Abstract
Mutations in Parkin are one of the predominant hereditary factors found in patients suffering from autosomal recessive juvenile Parkinsonism. Parkin is a member of the E3 ubiquitin ligase family that is defined by a tripartite RING1-in-between-ring (IBR)-RING2 motif. In Parkin, the IBR domain has been shown to augment binding of the E2 proteins UbcH7 and UbcH8, and the subsequent ubiquitination of the proteins synphilin-1, Sept5, and SIM2. To facilitate our understanding of Parkin function, the solution structure of the Parkin IBR domain was solved by using NMR spectroscopy. Folding of the IBR domain (residues M327–S378) was found to be zinc dependent, and the structure reveals the domain forms a unique pair scissor-like and GAG knuckle-like zinc-binding sites, different from other zinc-binding motifs such as the RING, LIM, PHD, or B-box motifs. The N terminus of the IBR domain, residues E307–E322, is unstructured. The disease causing mutation T351P causes global unfolding, whereas the mutation R334C causes some structural rearrangement of the domain. In contrast, the protein containing the mutation G328E appears to be properly folded. The structure of the Parkin IBR domain, in combination with mutational data, allows a model to be proposed where the IBR domain facilitates a close arrangement of the adjacent RING1 and RING2 domains to facilitate protein interactions and subsequent ubiquitination.
Keywords: ubiquitination, zinc-binding, NMR spectroscopy, protein folding, protein interactions
Parkinson's disease (PD) is a common neurodegenerative disease characterized by damage to the dopaminergic neurons of the substantia nigra of the midbrain manifesting itself with symptoms such as tremors, rigidity, and bradykinesia (1, 2). Autosomal recessive juvenile PD (ARJP) is an early-onset (<40 years of age) form of the disease that is caused by hereditary factors including mutations in the genes DJ-1, PINK1, and in about half of the cases, parkin. The protein Parkin has been identified as an E3 ubiquitin–protein ligase of the ubiquitin-proteosome system that is required to maintain cellular protein quality control by removing misfolded or damaged proteins (3, 4). Ubiquitin mediated proteolysis occurs through a cascade of ubiquitin transfers from an E1 activating enzyme, to an E2 conjugating enzyme and finally in complex with an E3 ligase to the target substrate (5, 6). At least 40 different missense and deletion mutations in the parkin gene have been identified and correlated to dysfunction of the ubiquitination system, manifesting itself as ARJP due to the loss of the dopaminergic neurons (7).
Parkin is a 465-residue protein comprising an N-terminal ubiquitin-like domain, a unique Parkin-specific domain in the central region, and two RING finger domains (RING1, RING2) separated by an in-between ring (IBR) or double ring linked domain near its C terminus. The cysteine and histidine rich RING-IBR-RING (RBR) domain architecture is highly conserved and found only in eukaryotes. To date, all characterized proteins containing the RBR supradomain have been found to possess E3 ligase activity (8). Based on sequence analysis, each RING domain in Parkin is expected to bind two zinc ions in a cross-brace fashion using a C3HC4 motif. However, the recently solved structure of the human homologue of Drosophila Ariadne (HHARI) RING2 domain indicates only a singly bound zinc ion (9), suggesting that some diversity exists within these domains. The RING fingers in Parkin have been shown to mediate protein–protein interactions with the E2 conjugating proteins UbcH7 and UbcH8 that are required for ubiquitination. Consistent with its role in ARJP, Parkin is able to ubiquitinate several substrates required for the proper functioning of dopaminergic neurons. These include unfolded Pael-R (10), O-glycosylated α-synuclein (11), synphilin-1 (12), α-synaptotagmin XI (13), Sept5 (4), and the transcription factor SIM2 (14) among others. Furthermore, ubiquitination of these proteins has been shown to require the C terminal RBR domain of Parkin.
The Parkin IBR domain assists in the recruitment of proteins involved in the ubiquitination pathway. For example, Parkin complexes formed with either of the E2 proteins UbcH7 or UbcH8 are greatly enhanced by the presence of the IBR domain. In the ubiquitination of Sept5, a protein associated with synaptic vesicles, the IBR-RING2 domain accentuates UbcH8 binding and activity (4). Similarly, the same effect is observed in the binding of synphilin-1 to Parkin (12). In addition to stabilizing and perhaps controlling E2 and substrate interactions, the IBR domain is required to maintain proper RBR domain geometry crucial in recruiting the chaperone Hsp70 (15), a necessary component in many ubiquitination complexes for the presentation of unfolded substrates. In vitro studies with the RBR E3 ligase HHARI have found that UbcH7 binding and subsequent ubiquitination requires both the RING1 domain and the N-terminal portion of the IBR (16).
Despite the importance of Parkin's IBR domain in the ubiquitin-mediated proteolysis pathway, there is little known about its structure and how this relates to ubiquitination or interactions with E2 conjugation enzymes or substrates. In this work, we determined the three-dimensional structure of the isolated IBR domain from Parkin using NMR spectroscopy. The structure shows that the IBR possesses two zinc-binding sites that adopt a dual scissor-like and GAG knuckle-like fold. Furthermore, zinc binding is required for the correct folding of the domain as substitution of zinc coordinating residues (C332S, C365S) causes its global unfolding. The missense T351P mutation found in patients suffering from ARJP (17) causes the global unfolding of the IBR domain, whereas the mutation R334C (18) causes some structural rearrangement. Together with previous biochemical data, the structure of the IBR domain has allowed a model to be proposed where the IBR domain orients the adjacent RING1 and RING2 domains to facilitate protein interactions and subsequent ubiquitination.
Results and Discussion
There have been no previous reports of a successful purification of recombinant full-length Parkin or any of its RING1, IBR or RING2 domains. To identify the possible boundaries of the IBR domain, Pfam and SMART databases were explored. Using these as a starting point, a 78-residue stretch from Parkin (residues M307–S384) comprising the IBR domain (IBR307–384) was successfully expressed from Escherichia coli as a soluble protein (2–5 mg per liter of culture). Other constructs of various lengths and starting points either did not express at high levels or were found to be insoluble upon purification.
Parkin IBR Folding Depends on Zinc Binding.
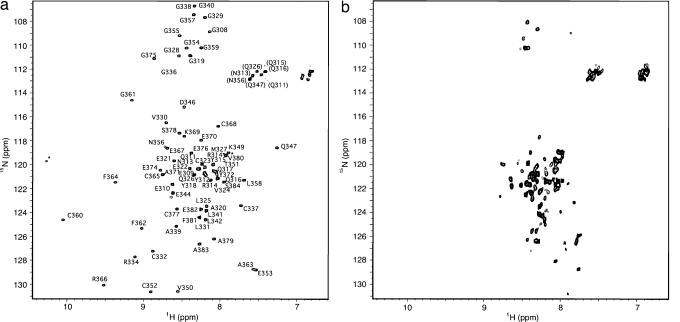
The 1H-15N HSQC of freshly purified IBR307–384 showed distinct and well dispersed peaks (Fig. 1a) indicative of a well folded domain for most of this sequence. Size exclusion chromatography indicated the protein was monomeric when compared with molecular weight standards [supporting information (SI) Fig. 6]. NMR assignments by conventional triple-resonance techniques showed that residues M307-Y318 of IBR307–384 clustered near the center of the 1H-15N HSQC spectrum (15N ≈ 120 ppm, 1H ≈ 8.2 ppm), typical of an unfolded or highly disordered polypeptide. In addition, these residues lacked any long-range nOes in either the 15N-NOESY and 13C-NOESY experiments.
Fig. 1.
Shown are 600 MHz 1H-15N HSQC spectra of Parkin IBR307–384. (a) Folded 0.2 mM IBR307–384 in 25 mM phosphate buffer (pH 6.95), 100 mM NaCl, 1 mM DTT at 25°C. (b) Spectrum of IBR307–384 from a upon EDTA addition showing the protein becomes unfolded. Peaks resulting from the leader sequence, an artifact of the cloning process, are indicated by an asterisk.
Analysis of IBR307–384 by inductively coupled plasma mass spectrometry and electrospray mass spectrometry indicated that the IBR domain (Fig. 1a) contained two bound zinc ions. This finding was confirmed by competitive binding experiments using media supplemented with 200 μM CdCl2 and showed that all three metal-bound species (2 Zn, 1 Zn/1 Cd, 2 Cd) could be identified by mass spectroscopy. However, attempts to introduce 113Cd into purified IBR307–384 by a variety of methods to perform 113Cd-NMR spectroscopy were not successful. Addition of EDTA to IBR307–384 caused the 1H-15N HSQC spectrum to collapse into a group of poorly dispersed resonances (Fig. 1b). Similar observations have been made for the zinc-binding RING domains from HHARI (9) and the RING-H2 domain from the rice protein EL5 (19), which are also unfolded in the absence of zinc. This indicated that binding of two Zn2+ ions is required for the correct folding of the IBR307–384 domain.
Parkin IBR Contains Two Zinc-Binding Domains.
A total of 95.9% of the proton, nitrogen, and carbon chemical shifts were assigned and used to calculate the three-dimensional structure of IBR307–384. This resulted in a total of 1,263 distance restraints derived from 13C-edited, 15N-edited, and 1H-1H NOESY spectra (SI Table 1). Initial structures were calculated in the absence of Zn2+ ion restraints to determine the overall fold of the domain. In agreement with our inductively coupled plasma mass spectrometry results, examination of the lowest energy structures from these calculations showed two separate well formed zinc sites, indicating that the side chains of C332, C337, C352, and C360 (site I), and C365, C368, H373, and C377 (site II) were involved in zinc coordination. The structures showed these two sites were well separated so that no other coordination possibilities existed. In addition, the algorithm by Kornhaber et al. (20) showed that six of the seven coordinating cysteine residues had a zinc-ligation probability >90% based on their Cα and Cβ chemical shifts. A final family of 20 structures was subsequently calculated by using additional distance restraints between the zinc ions and coordinating residues. None of the residues in the set of final structures were found in the disallowed region of the Ramachandran plot.
The IBR307–384 domain from Parkin adopts a bilobal fold about the two zinc-binding sites (Fig. 2). This arrangement brings the N terminus of the domain within close proximity to its C terminus. Although the structure shows no obvious α-helical or β-sheet secondary structural elements, as supported by chemical shift index calculations and circular dichroism experiments (not shown), the IBR domain was well defined between residues M327 and S378 (rmsd = 0.83 ± 0.17 Å). This fold extends just beyond the consensus sequence originally defined in IBR/double ring linked domain sequence alignments (21). The N-terminal zinc-binding lobe (site I) of the IBR structure is formed by four ribbons that form two elongated loops (L1: G329–L342; L2: V350–A363) arranged such that L1 and L2 lie roughly perpendicular to each other (Fig. 2a). The two loops are stabilized through the tetrahedral coordination of a Zn2+ ion sandwiched between them that utilizes C332 and C337 from L1 and C352 and C360 from L2 resulting in a “scissor-like” configuration with the metal ion at the fulcrum (Fig. 2b). The structure of the loops, excluding the tips beyond the coordinating cysteines, was significantly better defined (rmsd = 0.43 ± 0.10 Å) than the entire domain and was supported by nOes between the loops from the hydrophobic residues V330, A339, and L341 in L1 to C352 and V350 in L2. A large number of glycine residues in the L2 loop tip (G354, G355, G357, G359) provided minimal long-range contacts, indicating that this region was more flexible than the remaining portion of the site I zinc-binding site. The second Zn2+-binding region (site II) is considerably more compact than site I, encompassing only 14 residues (F364–C377). In this site, the polypeptide chain wraps around the metal ion, which is coordinated by the side chains of C365, C368, H373, and C377 toward the C terminus of the IBR domain. As with the first zinc-binding site, this site is considerably better defined (rmsd = 0.11 ± 0.04 Å) than the full domain. The structure of site II was supported by a large number of nOes observed between all of the zinc binding residues. The N-terminal 16 residues (M307–E322) and the C-terminal seven residues (S378–S384) of the IBR domain were poorly structured (not shown in Fig. 2), in agreement with the clustering of their resonances in the 1H-15N HSQC spectrum.
Fig. 2.
The structure of the Parkin IBR307–384 domain. (a) Backbone superposition of 20 structures having the lowest target function from CYANA. The superposition used residues G329–C332, C337–C352, and C360–S377 resulting in a backbone rmsd of 0.41 ± 0.09 Å. Zinc-binding site I comprises the extended loop regions L1 (blue) and L2 (green), and zinc site II is shown in red. (b) Ribbon diagram of Parkin IBR showing the position of the coordinating zinc residues. The unstructured N- (307–319) and C-terminal (378–384) regions are not shown.
Parkin IBR Is a Template for Other IBR-Containing Proteins.
The IBR domain is a common sequence found in hundreds of E3 ligase proteins in all branches of eukaryotes. It contains two bound zinc ions, bound to C4 and C2HC ligands, the most highly conserved residues in all IBR homologs (Fig. 3). Although reminiscent of the C5(C/H)H2 sequence used by the type I B-Box domain found in the RBCC/TRIM (RING–one or two B-boxes–coiled coil domains) E3 ligase family of proteins (22), the zinc sites in the Parkin IBR domain are notably different (described below). The high degree of similarity and location between the ligands in Parkin and other IBR domain proteins indicate these proteins share a common arrangement for zinc coordination. Some variation exists for the number (2–6) of intervening residues between the first two cysteines of L1 (C332, C337) across the IBR domain family (Fig. 3b). In addition, Parkin is unique amongst the IBR-containing proteins as it contains a linker region between the second pair of cysteine residues (C352, C360) within the L2 loop that extends for three to five more residues than other IBR domains.
Fig. 3.
Conserved zinc coordination and core regions of IBR domains. (a) A ribbon structure showing the packing of conserved core hydrophobic residues in Parkin. The structure shows residues V330, A339, and L341 (L1) pack with residues V350 and F364 (L2) allowing formation of a scissor-like arrangement. Two peripheral residues, P343 and R366 provide auxiliary interactions whereas Y372 helps stabilize site II to site I. (b) Multiple sequence alignment of IBR domains from several human paralogs constructed from ClustalW (43) and Jalview (44). Underlined proteins interact with human UbcH7 and UbcH8, whereas RNF14 interacts with UbcH7 alone. Conserved metal binding ligands (yellow) and hydrophobic core residues (green) are highlighted. Residues where mutations have been found in ARJP (open stars) and mutated in this study (filled stars) are indicated.
The structure of Parkin IBR307–384 shows that a unique hydrophobic core is maintained (Fig. 3a) that is different from other domains such as RING, PHD, or LIM that typically coordinate two zinc ions (23). The Parkin structure shows that V330 near the N terminus of L1 makes hydrophobic contacts with L341 and P343 on the opposite site of the loop, and with F364, R366, and K369 near the first two coordinating cysteines (C365, C368) of site II. Analogously, V350 near the N terminus of L2 has numerous contacts with F364 and R366 across this loop and with L341 and P343 from L1. These two hydrophobic residues (V330 and V350) upstream of the initial cysteine residues in IBR are not observed in RING, PHD, or LIM domains (23). In IBR domains, a bulky aromatic residue (Y372) adjacent to the coordinating histidine (H373) of the second zinc-binding site makes numerous contacts with L2 residues (V350, K349, A363). From this analysis, it is clear that many of the nonpolar core residues identified in the core packing region of the Parkin IBR domain (V330, A338, L341, V350, F364, R366, and Y372) fold are conserved across all RBR proteins and are expected to form the core regions and direct the fold of other IBR domains in proteins such as HHARI and Dorfin (Fig. 3b).
Parkin IBR Forms a Dual Scissor-Like and GAG Knuckle-Like Structure.
There are several features that indicate the Parkin IBR domain has a unique architecture that has been assembled from other well-characterized zinc-binding motifs. However, the sequential binding of the two zinc ions by Parkin's IBR remains distinct from RING, PHD, and B-box domains that coordinate two zinc ions in a cross-brace fashion (first and third pair, second and fourth pairs of ligands) (24, 25). In contrast, the IBR domain in Parkin takes on “scissor-like” and GAG knuckle-like zinc-binding structures for sites I and II respectively. Site I resembles that of a zinc ribbon, one of the largest groups of zinc fingers that includes several of the subunits (rpb1, rpb2, rpb9, rpb12) from RNA polymerase II (1I50), the transcriptional elongation factor SII (1TF1) and the transcription initiation factor TFIIB (1PFT, 1DL6). For example, superposition of the extended regions for L1 and L2 near the four coordinating cysteines of Parkin (V330–C332, C337–A339, V350–C352, C360–F362) results in a backbone rmsd of 1.69 Å with the analogous regions in the RNA polymerase II subunit rpb12, 1.83 Å with the transcription initiation factor TFIIB and 1.28 Å with the putative kinase CV3345 (SI Fig. 7a). Distinctly, the longer “tips” of the L1 and L2 loops are evident giving rise to the “scissor-like” arrangement of Site I (Fig. 2 and SI Fig. 7a). More importantly, several of the hydrophobes near the fulcrum of the scissor structure are conserved between the domains including V330 (L9 in CV3345, Y29 in rpb12, V13 in TFIIB), L341 (L18 in CV3345, L38 in rpb12, I22 in TFIIB) and V350 (L34 in CV3345, V46 in rpb12, M32 in TFIIB), further supporting the similarity of this portion of the structures.
Site II of the Parkin IBR domain closely resembles the GAG knuckle retroviral nucleocapsid proteins from MMTV (1DSV), HIV (1MFS), and MPMV (1CL4), which display backbone rmsds ranging from 1.42 to 1.97 Å for C365 to H373 of Parkin (SI Fig. 7b). The GAG knuckle is formed by two cysteines of a rubredoxin type turn followed by the third and fourth ligands flanking a one turn helix (26). The CX2C spacing is invariant in all IBR domains (Fig. 3b), and the Parkin IBR superimposes well with a backbone rmsd of 0.34 Å with rubredoxin (5RXN) in this region. The turn also possesses signature hydrogen bonds (E367HN and C368HN with the C365 thiol, and E370HN with the C368 thiol) that are typical of members of this zinc knuckle family. The Parkin IBR has HX3C spacing forming a short loop in place of the one-turn helix found in the nucleocapsid proteins, but this is unusual among most IBR family members that usually possess four residues between the last two ligands (Fig. 3b).
Mutations Observed in ARJP Cause Misfolding of the IBR Domain.
The onset of ARJP has been correlated with patients possessing one or more missense or truncation mutations in Parkin. In particular, several missense mutations, G328E, R334C, T351P, and V380L, in the IBR domain have been identified (17, 18). To probe the affect of mutations on the structure and folding of the IBR domain, we studied several proteins containing the disease correlated mutations (G328E, R334C, and T351P), and substitutions to the zinc-binding residues (C332S and C365S) by 1H NMR spectroscopy. Of these constructs, the protein with the C365S mutation was very poorly expressed and insoluble (not shown), suggesting that this protein adopts an unfolded structure. The C332S substitution also caused unfolding of the entire IBR domain based on analysis of 1H NMR spectra (Fig. 4, see below). Three representative regions of the spectra were chosen to analyze the affect of each mutation. In the parent IBR domain, the most downfield region (Fig. 4a) displays well resolved amide resonances for several of the zinc-coordinating residues (C332, C352, C360) and key residues in the hydrophobic core (F362, F364, R366). These are not evident in the unfolded C332S protein. The aromatic section of the spectrum (Fig. 4b) displays peaks for Y372, one of the anchoring residues in site II as well as the zinc-ligating residue H373. Finally, the upfield aliphatic region (Fig. 4c) shows the unique side chain resonances for K349(β1, γ1, γ2) and V350(γ1, γ2) in site I due to their proximity to the aromatic rings of Y372 and F364, respectively, in the IBR structure.
Fig. 4.
Selected regions of the 600 MHz 1H NMR spectra of Parkin IBR307–384 and missense mutations found in patients suffering from ARJP. Residues are labeled for the downfield amide region (a), aromatic region (b), and upfield aliphatic methyl proton region (c). Spectra were scaled to provide approximately equal peak intensities. A residual peak from the internal standard DSS is indicated with an asterisk.
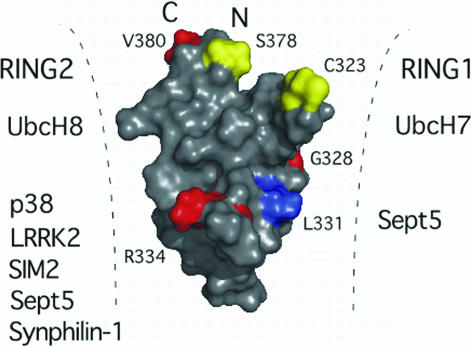
The mutation proteins G328E and R334C display many similar resonances as the parent IBR domain, indicating that the general IBR fold is preserved in these two proteins. This finding is consistent with the locations of these two residues in the IBR structure, as they do not contribute to the hydrophobic core of the protein. Interestingly, the R334C mutant has new resonances in the amide region of its spectrum (Fig. 4a) that are coincident with the disappearance of R334, and shifts of G361 and F362 amide protons. Further changes are noted for V350(γ1,γ2), one of the bridging residues between the two zinc sites. The remote nature of R334 from G361, F362, and V350 indicates that a structural rearrangement has occurred. We cannot discount the possibility that the R334C protein contributes an extra zinc ligand that might compete with the neighboring coordinating residue (C332). Nevertheless, both G328 and R334 are on the surface of the L1 loop of the IBR structure neighboring a highly conserved hydrophobic residue (L331) that is also exposed (Fig. 5). The alteration of this region (R334C), or change in its surface properties (G328E), likely interferes with protein interactions across the L1 face. In support of this idea, the G328E and R334C mutations result in decreased binding and ubiquitination of the Parkin substrates synphilin-1 and p38 (27). Furthermore, the binding of synaptic vesicle protein Sept5, synphilin-1, the transcription factor SIM2 and leucine-rich kinase 2 are each amplified with IBR-RING2 tandem domains compared with the RING2 domain alone (4, 12, 14, 28). Together with the Parkin IBR structure, this indicates that G328, R334, and likely L331 on the L1 loop form a portion of the interaction site for these substrates, which must be near the RING2 domain to facilitate interactions (Fig. 5).
Fig. 5.
Proposed Parkin interactions and arrangement of IBR and RING domains. Surface representation of the IBR structure showing the positions of ARJP mutations (G328E, R334C, V380L) is shown in red. Two of these mutations (G328E, R334C), and the conserved residue L331 (blue) lie along the L1 loop and result in decreased binding and ubiquitination of substrates such as synphilin-1 and p38. Other substrates are listed based on biochemical experiments that indicate interaction with either the RING1–IBR or IBR–RING2 domain pairs. Residues where S-nitrosylation is proposed (C323) or phosphorylation has been identified (S378) are shown in yellow. These residues, along with the missense mutation site V380 lie at the crown of the IBR domain where the N and C termini are close in space bringing the RING1 and RING2 domains within close proximity to each other and the IBR domain.
The mutations T351P and C332S resulted in 1H NMR spectra with many broad or missing peaks consistent with a global unfolding of the IBR domain. In particular, neither of these mutants had any downfield-shifted amide resonances for C332, C352, C360, or H373 indicative of zinc-binding to both sites in the IBR domain. Although T351 does not directly contact any residues on the interface with site II, the change to a proline may affect the backbone topology resulting in changes to the proximal core V350 and the zinc binding C352 residues. The T351 methyl also makes key contacts to E353 and G361 keeping the L2 loop intact. A global unfolding of the Parkin by the T351P mutant would dramatically alter its biological properties. For example, the Parkin IBR structure reveals a proximal arrangement of the N and C termini of the domain that would normally be preceded by the RING1 and followed by the RING2 domains, respectively. The proximity of the N and C termini in the IBR structure suggests a role for this domain is to bring the two flanking RING domains close together (Fig. 5), similar to that observed in the heterodimeric association of the BRCA/BARD1 domains that form an E3 ligase complex (29). This arrangement would facilitate protein interactions with either RING domain and/or the IBR domain with the E2-conjugating enzymes UbcH7 or UbcH8. In support for this model, coimmunoprecipitation experiments show the E2 proteins UbcH7 and UbcH8 bind to the RING1 and RING2 domains respectively facilitating ubiquitination. In both cases, the IBR domain greatly augments this interaction (4). For the RBR protein HHARI, the N-terminal region of the IBR domain and RING1 domain are required for interaction with UbcH7 (30). Based on the enhancement of UbcH7, UbcH8 and substrate binding (synphilin-1, Sept5, SIM2) in the presence of the IBR domain it is clear the T351P mutation would not only disrupt the binding site on the IBR domain surface but would spatially separate the RING1 and RING2 domains due to IBR unfolding. This idea, in combination with the Parkin IBR structure also agrees with the negative modulation of Parkin by either S-nitrosylation or phosphorylation (31–33). The only free thiol in the IBR domain (C323) a potential nitrosylation site, and the residue for phosphorylation (S378) are proximal to each other where the N and C termini meet near the edge of zinc site II. The Parkin mutant V380L is also located in this area (Fig. 5 Upper). It is clear that phosphorylation, nitrosylation, or mutation in this region would modify the arrangement of the N and C termini of the IBR domain and its geometry with respect to the RING1 and RING2 domains affecting its ubiquitination abilities.
In summary, this work has shown that the Parkin IBR domain is a zinc-dependent folding unit with preserved metal ion coordination and hydrophobic core. The IBR structure reveals that it is likely responsible for stabilizing the overall geometry and orientation of the RING domains within the Parkin protein. Furthermore, the sequence similarity of Parkin to other IBR domains should allow it to serve as a template for understanding the IBR role in ARJP and its function in other RING–IBR–RING containing proteins.
Methods
Protein Purification.
A human fetal brain serum cDNA library (ATCC, Manassas, VA) was used to clone the Parkin IBR domain (M307–S384) into a modified pET15b vector having a TEV cleavage site. Single site mutations were introduced by using the QuikChange (Stratagene, La Jolla, CA) protocol. Proteins for NMR experiments were expressed by using the E. coli BL21(DE3) bacterial strain in M9 media supplemented with 15NH4Cl (1 g/liter), 13C-glucose (2 g/liter) and ZnCl2 (200 μM) just before induction at 15°C. Cells were resuspended in buffer (25 mM Tris, 100 mM NaCl, 5% glycerol, and 5 mM imidazole, pH 7.5) and lysed by French press, and the resulting lysate incubated and then eluted from Ni2+-resin (Qiagen, Mississauga, ON, Canada). The IBR307–384 protein was purified through a second Ni2+-affinity column after hexahistidine tag removal leaving two amino acids (GH) as an artifact on the N terminus. Proteins were dialyzed against 25 mM Na2HPO4 pH 6.95, 100 mM NaCl, and 1 mM DTT and concentrated to 0.2–0.9 mM by ultrafiltration (Millipore, Mississauga, ON, Canada) for NMR experiments.
NMR Spectroscopy.
NMR experiments were acquired on a Varian Inova 600 MHz spectrometer using a xyz-gradient triple-resonance probe. Standard pulse sequences from the Varian BioPack package were used. NOE-derived distance restraints were obtained from 15N-NOESY (150 ms mixing time), 13C-NOESY (150 ms), and 2D 1H-1H NOESY (50 ms) spectra. HNHA experiments were used to measure 3JHNHα coupling constants to obtain φ angle restraints (34, 35). NMR data were processed by using NMRPipe (36) and analyzed by using NMRView (37).
NMR Structure Calculations.
Structures from manually assigned NOEs were determined by using the program CYANA (38). A total of seven iterations of refinement of 200 structures per cycle were completed by using distance calibrations and parameterization as described (39). The 20 structures with the lowest calculated target function from each cycle were used to seed the next round and used by CYANA for further automated NOE assignment. φ and ψ angular restraints, based on neighboring αCH, HN NOE intensities (40) and zinc ion distance and angular constraints (41, 42) were added once the initial protein fold was established. A CYANA amino acid library using a modified zinc-ligated cysteine residue was used to incorporate zinc into the structures. The final 20 structures with the lowest target function were chosen as representative of the calculation, although structures with higher target values had identical folds.
Acknowledgments
We thank Dr. Matt Revington (University of Western Ontario) for advice with NMR experiments and structure calculations, Dr. R. Jane Rylett (University of Western Ontario) for many helpful discussions, and Kathryn Barber (University of Western Ontario) for critical reading of this manuscript. This research was supported by the Canadian Institutes of Health Research and the Canada Research Chairs Program (to G.S.S.).
Abbreviations
- PD
Parkinson's disease
- ARJP
autosomal recessive juvenile PD
- IBR
in-between ring.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates reported in this study have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2JMO).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610548104/DC1.
References
- 1.Warner TT, Schapira AH. Ann Neurol. 2003;53(Suppl 3):S16–S23. doi: 10.1002/ana.10487. discussion S23–S25. [DOI] [PubMed]
- 2.Betarbet R, Sherer TB, Greenamyre JT. Exp Neurol. 2005;191(Suppl 1):S17–S27. doi: 10.1016/j.expneurol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka K, Suzuki T, Hattori N, Mizuno Y. Biochim Biophys Acta. 2004;1695:235–247. doi: 10.1016/j.bbamcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 7.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 8.Marin I, Lucas JI, Gradilla AC, Ferrus A. Physiol Genomics. 2004;17:253–263. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- 9.Capili AD, Edghill EL, Wu K, Borden KL. J Mol Biol. 2004;340:1117–1129. doi: 10.1016/j.jmb.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 10.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 11.Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 12.Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. Nat Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 13.Huynh DP, Scoles DR, Nguyen D, Pulst SM. Hum Mol Genet. 2003;12:2587–2597. doi: 10.1093/hmg/ddg269. [DOI] [PubMed] [Google Scholar]
- 14.Okui M, Yamaki A, Takayanagi A, Kudoh J, Shimizu N, Shimizu Y. Exp Cell Res. 2005;309:220–228. doi: 10.1016/j.yexcr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Tsai YC, Fishman PS, Thakor NV, Oyler GA. J Biol Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 16.Moynihan TP, Ardley HC, Nuber U, Rose SA, Jones PF, Markham AF, Scheffner M, Robinson PA. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- 17.von Coelln R, Dawson VL, Dawson TM. Cell Tissue Res. 2004;318:175–184. doi: 10.1007/s00441-004-0924-4. [DOI] [PubMed] [Google Scholar]
- 18.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, et al. N Engl J Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 19.Katoh S, Hong C, Tsunoda Y, Murata K, Takai R, Minami E, Yamazaki T, Katoh E. J Biol Chem. 2003;278:15341–15348. doi: 10.1074/jbc.M210531200. [DOI] [PubMed] [Google Scholar]
- 20.Kornhaber GJ, Snyder D, Moseley HN, Montelione GT. J Biomol NMR. 2006;34:259–269. doi: 10.1007/s10858-006-0027-5. [DOI] [PubMed] [Google Scholar]
- 21.van der Reijden BA, Erpelinck-Verschueren CA, Lowenberg B, Jansen JH. Protein Sci. 1999;8:1557–1561. doi: 10.1110/ps.8.7.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meroni G, Diez-Roux G. BioEssays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 23.Matthews JM, Sunde M. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 24.Maret W. J Trace Elem Med Biol. 2005;19:7–12. doi: 10.1016/j.jtemb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Massiah MA, Simmons BN, Short KM, Cox TC. J Mol Biol. 2006;358:532–545. doi: 10.1016/j.jmb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Krishna SS, Majumdar I, Grishin NV. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. Hum Mol Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- 28.Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Proc Natl Acad Sci USA. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 30.Ardley HC, Tan NG, Rose SA, Markham AF, Robinson PA. J Biol Chem. 2001;276:19640–19647. doi: 10.1074/jbc.M011028200. [DOI] [PubMed] [Google Scholar]
- 31.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 32.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, et al. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto A, Friedlein A, Imai Y, Takahashi R, Kahle PJ, Haass C. J Biol Chem. 2005;280:3390–3399. doi: 10.1074/jbc.M407724200. [DOI] [PubMed] [Google Scholar]
- 34.Pardi A, Billeter M, Wuthrich K. J Mol Biol. 1984;180:741–751. doi: 10.1016/0022-2836(84)90035-4. [DOI] [PubMed] [Google Scholar]
- 35.Karplus M. J Chem Phys. 1959;30:11–15. [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BA, Blevins RA. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Guntert P, Mumenthaler C, Wuthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 39.Herrmann T, Guntert P, Wuthrich K. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 40.Gagne SM, Tsuda S, Li MX, Chandra M, Smillie LB, Sykes BD. Protein Sci. 1994;3:1961–1974. doi: 10.1002/pro.5560031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuhaus D, Nakaseko Y, Schwabe JW, Klug A. J Mol Biol. 1992;228:637–651. doi: 10.1016/0022-2836(92)90846-c. [DOI] [PubMed] [Google Scholar]
- 42.Dempsey BR, Wrona M, Moulin JM, Gloor GB, Jalilehvand F, Lajoie G, Shaw GS, Shilton BH. Biochemistry. 2004;43:9361–9371. doi: 10.1021/bi0493057. [DOI] [PubMed] [Google Scholar]
- 43.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clamp M, Cuff J, Searle SM, Barton GJ. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]