison, a similar staining of cells isolated from decidua is shown in the fourth column. Plots in C were gated at the time of analysis to display CD56+ NK cells that were either CD16− or CD16+. Plots in B show all purified peripheral NK cells or gated CD56+CD16− dNK cells. The fine dotted line in B represents staining with an isotype control mAb. (A, B, and C represent similar but distinct experiments.)
TGFβ1 Also Causes Peripheral NK Cells to Acquire Similarities to dNK Cells.
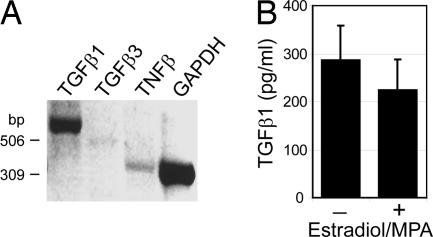
To identify factors responsible for the observed results, cultured decidual stromal cells were investigated for the expression of various cytokines. An initial exploratory gene array analysis of decidual stromal cells demonstrated high expression of TGFβ1 in addition to lower levels of TGFβ2 and TGFβ3 (data not shown). TGFβ1 expression by decidual stroma was confirmed by RT-PCR (Fig. 2A) and ELISA analysis (Fig. 2B). Concentrations of TGFβ1 in decidual stromal CM ranged from 81 to 419 pg/ml (n = 17). Production of IL-15 and several other cytokines was also observed (data not shown).
Fig. 2.
Decidual stromal cells, when isolated and cultured in vitro, produce TGFβ1. (A) RNA isolated from in vitro cultured decidual stromal cells was analyzed by RT-PCR. (B) TGFβ1 protein levels were examined by ELISA of CM from decidual stromal cells cultured in the absence or presence of 6α-methyl-17α-hydroxyprogesterone acetate (MPA) and estradiol. Shown are the mean TGFβ1 level and SD (n = 7 and 8), with signals from normal medium subtracted, of samples collected during weeks 3–4 after cell seeding (≈5–6 weeks after tissue isolation).
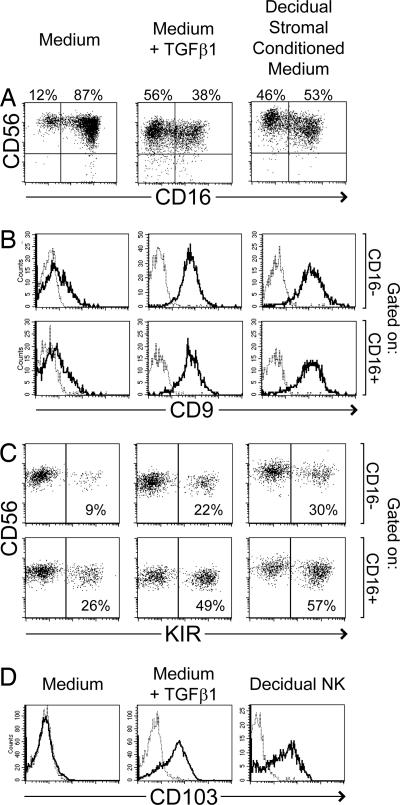
These data were collected after in vitro culture of stromal cells for ≈4–6 weeks and may not reflect the original stromal phenotype. However, TGFβ has been frequently reported to be expressed in the pregnant and nonpregnant uterus (reviewed in refs. 16 and 17). Therefore, the effect of TGFβ on differentiation of pNK cells was directly examined. After 2 weeks of culture with normal medium supplemented with 2 or 10 ng/ml TGFβ1, pNK cells again showed a much greater proportion of CD56+CD16− cells, compared with pNK cells cultured in medium alone (Fig. 3A and SI Table 1). This result was very similar to that obtained with decidual stromal CM. Compared with the experimental start, median fluorescence intensity of CD56 staining appeared to increase under all three culture conditions, although increases were smaller in the presence of TGFβ1 (data not shown). pNK cells treated with either TGFβ1 or decidual stromal CM also expressed high levels of CD9 (Fig. 3B and SI Table 1), and greater percentages of KIR compared with medium alone (Fig. 3C and SI Table 1). TGFβ1 cultures also accumulated substantial numbers of CD16−KIR+ cells. In addition, pNK cells incubated with TGFβ1 acquired CD103 (αE integrin) (Fig. 3D), which has been reported to be expressed by dNK cells but not by pNK cells (1, 18) (Fig. 3D).
Fig. 3.
Exogenously added TGFβ1 also causes peripheral NK cells to acquire certain similarities to dNK cells. NK cells isolated from peripheral blood were cultured in normal medium, medium supplemented with 2 ng/ml TGFβ1, or CM harvested from decidual stromal cells. All cultures were additionally supplemented with IL-15 and SCF. After 2–3 weeks, expression of CD56 and CD16 (A), CD9 (B), KIR (C), and CD103 (D) were analyzed by flow cytometry. KIR were detected with pooled HP3-E4, CH-L, and DX9 mAb. Plots in B and C were gated at the conclusion of the experiment to display CD56+ NK cells that were either CD16+ or CD16−. The fine dotted lines in B and D represent staining with an isotype control mAb. At the start of these experiments, enriched NK cells were 3% CD16− (A), completely CD9-negative (B), and completely CD103-negative (D). Initially, KIR were expressed by 5% and 54% of CD16− and CD16+ cells, respectively (C). Staining of dNK cells is included for comparison in D only. (A, B, C, and D represent similar but distinct experiments.)
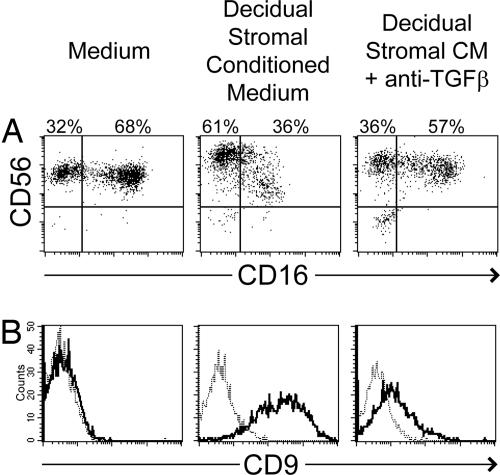
To clarify the role of TGFβ in decidual stromal CM, a neutralizing mAb which recognizes TGFβ1, TGFβ2, and TGFβ3 was used. Effects of decidual stromal CM on numbers of CD16− pNK cells were blocked by pretreatment of CM with this anti-TGFβ mAb (Fig. 4A and SI Table 1). Blocking of TGFβ also inhibited the induction of CD9 expression by decidual stromal CM (Fig. 4B and SI Table 1). Pretreatment with isotype control antibodies had minimal effect on pNK cell changes induced by decidual stroma CM (SI Table 1). TGFβ blocking experiments measuring KIR expression were inconclusive, opening the possibility that other factors in decidual stromal CM may be necessary to affect KIR expression and may synergize with TGFβ.
Fig. 4.
TGFβ is responsible for certain effects of decidual stromal CM on peripheral NK cells. NK cells isolated from peripheral blood were cultured in normal medium, decidual stromal cell CM, or decidual stromal CM pretreated with 10 μg/ml anti-TGFβ neutralizing antibody. All cultures were additionally supplemented with IL-15 and SCF. After 2 weeks, expression of CD56 and CD16 (A) and CD9 (B) were analyzed by flow cytometry.
TGFβ Directly Causes Conversion of CD16+ Peripheral NK Cells into CD16− NK Cells.
The effects of TGFβ1 or decidual stromal CM shown in Figs. 1, 3, and 4 could have resulted either from the conversion of CD16+ pNK cells to CD16− NK cells or from the preferential death of CD16+ pNK cells and/or selective proliferation of CD16− pNK cells. Depending on the context, TGFβ has been reported to have either proapoptotic or antiapoptotic effects (19). However, cultures supplemented with TGFβ1 did not display substantially larger numbers of dead or apoptotic cells compared with medium alone (SI Table 2). IL-15 and SCF may have had antiapoptotic effects (20, 21). However, fewer cells were recovered from TGFβ1 cultures, although this could represent lesser proliferation in this case (SI Table 2 and SI Fig. 7).
To more directly examine the fates of CD16+ and CD16− pNK, the two subsets present in enriched pNK cells were sorted by flow cytometry. After culture in the presence of TGFβ1, sorted CD16+ pNK cells produced a substantial fraction of CD16− NK cells, whereas the sorted CD16− pNK cells remained relatively unaltered in CD16 expression (Fig. 5). Moreover, both groups of pNK cells acquired CD9 in the course of incubation with TGFβ1 (Fig. 5 A and B). Both subsets were CD94 positive on isolation and remained so during the experiment (data not shown). Therefore, TGFβ1 appeared to induce a transition from the CD16+ to the CD16− NK cell phenotype, coupled with induction of CD9 expression. Decidual stromal CM substituted for TGFβ1 in inducing these effects. CD16+ pNK cells were sorted to very high purity (i.e., the contamination with CD16− pNK cells was <0.1%) (Fig. 5C), making it highly improbable that these results were caused by selective outgrowth of small numbers of contaminating cells of the other subset during the 2-week incubation. This transition of phenotype was accompanied by cell division, although not at levels sufficient to suggest outgrowth of very rare cells. After CFSE labeling of sorted cells, both CD16+ and CD16− pNK cells proliferated under these conditions (although proliferation appeared to be lesser in the presence of TGFβ1) (SI Fig. 7). The data are most consistent with a conversion of cells from CD16+ to CD16−, under the influence of TGFβ, accompanied by proliferation. Whether all CD16+ pNK, or only a subset of CD16+ cells, can respond in this way to TGFβ remains to be determined.
Fig. 5.
TGFβ1 causes conversion of CD16+CD9− peripheral NK cells into CD16−CD9+ NK cells. (A and B) CD56+ NK cells from peripheral blood were sorted by flow cytometry into CD16+ (A) and CD16− (B) populations, which were subsequently cultured in normal medium, medium supplemented with 2 or 10 ng/ml TGFβ1, or CM from decidual stromal cells. All cultures were additionally supplemented with IL-15 and SCF. After 2 weeks, expression of CD56, CD16, and CD9 were analyzed by flow cytometry. At the start of this experiment, both CD16+ and CD16− NK cells were CD9-negative. (C) A similar experiment is shown, showing the purity of sorted CD16+ and CD16− NK cells immediately after sorting, and the subsequent change of phenotype upon culture in the presence of 2 ng/ml TGFβ1.
Decidual Tissue Also Contains HPCs That Can Differentiate into NK Cells.
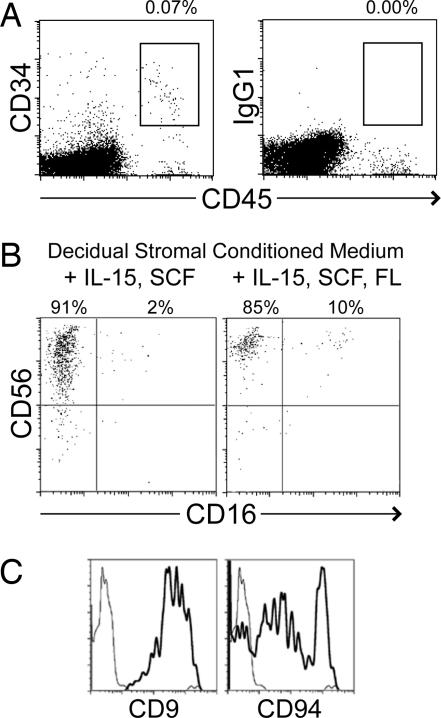
Cells with an HPC phenotype (Lin−CD34+CD45+) were also identified in human decidual lymphocyte preparations (Fig. 6A). The frequency of such cells in decidua (≈0.1%) was equivalent or greater to that found in human peripheral blood. As expected, these decidual HPC were negative for NK cell markers (CD56−CD94−KIR−) (data not shown).
Fig. 6.
Decidual tissue also contains Lin−CD34+CD45+ progenitors, which can differentiate into NK cells. Leukocyte preparations from decidual tissue were stained with anti-CD34, CD45, and Lin (CD3, CD14, CD16, CD19, CD20, and CD56) mAb. Gated Lin− cells are shown in A. Percentages show boxed cells as a proportion of all gated live cells. Decidual Lin−CD34+CD45+ cells were isolated by flow cytometry sorting and cultured in decidual stromal CM supplemented with IL-15 and SCF with or without Flt-3 ligand. After ≈30 days, expression of CD56, CD16, CD9, and CD94 on the resulting cells was examined by flow cytometry (B and C). Plots in C are gated on resulting CD56+CD16− cells that resemble dNK cells. Similar cells were observed in three of four experiments.
Decidual HPC were examined for their capacity to generate cells of NK lineage. Lin−CD34+CD45+ cells were sorted from decidual tissue by flow cytometry and incubated in decidual stromal CM supplemented with IL-15 and SCF (with and without Flt3 ligand). After ≈30 days, decidual HPC cultures produced NK cells showing some characteristics of dNK. The majority of these cells were CD56superbrightCD16− with essentially all expressing CD9 and many expressing CD94 (Fig. 6 B and C).
Discussion
Regardless of their origin, it is likely that dNK cells are influenced by factors in their local environment. When pNK cells were cultured for 2 weeks in medium supplemented with IL-15 and SCF, the percentage of CD56+CD16− NK cells was increased to some extent. This was likely because of a greater proliferation of CD16− pNK cells, which are known to respond to these cytokines with greater proliferation, compared with CD16+ pNK cells (22, 23) (SI Fig. 7). However, when medium was additionally supplemented with TGFβ1, the percentage of CD56+CD16− NK cells was increased much more. Sorting of highly purified cell populations suggested that this was because of conversion of CD16+ pNK cells to CD16− cells under the influence of TGFβ. Culture of pNK cells with TGFβ1 also caused an up-regulation of expression of CD9, a tetraspanin protein that has been reported to organize integrins and various other molecules into membrane microdomains (24). Activation of NK cells with IL-2 and PHA has also been reported to trigger CD9 expression (25). However, CD9 levels induced by TGFβ1 were substantially greater than those caused by culture in the presence of IL-2 (data not shown). Treatment of pNK cells with TGFβ1 was also found to up-regulate CD103 (αE integrin). CD103 induced by TGFβ could affect NK cell trafficking in tissues expressing E-cadherin ligands of CD103. Furthermore, after exposure to TGFβ1, cultures of pNK cells contained substantial numbers of CD16−KIR+ cells, presumably the result of CD16+KIR+ cells losing CD16 expression. [Sorted KIR− pNK cells did not acquire KIR during culture with TGFβ1 (data not shown)]. All four of these features (CD16−, CD9+, CD103+, and KIR+) are characteristic of dNK, suggesting that TGFβ may play a role in defining the dNK phenotype. However, many factors in the decidua are likely to play a role in influencing NK cell development, in addition to TGFβ.
The effects of TGFβ exposure are often context-dependent, varying with the local environment and presence of other factors and cytokines (26). Thus, the observed changes of pNK cells may depend on the culture conditions used, especially the inclusion of IL-15 and SCF in all media. However, similar TGFβ1-mediated effects on CD16, CD9, and KIR expression were also observed in pNK cell cultures containing IL-15 but lacking SCF (data not shown). IL-15 has previously been detected in endometrium and decidua (27, 28), including stromal cells (2, 29, 30).
After isolation and culture in vitro, decidual stromal cells were shown to produce TGFβ1. Cultures of pNK cells in CM from these stromal cells closely mimicked cultures with added TGFβ1. The effects of decidual stromal CM on numbers of CD16− and CD9+ pNK cells were clearly due to TGFβ because they could be blocked with a neutralizing anti-TGFβ1/2/3 mAb. The decidualization of stromal cells is driven by progesterone and estradiol, and this process has been extensively studied in vitro (2). Effects on pNK cells were similar when stromal CM was harvested in the absence or presence of β-estradiol and 6α-methyl-17α-hydroxyprogesterone acetate (data not shown), although no specific decidual products were examined to verify the decidualization process. Of course, these findings were obtained in an in vitro system, in which stromal cells likely differed from their original phenotype. Indeed, it has been reported that cells may up-regulate TGFβ production in response to adherence to tissue culture plastic (31). Although decidual stromal cells in vitro may behave differently from those in vivo, a large number of publications have identified TGFβ isoforms in human endometrial, decidual and placental tissues, using RT-PCR, immunohistochemistry, and ELISA methodologies (reviewed in refs. 16 and 17). Similar observations have been made in several other species (16). Thus, the observed effects of TGFβ many be relevant to uterine NK cells in normal cycling endometrium and during pregnancy.
These results suggest a possible differentiation route for the origin of dNK cells. CD16+ pNK cells may migrate into uterine tissue, then differentiate locally under the influence of TGFβ, and other factors, and thereby acquire the dNK phenotype. This hypothesis is attractive because CD16+ pNK cells represent the overwhelming majority of pNK cells and these cells share the presence of extensive cytotoxic granules with dNK cells (1). Previous hypotheses of dNK cell precursors have focused on the much less abundant CD16− pNK cell subset, primarily because both cell types lack CD16. CD16− pNK cells also express chemokine receptors which could favor recruitment into decidua (10–13). When CD16− pNK cells were isolated and cultured in vitro with TGFβ1, CD9 was also acquired. The present data do not exclude the possibility that they may be a source of dNK cells.
Because TGFβ is also expressed in many other contexts, the changes observed may not be specific to the decidual tissue but may occur when NK cells enter any tissue expressing TGFβ. Notably, NK cells accumulating in tissues during inflammation have also been described as CD16−, although they appear to resemble the CD16− cells from blood more than dNK cells (32). It will be interesting to examine the expression of CD9 and CD103 on tissue NK cells to determine whether these markers are specific to decidua or are acquired in other tissues.
CD16 (FcγRIII) is an important receptor mediating antibody-dependent cell-mediated cytotoxicity by NK cells. Thus, exposure to TGFβ may direct NK cell responses away from antibody-dependent cell-mediated cytotoxicity. This could be relevant in the decidua, given that maternal antifetal antibodies can be elicited.
Certain effects of TGFβ on NK cells have been previously studied, including an influence on cytokine secretion (33) and expression of activating receptors (34). Culture of NK cells in the presence of TGFβ is reported to partially inhibit NK cell cytotoxic activity (34, 35). Interestingly, dNK cells have also been shown to have low cytotoxic activity (7) which could possibly be related to the effects of TGFβ in decidual tissue. However, in the culture conditions of this study, TGFβ1 treatment induced only negligible to modest inhibition of pNK cell cytotoxicity (data not shown).
Last, it has been demonstrated that decidual tissue contains Lin−CD34+CD45+ cells with a HPC phenotype. This population may include hematopoietic stem cells as well as progenitors already committed to various lineages including NK cells. In the presence of decidual stromal CM supplemented with IL-15 and SCF, these HPC proliferated and differentiated into CD56+CD16−CD94+CD9+ NK cells that are characteristic of dNK. The percentage of cells with HPC phenotype in decidual tissue was equivalent or greater than seen in blood, indicating that such cells could not have been derived from small levels of contamination with peripheral blood. However, it cannot be eliminated that rare functional progenitors may have resulted from contamination with blood or with fetal HPC. Nonetheless, these results open the possibility that dNK cells may derive from progenitors which constitutively reside in uterine tissue or are periodically repopulated by blood progenitor cells. Rapid proliferation and differentiation from these progenitors in the uterus during the menstrual cycle and during early pregnancy could result in the large numbers of uterine and decidual NK cells observed. It is of interest that decidual/endometrial stromal cells also produce IL-15 (refs. 29 and 30 and data not shown), which supports NK cell development. SCF has also been reported in decidual tissue (36–38). Lately, there has been strong interest in stem cell niches: specific locations in a tissue where stem cells can reside for an indefinite period and produce progeny cells while self-renewing (14). Thus, the observed decidual HPC could reside in a uterine stem cell niche. Interestingly, similar CD34+ progenitor cells are present in lymph nodes and can differentiate into CD56+CD16− NK cells (39, 40). Subsets of Lin−CD34+ cells isolated from decidua also appear to express other markers associated with NK progenitor cells including integrin β7, and CD117 (data not shown). However, in mice, elegant cell transfer studies into NK cell-deficient animals suggested that dNK cell precursors were not present in uterine grafts, but could be found in secondary lymphoid tissues (41, 42).
In summary, on one hand we show that TGFβ favors conversion of CD16+CD9− pNK cells to CD16−CD9+ cells resembling dNK cells. On the other hand, decidua also contains progenitor cells that can develop into cells with characteristics of dNK. Thus, CD16+ pNK cells or decidual HPC, or both, are possible precursors of dNK cells. Although it is still not possible to draw conclusions on the nature of the in vivo precursor cells of dNK, this study suggests that TGFβ may play an important role in influencing the dNK phenotype.
Materials and Methods
Decidual Samples.
Preparation of dNK cells and stromal cells from decidual samples of patients undergoing elective pregnancy termination between 6 and 12 weeks of gestation was described previously (9). Freshly isolated adherent decidual stromal cells were cultured in DMEM/F12 medium (Gibco) supplemented with 10% charcoal-filtered FCS without added cytokines to favor survival and proliferation of stromal cells over contaminating decidual macrophages and lymphocytes. Cells were washed twice a week to remove nonadherent cells. Decidual stromal cells were used for further experiments only when they were free of CD14-positive and CD45-positive cells, as detected by flow cytometry (data not shown).
Preparation of Decidual Stromal CM.
A total of 2 × 104 decidual stromal cells were seeded on 100-mm tissue culture plates in Myelocult medium (StemCell Technologies) supplemented with 10% human serum and 5% FCS. Further experiments were performed with CM collected at 2–4 weeks of these cultures. Certain aliquots of CM were generated in the presence of 10−7 M 6α-methyl-17α-hydroxyprogesterone acetate and 10−8 M β-estradiol. Concentration of TGFβ1 in CM was measured with a BioSource multispecies TGFβ1 Immunoassay Elisa kit. RT-PCR analysis of TGFβ was performed by using a MultiGene-12 Human Cytokine RT-PCR Profiling Kit (Superarray Bioscience) and total RNA isolated from confluent decidual stromal cell cultures.
Differentiation of pNK.
NK cells were isolated from peripheral blood of healthy donors by using RosetteSep human NK cell enrichment mixture (Stem Cell Technologies). pNK cells were cultured for 2–3 weeks in 24-well plates (106 cells in 1 ml) under the following conditions: (i) normal medium (Myelocult Medium plus 10% human serum, 5% FCS, 20 ng/ml IL-15, and 20 ng/ml SCF), (ii) normal medium additionally supplemented with 2 or 10 ng/ml TGFβ1, or (iii) decidual stromal CM plus 20 ng/ml IL-15 and 20 ng/ml SCF. All cytokines were purchased from PeproTech. In certain experiments, pNK cells were separated into CD56+CD16+ and CD56+CD16− populations by flow cytometry sorting before similar culture in vitro. Where indicated, blocking of TGFβ was performed by pretreating CM with 10 μg/ml anti-TGFβ1, β2, β3 neutralizing mAb 1D11 (R & D Systems) or isotype control for several hours at 4°C before addition to pNK cells.
Isolation and Differentiation of Decidual CD34+ Progenitor Cells.
Decidual lymphocytes were prepared as described previously (9). Nonadherent cells were washed twice and stained with CD34, CD45, and Lin (CD3, CD14, CD16, CD19, CD20, and CD56) fluorochrome-conjugated mAb (BD Pharmingen). Lin−CD34+CD45+ cells were isolated by flow cytometry sorting and cultured (at 250–500 cells per well in 96-well round-bottom plates) with decidual stroma CM supplemented with IL-15 (20 ng/ml) and SCF (20 ng/ml) for ≈30 days, refreshing medium weekly. Some cultures were additionally supplemented with Flt-3 ligand (20 ng/ml).
Supplementary Material
Acknowledgments
We thank Marivic Mapa for help with references. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants AI053330 and AI50207.
Abbreviations
- NK
natural killer
- pNK
peripheral blood NK
- dNK
decidual NK
- CM
conditioned medium
- KIR
killer Ig-like receptor
- HPC
hematopoietic progenitor cell
- SCF
stem cell factor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611098104/DC1.
References
- 1.Moffett-King A. Nat Rev Immunol. 2002;2:656–663. doi: 10.1038/nri886. [DOI] [PubMed] [Google Scholar]
- 2.Dunn CL, Kelly RW, Critchley HO. Reprod Biomed Online. 2003;7:151–161. doi: 10.1016/s1472-6483(10)61745-2. [DOI] [PubMed] [Google Scholar]
- 3.King A. Hum Reprod Update. 2000;6:28–36. doi: 10.1093/humupd/6.1.28. [DOI] [PubMed] [Google Scholar]
- 4.Cooper MA, Fehniger TA, Caligiuri MA. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, Sykora KW, Schmidt RE. Eur J Immunol. 2001;31:3121–3127. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.King A, Allan DS, Bowen M, Powis SJ, Joseph S, Verma S, Hiby SE, McMichael AJ, Loke YW, Braud VM. Eur J Immunol. 2000;30:1623–1631. doi: 10.1002/1521-4141(200006)30:6<1623::AID-IMMU1623>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 7.Kopcow HD, Allan DS, Chen X, Rybalov B, Andzelm MM, Ge B, Strominger JL. Proc Natl Acad Sci USA. 2005;102:15563–15568. doi: 10.1073/pnas.0507835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma S, King A, Loke YW. Eur J Immunol. 1997;27:979–983. doi: 10.1002/eji.1830270426. [DOI] [PubMed] [Google Scholar]
- 9.Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, Lockwood CJ, Schachter AD, Park PJ, Strominger JL. J Exp Med. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell JJ, Qin S, Unutmaz D, Soler D, Murphy KE, Hodge MR, Wu L, Butcher EC. J Immunol. 2001;166:6477–6482. doi: 10.4049/jimmunol.166.11.6477. [DOI] [PubMed] [Google Scholar]
- 11.Hanna J, Wald O, Goldman-Wohl D, Prus D, Markel G, Gazit R, Katz G, Haimov-Kochman R, Fujii N, Yagel S, et al. Blood. 2003;102:1569–1577. doi: 10.1182/blood-2003-02-0517. [DOI] [PubMed] [Google Scholar]
- 12.Kitaya K, Nakayama T, Okubo T, Kuroboshi H, Fushiki S, Honjo H. J Clin Endocrinol Metab. 2003;88:1809–1814. doi: 10.1210/jc.2002-020980. [DOI] [PubMed] [Google Scholar]
- 13.Sentman CL, Meadows SK, Wira CR, Eriksson M. J Immunol. 2004;173:6760–6766. doi: 10.4049/jimmunol.173.11.6760. [DOI] [PubMed] [Google Scholar]
- 14.Ohlstein B, Kai T, Decotto E, Spradling A. Curr Opin Cell Biol. 2004;16:693–699. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Colucci F, Caligiuri MA, Di Santo JP. Nat Rev Immunol. 2003;3:413–425. doi: 10.1038/nri1088. [DOI] [PubMed] [Google Scholar]
- 16.Godkin JD, Dore JJ. Rev Reprod. 1998;3:1–6. doi: 10.1530/ror.0.0030001. [DOI] [PubMed] [Google Scholar]
- 17.Jones RL, Stoikos C, Findlay JK, Salamonsen LA. Reproduction. 2006;132:217–232. doi: 10.1530/rep.1.01076. [DOI] [PubMed] [Google Scholar]
- 18.Burrows TD, King A, Loke YW. Cell Immunol. 1993;147:81–94. doi: 10.1006/cimm.1993.1050. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Capelo A. Cytokine Growth Factor Rev. 2005;16:15–34. doi: 10.1016/j.cytogfr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Carson WE, Fehniger TA, Haldar S, Eckhert K, Lindemann MJ, Lai CF, Croce CM, Baumann H, Caligiuri MA. J Clin Invest. 1997;99:937–943. doi: 10.1172/JCI119258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carson WE, Haldar S, Baiocchi RA, Croce CM, Caligiuri MA. Proc Natl Acad Sci USA. 1994;91:7553–7557. doi: 10.1073/pnas.91.16.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson WE, Giri JG, Lindemann MJ, Linett ML, Ahdieh M, Paxton R, Anderson D, Eisenmann J, Grabstein K, Caligiuri MA. J Exp Med. 1994;180:1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matos ME, Schnier GS, Beecher MS, Ashman LK, William DE, Caligiuri MA. J Exp Med. 1993;178:1079–1084. doi: 10.1084/jem.178.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemler ME. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 25.Hanna J, Bechtel P, Zhai Y, Youssef F, McLachlan K, Mandelboim O. J Immunol. 2004;173:6547–6563. doi: 10.4049/jimmunol.173.11.6547. [DOI] [PubMed] [Google Scholar]
- 26.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 27.Verma S, Hiby SE, Loke YW, King A. Biol Reprod. 2000;62:959–968. doi: 10.1095/biolreprod62.4.959. [DOI] [PubMed] [Google Scholar]
- 28.Kitaya K, Yasuda J, Yagi I, Tada Y, Fushiki S, Honjo H. Biol Reprod. 2000;63:683–687. doi: 10.1095/biolreprod63.3.683. [DOI] [PubMed] [Google Scholar]
- 29.Okada H, Nakajima T, Sanezumi M, Ikuta A, Yasuda K, Kanzaki H. J Clin Endocrinol Metab. 2000;85:4765–4770. doi: 10.1210/jcem.85.12.7023. [DOI] [PubMed] [Google Scholar]
- 30.Dunn CL, Critchley HO, Kelly RW. J Clin Endocrinol Metab. 2002;87:1898–1901. doi: 10.1210/jcem.87.4.8539. [DOI] [PubMed] [Google Scholar]
- 31.Streuli CH, Schmidhauser C, Kobrin M, Bissell MJ, Derynck R. J Cell Biol. 1993;120:253–260. doi: 10.1083/jcb.120.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalbeth N, Gundle R, Davies RJ, Lee YC, McMichael AJ, Callan MF. J Immunol. 2004;173:6418–6426. doi: 10.4049/jimmunol.173.10.6418. [DOI] [PubMed] [Google Scholar]
- 33.Eriksson M, Meadows SK, Wira CR, Sentman CL. J Leukocyte Biol. 2004;76:667–675. doi: 10.1189/jlb.0204090. [DOI] [PubMed] [Google Scholar]
- 34.Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R, Biassoni R, Bottino C, Moretta L, Moretta A. Proc Natl Acad Sci USA. 2003;100:4120–4125. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellone G, Aste-Amezaga M, Trinchieri G, Rodeck U. J Immunol. 1995;155:1066–1073. [PubMed] [Google Scholar]
- 36.Kauma S, Huff T, Krystal G, Ryan J, Takacs P, Turner T. J Clin Endocrinol Metab. 1996;81:1261–1266. doi: 10.1210/jcem.81.3.8772609. [DOI] [PubMed] [Google Scholar]
- 37.Umekage H, Saito S, Morikawa H. J Reprod Immunol. 1998;40:1–24. doi: 10.1016/s0165-0378(98)00028-x. [DOI] [PubMed] [Google Scholar]
- 38.Sharkey AM, Jokhi PP, King A, Loke YW, Brown KD, Smith SK. Cytokine. 1994;6:195–205. doi: 10.1016/1043-4666(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 39.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, Hughes TL, Marburger TB, Sung J, Baiocchi RA, et al. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA. J Exp Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chantakru S, Miller C, Roach LE, Kuziel WA, Maeda N, Wang WC, Evans SS, Croy BA. J Immunol. 168:22–28. doi: 10.4049/jimmunol.168.1.22. [DOI] [PubMed] [Google Scholar]
- 42.van den Heuvel MJ, Chantakru S, Xuemei X, Evans SS, Tekpetey F, Mote PA, Clarke CL, Croy BA. Immunol Invest. 34:273–293. doi: 10.1081/imm-200064488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.