Abstract
A large fraction of pediatric pre-B acute lymphoblastoid leukemias (ALL) consistently contain a t(1;19) chromosomal translocation. The t(1;19) translocation results in the production of a chimeric transcription factor containing the N-terminal transactivation domain of E2A fused to the C-terminal DNA-binding homeodomain of Pbx1. Here, we show that the E2A-Pbx1 fusion protein activates the expression of a novel WNT gene, WNT-16. WNT-16 normally is expressed in peripheral lymphoid organs such as spleen, appendix, and lymph nodes, but not in bone marrow. In contrast, high levels of WNT-16 transcripts are present in bone marrow and cell lines derived from pre-B ALL patients carrying the E2A-Pbx1 hybrid gene. Inhibition of E2A-Pbx1 expression leads to a significant decrease in WNT-16 mRNA levels, suggesting that WNT-16 is a downstream target of E2A-Pbx1. Three putative WNT receptors, FZ-2, FZ-3, and FZ-5, are expressed in cells of the B lineage, including pre-B ALL cells aberrantly expressing WNT-16. We propose that a WNT-16-mediated autocrine growth mechanism contributes to the development of t(1;19) pre-B ALL.
Human acute leukemias frequently contain chromosomal translocations that involve transcription factors (1). In many cases, these transcription factors are homologs of evolutionarily conserved proteins that have been shown to regulate the expression or target specificity of the homeotic selector (Hox) proteins. Further evidence for a role of Hox proteins in hematopoiesis comes from studies in which mice were reconstituted with bone marrow stem cells programmed to overexpress individual Hox proteins (2, 3). Each Hox protein showed dramatically different, lineage-specific effects on the proliferation and differentiation of hematopoietic stem cells or committed progenitors (2, 3). Together, these findings indicate that aberrant regulation of Hox genes may play a critical role in many types of acute leukemia.
In pediatric acute lymphoblastoid leukemia (ALL) containing the t(1;19) chromosomal translocation, the N-terminal transactivation domain of the basic helix–loop–helix transcription factor E2A is fused to the homeodomain of Pbx-1 to create a chimeric transcription factor, E2A-Pbx1 (4, 5). There are three closely related Pbx genes in mammals: Pbx-1, Pbx-2, and Pbx-3 (6). During lymphoid development Pbx-2 and Pbx-3 are expressed at all stages. In contrast, Pbx-1 is not expressed in the B and T cell lineages. The Pbx proteins belong to the TALE (three-amino acid extension) family of homeodomain proteins, which is characterized by a 3-aa insertion between helix 1 and helix 2 of the homeodomain (7).
Pbx proteins have the ability to form complexes with Hox proteins, thereby increasing their DNA-binding affinity and specificity (8–11). Hox and Pbx proteins act in concert to regulate common downstream target genes (12), as has been shown for autoregulation of the Hoxb-1 gene in rhomobomere 4 of the mammalian hindbrain and activation of the somatostatin gene in pancreatic islet cells by the orphan homeobox protein STF-1 (12, 13).
Mutations in extradenticle (exd), the Drosophila homolog of the Pbx family, result in homeotic transformations (14, 15). Two target genes that are regulated by exd encode secreted intercellular signaling proteins, wingless/Dwnt-1 (wg) and decapentaplegic (dpp), which belong to the Wnt and TGF-β/BMP gene families, respectively (16). The Wnt family has grown to 21 vertebrate members, 13 of which have been identified in humans (17). Wnt genes encode cysteine-rich proteins with typical hydrophobic signal sequences at their amino-terminal ends. It is likely that all Wnt proteins are secreted and mediate interactions between cells. The first member of this family, Wnt-1, originally was identified as a common site of integration in mouse mammary tumor virus-induced mouse mammary carcinomas (18). Intracellular components of the Wnt-signaling pathway also have been implicated in human cancers such as colon carcinoma and melanoma (19–22).
E2A-Pbx1 exhibits multiple transforming properties. In NIH 3T3 cells, it has the ability to induce the formation of foci, anchorage-independent growth, and tumor formation in nude mice (23). In thymocytes and myeloid cells, aberrant expression of E2A-Pbx1 promotes the rapid development of T cell lymphomas and myeloid leukemias (24, 25). Although Pbx1 has no detectable transactivating activity by itself, E2A-Pbx1 has strong transactivation potential (26, 27). This has led to the proposal that the oncogenic activity of E2A-Pbx-1 is caused by the inappropriate activation of genes normally regulated by Pbx/Hox complexes (28).
Here, we have used representational difference analysis (RDA) to identify genes up-regulated by E2A-Pbx1 in pre-B ALL. One of these genes encodes a novel member of the WNT family. We propose that the aberrant expression of WNT-16 in pre-B cells is a key step toward the development of t(1;19)-positive leukemia.
MATERIALS AND METHODS
Cell Culture.
Human leukemic cell lines were grown in Opti-MEM I (GIBCO/BRL) supplemented with 10% FBS/50 units/ml penicillin/50 μg/ml streptomycin/2 mM l-glutamine/50 μM 2-mercaptoethanol.
RDA of cDNAs.
Total RNA was isolated from 697, KOPN-36, and Nalm-6 cells by using Trizol reagent (GIBCO/BRL). Poly(A)-positive RNA was purified from total RNA by using oligo(dT)-conjugated magnetic beads (Dynal). Double-stranded cDNAs were synthesized by using the Superscript II cDNA synthesis kit according to the manufacturer’s protocol (GIBCO/BRL). First-strand cDNA synthesis was primed with oligo(dT) by using 5 μg of poly(A)+ RNA. RDA of cDNAs was performed as described by Hubank and Schatz (29). The third difference products were isolated on a 2.5% agarose gel and cloned into vector pGEM-T (Promega).
Synthesis of cDNA Probes.
Double-stranded cDNA probes were labeled with [α-32P]dCTP by using Klenow fragment and random primers. The asymmetric PCR probe for human WNT-16 was made by using an 848-bp fragment of the WNT-16 cDNA (nucleotides 573-1421) as template and the primer 5′-GCACCTCTCTGGTTGCAATGC-3′. The asymmetric PCR probe for mouse WNT-16 was made by using a 647-bp fragment of mouse WNT-16 genomic DNA (corresponding to the 5′ end of exon 4) as template and the primer 5′-GCAATCCTCTGGATCAGCTTGTG-3′. The asymmetric PCR probe for E2A-Pbx1 was made by using an 826-bp fragment of the Pbx1 cDNA (GenBank accession no. M86546, nucleotides 578-1404) as template and the primer 5′-ATCAGAGTGAACACTGCCAG-3′. Asymmetric PCRs contained 50 μM each of dATP, dGTP, and dTTP, 250 μCi [α-32P]dCTP, 50 ng of DNA template, 1 μM primer, 2.5 units of Taq DNA polymerase, and 1× PCR buffer (50 mM KCl/10 mM Tris⋅HCl, pH 8.3/1.5 mM MgCl2) in a volume of 25 μl. PCR conditions were 1 cycle at 2 min/91°C, 30 cycles at 1 min/91°C, 1 min/58°C, 1.5 min/72°C, and 1 cycle at 10 min/72°C. Probes were purified over G-50 sephadex spin columns.
Southern and Northern Blotting.
PCR products were separated on 2.5% agarose gels and denatured. RNAs were separated on denaturing formaldehyde-agarose gels. The PCR products or RNAs were transferred to nylon membranes and immobilized by UV crosslinking. Human and mouse Multiple Tissue Northern blots were purchased from CLONTECH. Hybridizations were done overnight at 42°C in 5× SSPE buffer, 50% formamide/10% dextran sulfate/1× Denhardt’s reagent/0.15% SDS/100 μg/ml denatured salmon sperm DNA. Washes were done at 65°C in 0.2× SSPE buffer/0.1% SDS.
Reverse Transcription–PCR (RT-PCR).
To analyze expression of WNT-16 in bone marrow samples, RT was performed according to the manufacturer’s protocol in a 25-μl reaction containing 50 ng of random hexamers, 3 μg of total RNA, and 200 units of Superscript II RNase H− reverse transcriptase (GIBCO/BRL). For WNT-16, the PCR was performed by using forward primer 5′-CAGGGACACAAGGCAGAGAATG-3′ and reverse primer 5′-GCTGGATGGAGTGGTTACTT-3′. PCR conditions were 1 cycle at 5 min/94o, 18 cycles at 45 sec/94o, 45 sec/58o, 45 sec/72o, and 1 cycle at 10 min at 72o. GAPDH primers for RT-PCR were purchased from Stratagene, and PCR conditions were 1 cycle at 5 min/94o, 18 cycles at 45 sec/94o, 45 sec/60o, 90 sec/72o, and 1 cycle at 10 min/72o.
Isolation of the WNT-16 cDNA.
A cDNA library was prepared in vector Uni-ZAP XR (Stratagene) by using poly(A)+ RNA isolated from 697 cells. The library was screened with a 400-bp DpnII fragment (nucleotides 1104–1504 of cDNA) of WNT-16 cDNA as probe. Sixteen positive clones were isolated, and one, a 3,129-bp, full-length cDNA, was sequenced on both strands.
Isolation of Genomic Clones for Mouse WNT-16.
A 129/SVJ mouse BAC genomic library (Genome Systems, St. Louis) was screened by hybridization with a 185-bp fragment of the human WNT-16 cDNA (nucleotides 1165–1350). Two positive clones were isolated. Restriction fragments that cross-hybridized with the human WNT-16 cDNA on Southern blots were subcloned into pBluescript SK (Stratagene) and sequenced to identify the putative mouse WNT-16 exons.
Isolation of Frizzled (Fz) cDNA Clones and RNase Protection.
Fz homologs expressed in the t(1;19) pre-B ALL cell line 697 were identified by degenerate PCR amplification as described by Wang et al. (30). Briefly, degenerate oligonucleotide primers corresponding to the conserved Fz amino acid sequences YPERPI and WFLAA were used to amplify cDNA prepared from the cell line 697. The PCR products were cloned into vector pGEM-T (Promega) and screened by sequencing. Four Fz homologues were identified. Antisense riboprobes were synthesized by transcribing the pGEM-T Fz constructs in vitro with [α-32P]CTP and SP6 RNA polymerase. Antisense RNA probe (2 × 105 cpm) was annealed to 10 μg of total RNA from the indicated cell lines or to 10 μg of yeast tRNA as a negative control. Unannealed RNA was removed by digestion with RNase A and RNase T1. The protected fragments were separated by electrophoresis on a 5% denaturing polyacrylamide gel and detected by autoradiography.
Antisense Oligonucleotides.
The antisense oligonucleotides used in this study are chimeric 2′-methoxyethyl phosphorothioate oligonucleotides in which the first six and last six nucleosides are modified with 2′-methoxyethyl substituents and the eight, centered nucleosides contain 2′-deoxy residues (31, 32). The centered 2′-deoxy residues support RNase H activity in cells, which serves as the terminating mechanism for these molecules. Oligonucleotide sequences are as follows: ISIS16123, 5′-ACCAGGCTGACAGCTGGAGG-3′; ISIS18070 (4 mismatches), 5′-ACCTGGCAGTCAGCAGGAGG-3′; ISIS16124, 5′-CCTTCAGTGATATGAGAGAC-3′. 697 cells (8 × 106) were resuspended in 0.36 ml of culture medium plus 40 μl of oligonucleotide (200 μM in PBS) and electroporated in a 0.2-cm cuvette electrode at 1,125 V/cm and 500 μF. The cells were diluted into 10 ml of culture medium and incubated at 37o. Twenty hours after electroporation total RNA was isolated from the cells by using Trizol reagent (GIBCO/BRL) and analyzed by Northern blotting.
RESULTS
Identification of Downstream Target Genes Induced by E2A-Pbx1.
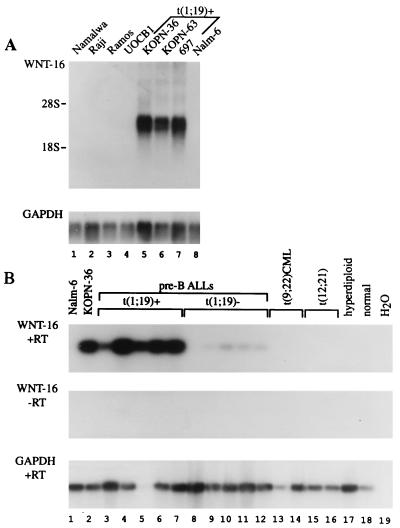
To identify genes that are differentially expressed in t(1;19)-positive pre-B ALL, mRNA was isolated from a pre-B cell line containing the E2A-Pbx1 fusion protein and Nalm-6, a pre-B cell line lacking E2A-Pbx1 expression. Double-stranded cDNAs were prepared from the two cell populations, and RDA, a process of subtractive hybridization coupled to PCR amplification, was performed on two pools of cDNAs to isolate transcripts that were expressed in cells containing E2A/Pbx-1 but not in pre-B cells lacking this fusion protein (28, 29). Two independent RDAs were performed by using two different t(1;19) cell lines: 697 and KOPN-36. When the Nalm-6 cDNA representation was used as driver and the 697 cDNA representation was used as tester in the RDA, the third difference product consisted of a number of fragments visible on an ethidium bromide-stained agarose gel. The fragments were isolated, cloned, and analyzed by DNA sequencing. The sequences were compared with the National Center for Biotechnology Information database by using the blast algorithm (33). Forty-two distinct cDNA fragments were identified. Comparison of the 697 and KOPN-36 difference products by sequence alignment revealed that six of the 42 fragments from 697 also were expressed in KOPN-36. To verify that the six genes were true difference products, the cDNA fragments were labeled with 32P and used to probe Northern blots. As expected, each of the genes was detectable in 697 and KOPN-36 but not in Nalm-6 (Fig. 1A and Table 1).
Figure 1.
WNT-16 expression in pre-B cell lines and bone marrow samples derived from leukemia patients. (A) Northern blot analysis of WNT-16 expression in B cell lines. Each lane contains 2 μg of poly(A)+ RNA. KOPN-36, KOPN-63, and 697 carry the E2A-Pbx1 fusion gene, whereas Namalwa, Raji, Ramos, UOCB-1, and Nalm-6 lack the t(1;19) chromosomal translocation. The probe was a human WNT-16 cDNA. (B) RT-PCR analysis of WNT-16 expression in bone marrow samples derived from patients. WNT-16-specific primers were used for PCR. PCR products were analyzed by Southern blotting and hybridization by using a WNT-16 cDNA as a probe. E2A-Pbx1-positive samples are indicated. (Middle) Minus RT controls. (Bottom) Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) controls.
Table 1.
Summary of gene expression profiles in t(1;19) vs. non-t(1;19) human leukemias
| RNA | Type of leukemia | Gene function
|
|||||
|---|---|---|---|---|---|---|---|
| WNT-16 | RZR-β | PKC-ξ | FAT | EphA3 | IRX-related | ||
| Intercellular signaling molecule | Nuclear receptor for melatonin | Serine/threonine protein kinase | Cadherin-related cell adhesion molecule | Receptor protein tyrosine kinase | TALE homeo domain protein | ||
| 697 | t(1;19) pre-B-ALL | + | + | + | + | + | + |
| KOPN-36 | t(1:19) pre-B-ALL | + | + | + | + | + | + |
| KOPN-54 | t(1;19) pre-B-ALL | + | + | + | + | + | − |
| KOPN-63 | t(1;10) pre-B-ALL | + | + | + | + | + | + |
| Reh | Non-t(1;19) pre-B-ALL | − | − | − | − | − | − |
| Nalm-6 | Non-t(1;19) pre-B-ALL | − | − | − | − | − | − |
| Namalwa | Burkitt’s lymphoma | − | − | − | − | − | − |
| Raji | Burkitt’s lymphoma | − | − | − | − | − | − |
| Ramos | Burkitt’s lymphoma | − | − | − | − | − | − |
| UOCB-1 | t(17;19) Pro-B-ALL | − | − | − | − | − | − |
| Jurkat | T-ALL | − | + | − | − | + | − |
| BM 1 | t(1;19) pre-B-ALL | ++ | + | ± | ++ | − | − |
| BM 2 | t(1;19) pre-B-ALL | + | + | ++ | ++ | + | + |
| BM 3 | t(1;19) pre-B-ALL | ++ | + | ± | + | − | + |
| BM 4 | t(1;19) pre-B-ALL | ++ | + | ++ | ++ | − | − |
| BM 5 | t(1;19) pre-B-ALL | ++ | + | ++ | ++ | ++ | − |
| BM 6 | Non-t(1;19) pre-B-ALL | − | − | ± | ± | − | + |
| BM 7 | Non-t(1;19) pre-B-ALL | ± | ± | ± | + | − | + |
| BM 8 | Non-t(1;19) pre-B-ALL | ± | − | ± | − | − | − |
| BM 9 | Non-t(1;19) pre-B-ALL | ± | − | ± | ± | − | − |
| BM 10 | Non-t(1;19) pre-B-ALL | ± | − | ± | − | − | + |
| BM 11 | t(9;22) CML | − | − | ± | − | − | − |
| BM 12 | t(9,22) CML | − | − | ± | − | − | − |
| BM 13 | t(12;21) pro-B-ALL | − | − | ± | − | − | − |
| BM 14 | t(12;21) pro-B-ALL | − | − | ± | − | − | − |
| BM 15 | Hyperdiploid | − | − | ± | ± | − | + |
| BM 16 | Normal cytogenetics | − | − | ± | − | − | − |
Gene expression was detected in leukemic cell lines by Northern blot analysis of poly(A)+ RNa and in patient bone marrow (BM) samples by RT-PCR analysis. Low levels of expression detectable by PCR but not by Northern or RNAse protection are indicated by ±. High levels of expression detected by PCR are indicated by + (10- to 20-fold above background) or ++ (20- to 100-fold above background).
To examine whether the six genes are expressed consistently in cells containing the E2A-Pbx1 fusion protein, mRNA was isolated from additional cell lines containing E2A-Pbx1. These include KOPN-54 and KOPN-63. Control lymphoid cell lines lacking E2A-Pbx1 include Reh, Namalwa, Raji, Ramos, UOCB-1, and Jurkat. This analysis showed that five of six genes were expressed in all four t(1;19) cell lines, but not in B cell lines lacking E2A-Pbx1 (Table 1, Fig. 1A, and data not shown). These included: RZR-β, a putative nuclear receptor for the hormone melatonin (34); protein kinase C-ζ (PKC-ζ) (35); FAT, a cadherin-related cell adhesion molecule (36); EphA3, a receptor protein–tyrosine kinase (37); and a novel WNT-related gene, which we have named WNT-16. One gene, encoding a novel TALE homeobox gene related to the Drosophila Iroquois complex (Iroc/Irx) family (38), was expressed in three of four t(1;19) cell lines (Table 1).
To analyze the expression patterns of these six genes in primary tumors, RNA was isolated from bone marrow with abnormal cytogenetics, including t(1;19) pre-B ALL, non-t(1;19) pre-B ALL, t(9;22) CML, and t(12;21) pro-B ALL as well as from bone marrow derived from a healthy patient (Table 1). The mRNA was analyzed by RT-PCR. Interestingly, the cDNAs encoding WNT-16 and RZR-β both were expressed consistently in pre-B ALL containing the t(1;19) translocation (Table 1 and Fig. 1B). We note that very low levels of WNT-16 could be detected by RT-PCR in some non-t(1;19) pre-B cell leukemias (Fig. 1B). Taken together, the data indicate that RZR-β and a novel WNT gene are aberrantly expressed in pre-B cells that contain the E2A-Pbx1 fusion protein.
Molecular Cloning and Analysis of the WNT-16 Gene.
A full-length WNT-16 cDNA was isolated from a cDNA library derived from 697. The sequence contained a 1,095-bp ORF that would encode a 365-aa protein (Fig. 2). A putative mouse homolog of WNT-16 also was identified by screening a mouse genomic DNA library with the human cDNA (Fig. 2).
Figure 2.
Amino acid alignment of human WNT-16, mouse WNT-16, human WNT-1, mouse WNT-7b, and mouse WNT-4.
The deduced amino acid sequence of human and mouse WNT-16 was compared with the sequences of other mammalian WNT proteins (Fig. 2). The WNT-16 sequence is 42–48% identical to those of mouse WNT-7b, mouse WNT-4, and human WNT-13. Twenty-three cysteine residues in WNT-16 are particularly well conserved, in nearly identical positions, when compared with other members of the WNT protein family.
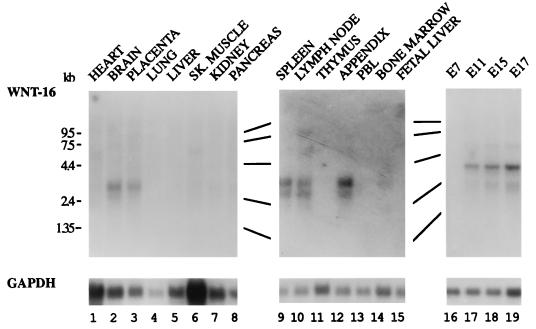
The predicted sequence of WNT-16 shows many of the characteristics of a secreted protein: a hydrophobic signal peptide, the lack of an obvious transmembrane domain, and abundant and highly conserved cysteine residues located throughout the protein. We note that upon ectopic expression in Drosophila S2 cells, WNT-16 secretion is detected, as has been reported for other WNT proteins (39). However, we have not been able to demonstrate secreted WNT-16 protein in supernatants of pre-B ALL cells (J.R.M., unpublished observations). To determine the expression pattern of WNT-16 in human tissues, a Northern blot, containing mRNA derived from various tissues, was probed by using a radiolabeled WNT-16 probe (Fig. 3). A 3.2-kb transcript was detectable in brain and placenta (Fig. 3). Additionally, two distinct mRNAs of 3 and 3.5 kb were present in mRNA derived from peripheral lymphoid organs, including spleen, lymph node, and appendix. No expression was detectable in bone marrow, thymus, fetal liver, or peripheral blood leukocytes.
Figure 3.
Northern blot analysis of WNT-16 expression in human tissues and during mouse embryonic development. Northern blots were purchased from CLONTECH. Transcripts were detected with antisense asymmetric PCR probes. Molecular weight markers are indicated on the left.
To determine whether the WNT-16 gene was expressed during embryonic development, a mouse WNT-16 probe was hybridized to a Northern blot of poly(A)+ RNA isolated from mouse embryos at several stages (Fig. 3). No expression was detectable at embryonic day 7. However, a 3.5-kb transcript was present at days 11, 15, and 17. Taken together, these data indicate that WNT-16 is normally expressed in peripheral lymphoid organs but not in thymus, bone marrow, or fetal liver. In contrast, high levels of WNT-16 transcripts are present in pre-B ALL cells containing E2A-Pbx1.
Expression of WNT Receptors in B Lineage Cells and in pre-B ALL Containing E2A-Pbx1.
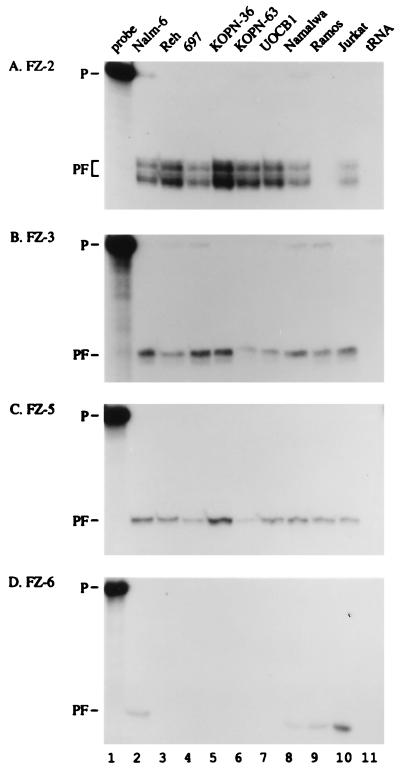
Recently, a number of genetic and biochemical studies have provided evidence that members of the Fz family function as receptors for WNT proteins (40, 41). Fz proteins are seven-transmembrane-receptor-related proteins with cysteine-rich extracellular domains (40). Fz homologs expressed in the t(1;19) pre-B ALL cell line 697 were identified by degenerate PCR amplification as described by Wang et al. (30). Four Fz homologues were found (data not shown). One is identical to human Fz-5, and the other three appear to be the human homologues of the rat Fz-2, mouse Fz-3, and mouse Fz-6 previously identified by Wang et al.(30). To determine the expression pattern of the various Fz genes, mRNA was isolated from a wide variety of B lineage cells and analyzed by RNase protection (Fig. 4). Interestingly, Fz-2, Fz-3, and Fz-5 were expressed in most B lineage cells including pre-B cells containing the t(1;19) chromosomal translocation (Fig. 4 A–C). Fz-6 expression, using RNase protection, was detectable only in Nalm-6, Namalwa, Ramos, and Jurkat cells (Fig. 4D).
Figure 4.
Expression of Frizzled genes in human B cell lines. RNA was derived from human pre-B cells carrying the t(1;19) translocation (697, KOPN-36, KOPN-63), pro-B cells (UOCB-1), pre-B cells (Nalm-6, Reh), mature B cells (Namalwa and Ramos), and a T lineage cell (Jurkat). Yeast tRNA is included as a control. RNA was analyzed by RNase protection by using antisense transcripts of FZ-2, FZ-3, FZ-5, and FZ-6 as probes. P, labeled probe; PF, protected fragment.
These data raise the possibility that WNT-16 may function in an autocrine mechanism to promote the growth of pre-B ALL cells by activating WNT-signaling pathways through Fz receptors.
The Presence of E2A-Pbx1 Is Required for Aberrant Expression of WNT-16.
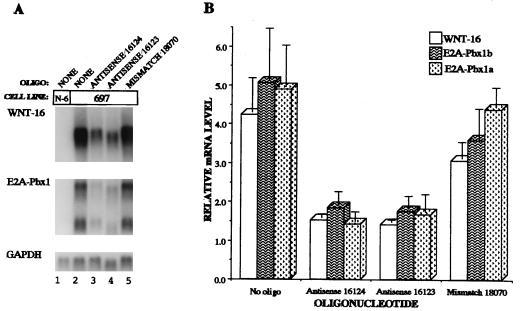
The data described above demonstrate the aberrant expression of WNT-16 in pre-B ALL containing the E2A-Pbx1 fusion protein. To determine whether E2A-Pbx1 is required for WNT-16 transcription, antisense oligonucleotides were designed with specificity for the E2A-Pbx1 fusion gene. Two antisense oligonucleotides, 16123 and 16124, were shown to inhibit both E2A-Pbx1 and WNT-16 mRNA expression upon electroporation into 697 cells (Fig. 5). A mismatch oligonucleotide, 18070, did not significantly affect E2A-Pbx1 or WNT-16 expression (Fig. 5). These data suggest that E2A-Pbx1 regulates the expression of WNT-16 in pre-B ALL cells.
Figure 5.
Treatment of t(1;19)-positive pre-B acute lymphoblastoid cell lines with antisense 2′-methoxyethyl phosphorothioate oligonucleotides to E2A-Pbx1 reveals that WNT-16 transcript levels are dependent on the presence of E2A-Pbx1. 697 cells were electroporated with antisense oligonucleotides with specificity for E2A-Pbx1. Twenty hours later, total RNA was derived from the cells and analyzed by Northern blotting by using asymmetric PCR probes for E2A-Pbx1 and WNT-16 and a random primer-labeled cDNA probe for GAPDH. (A) Northern blots. Nalm-6 (N-6) RNA was included as a control. (B) Graphical representation of Northern blot results. Relative mRNA levels detected on Northern blots were quantitated by using a PhosphorImager. Bar = mean ± SD from three independent experiments.
DISCUSSION
Our findings indicate that the E2A-Pbx1 fusion protein consistently activates transcription of a novel WNT gene in pre-B ALL. This raises the possibility that WNT signaling may contribute to the initiation or progression of this disease. It is also striking that Pbx/exd has been shown to be required for regulation of the wg/Dwnt-1 gene in Drosophila, suggesting that Pbx/exd proteins control similar downstream targets in both flies and mammals (15, 16).
What is the potential role of WNT-16 in pre-B ALL and in normal lymphocyte development? We show that WNT receptors belonging to the Fz family are expressed in B lineage cells. Their presence is not limited to pre-B cells carrying the E2A-Pbx1 fusion gene, but also can be detected in pro-B, pre-B, and mature B cell lines. Additionally, WNT-5a and WNT-10b gene expression has been detected in early hematopoietic progenitors and stromal cells of the yolk sac, fetal liver, and fetal bone marrow, and WNT-16 is expressed in peripheral lymphoid organs (refs. 42 and 43; Fig. 3). WNT proteins also have been shown to promote expansion of early hematopoietic progenitors in vitro (42, 43). These data are strongly suggestive of a role for WNT signaling during hematopoiesis. Thus, it is conceivable that the presence of WNT proteins is required at certain stages of normal development for proper expansion, survival, and/or differentiation of early hematopoietic cells, including those of the B cell lineage. Pre-B cells harboring the E2A-Pbx1 fusion protein would constitutively express WNT-16, perhaps promoting aberrant proliferation and/or survival of B lineage cells.
In conclusion, we would like to propose that members of the Pbx family normally repress WNT-16 transcription in pre-B lymphocytes. Upon a t(1;19) chromosomal translocation, Pbx-1 is converted into a transcriptional activator, promoting the aberrant activation of downstream target genes, including WNT-16. WNT-16 then would act in an autocrine fashion to perturb the normal development, expansion, or death of pre-B lineage cells, ultimately leading to the development of an acute leukemia.
Acknowledgments
We thank Paul Conn for technical assistance. J.R.M. was supported by a grant from the American Cancer Society, California Division. The work was supported by grants from the National Institutes of Health to C.M. and J.D. (RO1 CA65797 05 and PO1 CA71907-03).
ABBREVIATIONS
- ALL
acute lymphoblastoid leukemia
- RDA
representational difference analysis
- RT-PCR
reverse transcription–PCR
- Fz
Frizzled
- GAPDH
glyceraldehyde-3-phosphate-dehydrogenase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. 282013 and 282020).
References
- 1.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 2.Sauvageau G, Thorsteinsdottir U, Hough M R, Hugo P, Lawrence H J, Largman C, Humphries R K. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 3.Kroon E J, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. EMBO J. 1998;13:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamps M P, Murre C, Sun X, Baltimore D. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 5.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M L. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 6.Monica K, Galili N, Nourse J, Saltman D, Cleary M L. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bürglin T. Nat Genet. 1992;1:319–320. doi: 10.1038/ng0892-319. [DOI] [PubMed] [Google Scholar]
- 8.Knoepfler P S, Kamps M P. Mol Cell Biol. 1995;15:5811–5819. doi: 10.1128/mcb.15.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berthelsen J, Zappavigna V, Ferreti E, Mavillo F, Blasi F. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C-P, Shen W-F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 11.Neuteboom S T C, Peltenburg L T C, van Dijk M A, Murre C. Proc Natl Acad Sci USA. 1995;92:9166–9170. doi: 10.1073/pnas.92.20.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popperl H, Bienz M, Studer M, Chan S-K, Aparicio S, Brenner S, Mann R S, Krumlauf R. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 13.Peers B, Sharma S, Johnson M, Kamps M, Montminy M. Mol Cell Biol. 1995;15:7091–7097. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peifer M, Wieschaus E. Genes Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- 15.Rauskolb C, Peifer M, Wieschaus E. Cell. 1993;74:1101–1112. doi: 10.1016/0092-8674(93)90731-5. [DOI] [PubMed] [Google Scholar]
- 16.Rauskolb C, Wieschaus E. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 18.Nusse R N, Varmus H. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 19.Korinek V, Barker J, Morin D, Kinzler K, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 20.Morin P J, Sparks A B, Korinek V, Barker J, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 21.Morin P J, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubenfield B, Albert E, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1997;275:1790–1792. [Google Scholar]
- 23.Kamps M, Look T, Baltimore D. Genes Dev. 1991;5:358–369. doi: 10.1101/gad.5.3.358. [DOI] [PubMed] [Google Scholar]
- 24.Kamps M P, Baltimore D. Mol Cell Biol. 1993;13:351–357. doi: 10.1128/mcb.13.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dedera D A, Waller E K, LeBrun D P, Sen-Majumbar A, Stevens M E, Barsh G S, Cleary M L. Cell. 1993;674:833–843. doi: 10.1016/0092-8674(93)90463-z. [DOI] [PubMed] [Google Scholar]
- 26.van Dijk M A, Voorhoeve P M, Murre C. Proc Natl Acad Sci USA. 1993;90:6061–6065. doi: 10.1073/pnas.90.13.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Q, Wright D D, Kamps M P. Mol Cell Biol. 1994;14:3938–3948. doi: 10.1128/mcb.14.6.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McWhirter J R, Goulding M, Weiner J A, Chun J, Murre C. Development (Cambridge, UK) 1997;124:3221–3232. doi: 10.1242/dev.124.17.3221. [DOI] [PubMed] [Google Scholar]
- 29.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Macke J P, Abella B S, Andreasson K, Worley P, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. J Biol Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 31.Monia B P. Anticancer Drug Design. 1997;12:327–339. [PubMed] [Google Scholar]
- 32.Freier S M, Altmann K H. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Carlberg C, Hooft van Huijsduinen R, Staple L, DeLamarter J F, Becker-Andre M. Mol Endocrinol. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 35.Kochs G, Hummel R, Meyer D, Hug H, Marme D, Sarre T F. Eur J Biochem. 1993;216:597–606. doi: 10.1111/j.1432-1033.1993.tb18179.x. [DOI] [PubMed] [Google Scholar]
- 36.Dunne J, Hanby A M, Poulson R, Jones T A, Sheer D, Chin W G, Da S M, Zhao Q, Beverly P C, Owen M J. Genomics. 1995;30:207–223. doi: 10.1006/geno.1995.9884. [DOI] [PubMed] [Google Scholar]
- 37.Wicks I P, Wilkinson D, Salvaris E, Boyd A W. Proc Natl Acad Sci USA. 1992;89:1611–1615. doi: 10.1073/pnas.89.5.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosse A, Zulch A, Becker M B, Torres M, Gomez-Skarmeta J L, Modolell J, Gruss P. Mech Dev. 1997;69:169–181. doi: 10.1016/s0925-4773(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 39.Van Leeuwen F, Harryman-Samos C, Nusse R. Nature (London) 1994;368:342–344. doi: 10.1038/368342a0. [DOI] [PubMed] [Google Scholar]
- 40.Rattner A, Hsieh J, Smallwood P M, Gilbert D J, Copeland N G, Jenkins N, Nathans J. Proc Natl Acad Sci USA. 1997;94:2859–2863. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhanot P, Brink M, Harryman Samos C, Hsieh J-C, Wang Y, Macke J P, Andrew D, Nathans J, Nusse R. Nature (London) 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 42.Austin T W, Solar G P, Ziegler F C, Liem L, Matthews W. Blood. 1997;89:3624–3635. [PubMed] [Google Scholar]
- 43.Van Den Berg D J, Sharma A K, Bruno E, Hoffman R. Blood. 1998;92:3189–3202. [PubMed] [Google Scholar]