Abstract
The rearrangement of antigen receptor genes is initiated by double-strand breaks catalyzed by the RAG1/2 complex at the junctions of recombination signal sequences and coding segments. As with some “cut-and-paste” transposases, such as Tn5 and Hermes, a DNA hairpin is formed at one end of the break via a nicked intermediate. By using abasic DNA substrates, we show that different base positions are important for the two steps of cleavage. Removal of one base in the coding flank enhances hairpin formation, bypassing a requirement for a paired complex of two signal sequences. Rescue by abasic substrates is consistent with a base-flip mechanism seen in the crystal structure of the Tn5 postcleavage complex and may mimic the DNA changes on paired complex formation. We have searched for a tryptophan residue in RAG1 that would be the functional equivalent of W298 in Tn5, which stabilizes the DNA interaction by stacking the flipped base on the indole ring. A W956A mutation in RAG1 had an inhibitory effect on both nicking and hairpin stages that could be rescued by abasic substrates. W956 is therefore a likely candidate for interacting with this base during hairpin formation.
Keywords: immunoglobulin gene, recombination, transposase
In many vertebrates, the vast antigen-binding repertoire of immunoglobulins and T cell receptors is created by combinatorial V(D)J recombination using an array of variable (V), diversity (D), and joining (J) gene segments in B and T cell genomic DNA (1). A complex of the RAG1 and RAG2 gene products is responsible for initiating DNA cleavage and probably for mediating the recruitment of nonhomologous end-joining factors to process and join the broken V, (D), and J segments.
The sites of DNA cleavage are directed by recombination signal sequences (RSSs) that flank each coding segment. The two types of RSSs (denoted 12RSS and 23RSS) consist of conserved heptamer and nonamer motifs separated by a spacer of 12 or 23 nonconserved base pairs. Normally, DNA cleavage occurs when the RAG complex recognizes and pairs a 12RSS and a 23RSS, thus ensuring the correct recombination of a V to a J or of a V to a D and of a D to a J segment (termed the “12/23 rule”) (2). Ordered assembly usually begins with RAG1/2 binding to the 12RSS, followed by capture of free 23RSS (3, 4). Changes in the sensitivity of a 12RSS to chemical modification occur upon RAG1/2 binding, consistent with some unwinding at the heptamer-coding border (5, 6).
Double-strand breaks at the coding–RSS border are produced by a nick 5′ of the heptamer; nucleophilic attack on the opposing strand by the 3′ hydroxyl group at the nick then creates a hairpin by a transesterification reaction (7). The resulting double-strand break consists of a hairpin on the coding flank and a blunt-ended signal sequence. In the presence of Mg2+, the complete reaction takes place only in the presence of an RSS pair (coupled cleavage) (8, 9). The mechanism of DNA cleavage has been characterized in vitro, mainly with truncated RAG proteins that retain activity but are more soluble than the full-length proteins, and with oligonucleotides containing RSSs. HMGB1 or -2 protein is also included in in vitro studies, because it aids the formation of 12/23 paired complexes, probably through DNA bending (10, 11). With a single RSS and Mg2+, the RAG proteins can only catalyze the nicking step. In Mn2+, hairpin formation is also allowed even with a single RSS (7) (Fig. 1a).
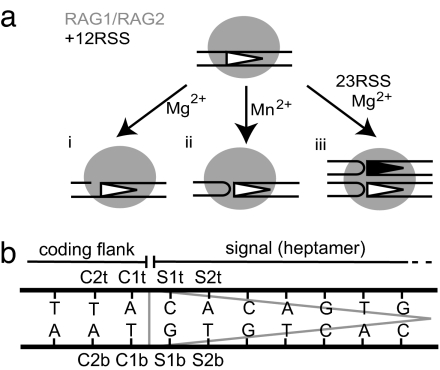
Fig. 1.
Schematic diagram of the outcomes of in vitro cleavage assays and notation of base positions. (a) (i) RAG1/2 catalyzes nicking 5′ of the heptamer in the presence of Mg2+. (ii) Mn2+ allows RAG1/2 to convert nicks to hairpin products at the coding flank by transesterification. (iii) Mg2+ is only able to catalyze hairpin formation when 12RSS and 23RSS are present in the RAG1/2 complex (coupled cleavage). (b) Positions of abasic nucleotides incorporated into the oligonucleotide substrates at the border (1) and penultimate (2) positions of the coding flank (C) or signal (S) on top (t) or bottom (b) strands.
After DNA cleavage in vitro, RAG1/2 is capable of transposing the signal end into a target DNA (12, 13). Evidence suggests that V(D)J recombination has evolved from an ancestral transposon (see e.g., ref. 2 for a review). Although no structural information exists for the core RAG regions, biochemical data has identified the binding site in RAG1 for the catalytic metal ions coordinated by three acidic residues (D600, D708, and E962) (14–16). These sites are likely to be held in an RNaseH-like fold also possessed by transposases (17). Much may be learned from RAG1's transposase relatives that also operate via a hairpin intermediate, e.g., Tn5, Tn10, and the eukaryotic hAT transposases (18). The structure of the Tn5 transposase complexed with two transposon ends in a postcleavage complex shows how the DNA interacts near the active site (19). In Tn5, the hairpin forms on the transposon end, and in the crystal, a flipped-out base is seen stabilized by a tryptophan (W298). Mutagenesis studies show that this interaction is important for efficient hairpin formation (20). Similar findings were made with mutagenesis of equivalent residues in Tn10 (W265) (21) and Hermes (W319) transposases (22). Thus, parallel mechanisms may operate in transposases in which the hairpin is on the transposon end (Tn5 and Tn10), and those in which the hairpin is on the flanking DNA (Hermes and RAG1/2) (18, 23). In this work, we used abasic substrates to investigate the role of bases around the coding–RSS border during the two steps of DNA cleavage. A critical base whose removal from the helix stimulates transesterification was identified, suggesting which DNA conformation changes are produced on synaptic complex formation to initiate coupled cleavage. By mutagenesis we identified Trp residues that affect the reaction, with and without alterations of DNA binding, one of which is likely to interact with the critical base.
Results
Abasic Substrates Form Normal Complexes with RAG1/2.
Single abasic nucleotides were incorporated into a 12RSS substrate at positions bordering the cleavage site (Fig. 1b). When the binding of RAG1/2 to the modified substrates was analyzed by EMSA, the amount of single complex (SC) formed on the abasic 12RSS substrates or of paired complexes with normal 23RSS was not significantly different from the normal 12RSS, within the range of variation [see supporting information (SI) Fig. 5].
Base Positions in the Coding Flank That Are Critical for Nicking.
The contribution of base–protein interactions and base pairing was assessed for both the nicking and hairpinning steps of cleavage. Nicking of the top strand (5′ of the heptamer) was analyzed by in vitro cleavage of the abasic substrates by core RAG1 and full-length RAG2. Several conditions were tested: Mg2+ or Mn2+ ion on a 12RSS substrate and coupled cleavage with added 23RSS in Mg2+ (Fig. 1a).
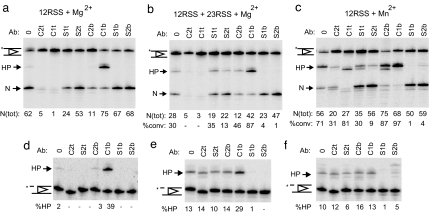
In Mg2+, abasic substitutions at bordering (1) and penultimate (2) nucleotides of the top (t) strands of the coding flank (C1t and C2t; see Fig. 1b legend for full notation) resulted in a marked inhibition of nicking (Fig. 2a). A decrease in nicking was also observed for substrates modified at C2b and S1t, albeit to a lesser extent. The inhibitory effects of the C1t and C2t abasic nucleotides were not overcome when 23RSS was present (coupled cleavage, Fig. 2b). Enhanced nicking was observed with abasic C1b under all conditions as deduced from the sum of hairpin and nick.
Fig. 2.
RSS cleavage assays with abasic substrate. Assays were performed with intact (a–c) or prenicked (d–f) 12RSS that was abasic at various positions (see Fig. 1b for notation). Cleavage products obtained from incubation with MR1 and full-length RAG2 were separated on a acrylamide/urea gel. Nick (N) and hairpin (HP) products are indicated and quantified for each condition: Mg2+ (a and d); 23RSS and Mg2+ (b and e); Mn2+ (c and f). N(tot), total nick, the percentage of nick plus hairpin in each lane; %conv, percentage HP/N(tot).
In Mn2+, the effects of abasic substitutions were different; abasic C1t and C2t were less inhibitory to the nicking step (Fig. 2c), and additional nicks inside the coding flank were more evident with all abasic substrates. Furthermore abasic C2b, which was poorly nicked in Mg2+, appeared to enhance nicking similarly to C1b. These data demonstrate a relaxed specificity for the nick site in Mn2+.
Removal of a Specific Base in the Coding Flank Potentiates Hairpin Formation.
With a single RSS, Mg2+ normally allows cleavage to proceed no further than nicking and only trace amounts of hairpin can be detected in the absence of partner DNA (Fig. 1a). Remarkably, abasic 12RSS(C1b) supported robust hairpin formation without a partner RSS (64% of substrate was converted to hairpin) (Fig. 2a). Similarly, a 23RSS with an abasic substitution at C1b forms hairpins in the absence of 12RSS (data not shown). Even in the presence of a 23RSS, an increase in hairpinning was observed with abasic 12RSS(C1b) compared with normal substrate (87% vs. 30% of nicks converted to hairpin on abasic and normal substrates, respectively; Fig. 3b). Abasic 12RSS(C1b) was also more efficient at hairpinning than normal 12RSS in Mn2+ (97% nicks converted to hairpin compared with 71%; Fig. 2c). When the neighboring base (C2b) was removed instead, an increase in transesterification was also seen in Mn2+ (from 71% to 87% of nicks converted).
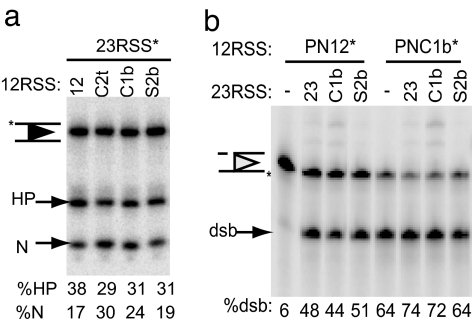
Fig. 3.
Coupled cleavage with abasic substrates. (a) Cleavage assays with radiolabeled 23RSS (5′ top strand) and unlabeled 12RSS abasic at the indicated position were performed in the presence of MR1/MR2 and Mg2+. Nick (N) and hairpin (HP) products were separated on an acrylamide/urea gel and quantified as a percentage of total signal in each lane. (b) Coupled-cleavage assay with prenicked 12RSS and prenicked 23RSS substrates.
Mg2+ could also support hairpinning on a single prenicked 12RSS abasic at C1b (Fig. 2d). Hairpin formation by prenicked abasic 12RSS(C1b) in Mn2+ showed little enhancement over normal prenicked substrate (13% vs. 10%, respectively; Fig. 2f). The stimulatory effect of abasic C1b therefore is most evident on intact substrate, which may be due to preferred processing after nicking.
The stimulation of transesterification by abasic (C1b) did not require HMGB1, although hairpin formation was further enhanced by HMGB1 (data not shown). Furthermore, hairpinning in Mg2+ occurs with all substrates abasic at C1b regardless of coding flank sequence, including “bad flank” sequences (reading TTT or TTC 5′ to 3′ adjoining the RSS) that are rarely found at RSS flanks in antigen receptor coding sequences (24) and are poor at transesterification in Mn2+ (refs. 25 and 26 and data not shown). The enhancement of transesterification by abasic C1b in Mg2+ was greater than that with a mismatched base (C1t/C1b) in the coding flank, which only led to minimal amounts of transesterification (ref. 27 and data not shown).
Bases in the RSS That Are Essential for Hairpinning.
Although the base at C1b appears to be involved in controlling transesterification, other bases around the cleavage site also have an effect on this step. Removal of RSS bases on the bottom strand (S1b or S2b) severely inhibited hairpinning in Mg2+-supported coupled cleavage or in cleavage of a single RSS in Mn2+, despite normal or elevated amounts of nicked product (Fig. 2 b and c). A partial inhibition of hairpin formation was observed with abasic S2t. Hairpin formation was also analyzed by using prenicked substrates, thus bypassing the nicking step (Fig. 2 e and f). The bases of the RSS bottom strand were still critical to this step, but abasic C2t (which inhibited the nicking step) produced near normal hairpin levels. In Mn2+, this substrate yielded at least two different hairpin products (≤32 nt) derived from different nick sites within the coding flank. A prenicked substrate still allowed inaccurate nicking at other sites, leading to the formation of a number of hairpin products (Fig. 2f).
Abasic Substrates That Block Cleavage Do Not Inhibit Hairpin Formation on Partner DNA.
The abasic substrates poor at nicking (C2t) and transesterification (S2b) were used to ask at which stage hairpin formation is stimulated by the second RSS: binding, nicking, or transesterification. Although synaptic complex formation with a 23RSS could not overcome the defects in nicking and hairpinning on abasic C2t or S2b 12RSS (Fig. 2b), cleavage of the 23RSS partner was not inhibited by these abasic substrates. Only small decreases in transesterification on the 23RSS were observed by blocking nicking and hairpinning of the 12RSS (Fig. 3a).
Similarly, we tested whether the enhanced cleavage in Mg2+ of abasic 12RSS(C1b) could be stimulated further by the addition of abasic 23RSS(C1b). A small increase in hairpin formation at the 12RSS was seen with a normal 23RSS partner, but no further stimulation was evident with abasic 23RSS(C1b) (Fig. 3b). Thus, hyper-transesterification at one site does not increase the amount of hairpin formed at the other. In all cases, simply binding to the partner site is able to induce changes at the other site to initiate transesterification. Communication between the 12RSS and 23RSS sites with abasic substrates is apparently transduced before nicking and hairpin formation.
Mutagenesis of Tryptophan Residues in RAG1.
We have shown that cleavage at an RSS can be induced by binding of a second RSS, presumably by inducing a conformational change in the DNA and/or protein. This requirement is bypassed by the removal of the base at C1b, which permits Mg2+-mediated transesterification to proceed even in the absence of a second RSS. Such a mechanism may be consistent with a base-flip event as seen with certain DDE transposases that form DNA hairpins. We explored the possibility that the C1b base may be stabilized in an extrahelical position by contacts similar to those seen in the crystal structure of the Tn5 postcleavage complex, in which the penultimate T of the Tn5 transposon “signal” sequence is stacked against the indole ring of tryptophan W298 (19). A second Trp in Tn5 transposase appears to intercalate between the strands of DNA. Because two Trp residues are also seen at the active site of the Hermes transposase (22), we mutated all six of the tryptophans found in murine core RAG1 to alanine to test whether a similar Trp–base interaction occurs in RAG-mediated cleavage.
Mutations of Conserved RAG1 Trp Residues Decrease in Vivo Recombination.
Standard in vivo recombination assays were used to determine whether RAG1-W519A, W760A, W829A, W893A, W956A, and W992A mutants affected formation of signal and coding joints. RAG1 mutants were introduced into the full-length RAG1 coding expression vector pJH548 and cotransfected with full-length RAG2 and the substrate plasmid (see Materials and Methods). Signal joints were reduced >10-fold in all but the W829A mutant compared with WT RAG1 (Table 1). Recombination of W893A was reduced 100-fold compared with WT, and W760A, W956A, and W992A were even more defective.
Table 1.
Recombination frequencies (R) of RAG1 tryptophan mutants
| RAG1 | RAG2 | No. screened | SJ R, % |
|---|---|---|---|
| WT | WT | 440,000 | 0.39 |
| W519A | WT | 240,000 | 0.01 |
| W760A | WT | 490,000 | <0.0002 |
| W829A | WT | 145,000 | 0.22 |
| W893A | WT | 410,000 | 0.004 |
| W956A | WT | 36,000 | <0.003 |
| W992A | WT | 36,000 | <0.003 |
| WT | — | 50,000 | <0.003 |
| — | WT | 77,000 | <0.002 |
Data in the “No. screened” column indicate the approximate number of plasmids recovered from transfected NIH 3T3 cells that were screened for recombination. The recombination frequency (R) of recombined signal joints (SJ) was confirmed by hybridization.
RAG–DNA Complexes of Trp Mutants.
WT and mutant RAG1 varieties were coexpressed with RAG2 and batch purified in parallel for optimal comparison and best activity for potentially unstable mutants. All mutants were able to bind RSS substrate to some extent as shown by EMSA (Fig. 4a). The main point of interest was that the complex formed by W760A appeared to have increased mobility compared with the other species. This complex, probably the same as stable complex 1 described in refs. 28 and 29, is believed to contain fewer RAG2 monomers than stable complex 2, which is observed for WT and the other mutants. In partial confirmation, an interaction assay using FLAG-tagged RAG2 showed that W760A had reduced affinity for RAG2 compared with WT (and W829A, W893A, and W956A, not shown) when copurified on anti-FLAG resin (SI Fig. 6).
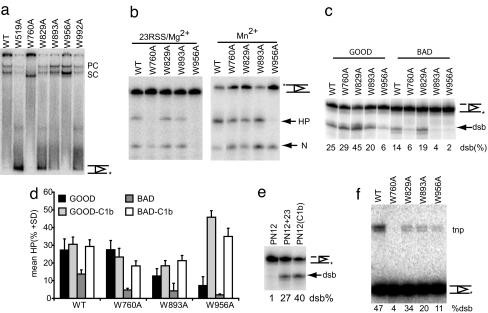
Fig. 4.
In vitro characterization of Trp mutations. (a) Complex formation was analyzed by EMSA, with approximately equal amounts of purified, coexpressed mutant MR1 and MR2 with labeled 12RSS and 23RSS substrates. Normal stable complexes (SC) with single substrate and paired complexes (PC) are formed with all mutants except W760A, which forms a faster species SC only. (b) Nick (N) and hairpin (HP) products from coupled and Mn2+ catalyzed cleavage reactions were analyzed on a 90 mM Tris, 90 mM boric acid, and 2 mM EDTA (pH 8.3) (TBE)-urea gel. (c) Good (TTA)- and bad (TTC)-flanked, prenicked 12RSS substrates were used in reactions containing 30 ng/μl copurified mutants and 1 mM Mn2+. The percentage of hairpin product was quantified on a TBE-urea gel. (d) Similar experiments with and without an abasic site at Clb. The bar chart displays the average from three or four experiments in which the error bars represent the SD. (e) Comparison of the rescue of hairpinning by W956A in Mn2+ by addition of prenicked 23RSS and by using an abasic substrate at C1b. Bad flank substrate (TTC) was used in this cleavage assay. (f) Intact substrate was used in transposition reactions analyzed on a 1% agarose gel. Transposition (tnp) of the bottom strand of the RSS into pBR322 after Mg2+-catalyzed cleavage can be compared with the amount of double-strand breaks (dsb) quantified in a parallel reaction (without plasmid DNA).
W760A and W956A Are Defective at Nicking.
Any reduction in hairpin formation would lower the amount of cleaved substrate available for subsequent recombination events. The RAG1 Trp mutants were tested for in vitro cleavage, to identify any changes in catalytic function. Under coupled cleavage conditions (including 23RSS and Mg2+), nicking was decreased by the W760A and W956A mutations (Fig. 4b). These mutants were further characterized with the abasic substrates. For both W760A and W956A, substrates abasic at C1b, S1b, and S2b greatly stimulated the nicking (data not shown). Nicking by W760A and W956A was also improved in Mn2+ (Fig. 4b). These results do not necessarily imply direct interactions of tryptophans with DNA bases during nicking. The active site of the mutated proteins may be distorted so that it can catalyze the nicking only in the presence of permissive substrates or metal ion.
W519A and W992A were analyzed separately and, despite poorer solubility, did not have severely reduced nicking or transesterification activities (SI Fig. 7).
Transesterification by RAG1 W956A Is Defective but Rescued by Abasic Substrates.
By using Mn2+ to catalyze transesterification, W956A was unable to efficiently convert nick to hairpin (Fig. 4b). With prenicked substrates, the ability of W956A to form hairpins in Mn2+ was still reduced (Fig. 4c), but the other mutants did not appear to have major defects in transesterification. Under coupled cleavage conditions, hairpinning was reduced by W760A for reasons of poor paired-complex formation (Fig. 4a). Mn2+-catalyzed transesterification on a single substrate was normal with this mutant.
Because W893 has been suggested to be the equivalent Trp to W298 of Tn5 (18, 30), we examined the effect of the W893A mutation on hairpin formation more closely. In Mn2+, only a minor decrease in conversion of nicks to hairpins was seen with normal substrates (a reduction of one-third). Under coupled cleavage conditions, the difference between W893A and WT was not significant. A major defect in hairpinning by W893A was only seen when using a “bad flank” substrate (ending TTC; Fig. 4c), which also decreased transesterification by WT RAG1/2, severely inhibited hairpin formation by W760A, and reduced transesterification W956A even further.
It has been shown in the Tn5 system that rescue of hairpin formation by a Trp mutant involved in base flipping can be achieved by using the appropriate abasic DNA substrate, because there is no base needing to be stabilized (20). We therefore tested for the rescue of transesterification by W760A, W893A, and W956A on both types of flanks in Mn2+. By using the optimal flank, hairpinning by WT, W760A, or W893A was not appreciably effected by removing C1b (Fig. 4d). However, abasic 12RSS(C1b) produced a five-fold stimulation in hairpinning by W956A. Furthermore 12RSS(C1b) was the only prenicked abasic substrate to have a major effect (data not shown). Removing the base at C1b in bad flank substrates overcame the inhibitory effect in all cases. Transesterification on good and bad flanks by W956A could also be rescued to WT levels by the addition of 23RSS (coupled cleavage) (Fig. 4e).
Transposition by Trp Mutants.
In Tn5, mutation of W298 affected strand transfer during transposition (20). However, mutation of W314 in the Hermes system, in which, as with RAG1/2, the hairpin is formed on the flank, had no effect on strand transfer (22). For analogy, the soluble RAG1 Trp mutants were tested for their ability to transpose RSS into donor plasmid DNA. Intact substrate was used because the concentration of mutant proteins was too low for transposition assays with precleaved substrate. Although transposition then depended on coupled cleavage in Mg2+ as well as on strand transfer, transposition products were detected for all mutants except W760A (which failed to form a paired complex efficiently). For the other mutants, transposition was somewhat decreased, corresponding to lower levels of cleavage (Fig. 4f), but in no case was there a specific defect in transposition.
Discussion
Hairpin formation is the most challenging step to understand in the reactions of RAG1/2 and the related transposases because of the large DNA distortion that is required. For Tn5, the crystal structure of the protein–DNA complex provides a satisfactory explanation by showing that a base is flipped out of the double helix and stabilized by a tryptophan, allowing the DNA chain to rotate. Lacking comparable structural information for RAG1/2, we have used biochemical tests to study whether a similar mechanism is used in this system. The abasic substrate studies described here provided relevant information about this question and other features of DNA cleavage.
Changes in Base Contacts and Complex Conformation During Cleavage.
In each step of cleavage, the two bases 5′ of the cleavage site are shown to be essential. Bases in the top strand coding flank are more critical for nicking, whereas bases in the bottom strand RSS are required for efficient hairpin formation. Contacts with these bases may be responsible for positioning the correct phosphodiester bond for attack but also for controlling the hydroxyl nucleophile of the coding flank that is released after strand cleavage for use in subsequent transesterification. The differences between reactions containing Mg2+ or Mn2+ are very apparent with abasic substrates. The relaxed requirement for coding flank bases and the variable position of nicking in Mn2+ suggest a greater flexibility of the enzyme–substrate complex in this condition. The contacts with RSS bases in both Mg2+ and Mn2+ remain stringent and result in an accurate site of transesterification.
The base on the other side (3′) of the site of transesterification (C1b) has, if anything, a negative influence on that reaction, because its removal enhances hairpin formation on a single RSS; this effect is most apparent in Mg2+. It would be reasonable to suppose that, with a normal 12RSS, the presence of 23RSS triggers a conformational change at the active sites such that a contortion in the coding flank DNA allows transesterification to occur only in the context of the 12/23 complex.
We found that only the binding of the partner RSS, rather than any later stage in the process, was necessary to stimulate hairpin formation. In one report, poor nicking at one RSS due to coding flank-sequence effects appeared to limit the transesterification at the other (31), but this effect did not appear in our studies with abasic substrates. An abasic 12RSS deficient at nicking had only minimal effects on the cleavage of the partner RSS. Our findings using hairpinning-deficient abasic substrates are consistent with those on phosphorothioate substrates (also unable to form hairpins) that do not block hairpinning at the partner site (32).
Evidence for Base-Flipping in RAG-Mediated Cleavage.
Our observations define the base at C1b as a critical obstacle to the transesterification reaction. Mismatched coding flanks stimulate transesterification on a single substrate in Mg2+ to a much lesser extent. In normal substrates, unpairing of the coding flank bases is likely to be required, but the C1b base may also have to move out of the helix by breaking base stacking interactions. Bases in the resulting hairpin cannot form base pairs because of contortion of the DNA backbone. Protein contacts with coding flank bases would presumably aid transesterification by stabilizing the product and/or positioning the scissile bond for attack. Base flipping is common to several enzymes that act on DNA (33), e.g., DNA-modifying enzymes (methyltransferases), DNA-repair enzymes, and hairpin-binding proteins (e.g., TnpA of an IS605 transposon) (34). These enzymes use a variety of hydrophobic, aromatic, and charged amino acids to recognize and stabilize the extrahelical base. The most relevant example is the Tn5 transposase, in which a Trp interacts with a base in the hairpin (the penultimate thymine, T2) at the transposon end by stacking of two planar rings.
C1b, like T2 of the Tn5 transposon, lies on the strand opposite the nick site and within the sequence that forms the hairpin, but unlike in Tn5, C1b is one of the bases in the tip of the hairpin. Nevertheless, C2b (the positional equivalent to T2) does show similar characteristics to C1b in Mn2+ and may adopt an extrahelical position as well. With RAG1/2, the flipped-out base is in the coding flank of the V, (D), or J segment and, by its variable nature, must be stabilized by nonsequence-specific contacts. Base stacking by Trp and backbone interactions by Tyr are sequence-independent methods for doing so. In the case of Tn5, T2 appears to have no base specific contacts either, so that abasic T2 also aids hairpin formation (19, 20). Removing the base from C1b has a similar effect on hairpin formation regardless of whether a purine or pyrimidine is present at C1t. Thus, the type of base at C1t has no influence on transesterification. Yet a purine at C1b is not as easily tolerated by RAG1/2 (25, 26) and particularly by the mutants studied here and elsewhere (27).
RAG1/2 was found to preferentially bind to substrates with a cis-glycol modified C1b base (6). Such oxidized bases tend to be displaced from the helix. Furthermore, cross-linking experiments using iodopyrimidine-modified bases across the coding–heptamer region identified C1b (and S2b) as forming crosslinks to RAG1 (35). Although specific amino acid–base contacts were not identified with these methods, it appears that coding flank interactions (at C2t) are occurring in the C-terminal domain between amino acids 889 and 974 (36). These biochemical experiments support the notion that C1b lies extrahelically during cleavage and interacts with RAG1 and that contacts with C2t and S2b are also occurring, as suggested by our abasic screen.
Evidence for a Hairpin-Binding Domain in RAG1.
It had been proposed that certain cut-and-paste transposases and the resolvase ResT (that can either make or open a DNA hairpin) possess a common motif for binding the hairpin intermediate (PXW-linker-YXXK) where Pro and Trp form a hydrophobic pocket and ring-stacking interaction, and Tyr and Lys or Arg make contacts with the phosphate backbone of the flipped-out base (30). Although W319 of Hermes has been implicated in hairpin formation (22) and appears to follow the first part of the motif, other members of the hAT transposase family do not conform to either part of this motif (37) (SI Fig. 8).
Even so, the functional Trp is conserved in practically all Tn5/Tn10 transposases and hAT family members, with the exception of Tag-1, in which a different aromatic residue, Phe, is present (38). The region around RAG1 W893 has been suggested to share the Tn5/Tn10/ResT hairpin-binding motif (18, 30); indeed, mutations in surrounding residues are deficient in hairpinning (39).
Our mutational studies show W893 to be functionally important for in vivo recombination; however, in vitro assays did not show a severe defect in cleavage when an optimal coding flank sequence (TTA) was used. In earlier work, some coding flank sequences directly adjoining the RSS were found to decrease V(D)J recombination when combined with a particular RAG1 mutation (40). A subclass of these bad flanks (e.g., TTT) even decreased recombination with WT RAG1/2 (24). Bad flank substrates also decreased hairpin formation by purified WT RAG1/2 proteins (25, 26). In our present work, the W893A or W956A mutations display an even greater in vitro defect with bad flank substrates. A comparable defect of W893A in the context of bad flank substrates has recently been reported (41). However, with substrates containing a good coding flank, RAG1 W893A has only a minimal defect in hairpinning. The RSSs in the in vivo substrate are also flanked by the preferred pyrimidine–purine alternation (CTG) (40); thus, the mutation at W893 may have affected a stage following cleavage in the recombination process.
In contrast, W956A is still defective in cleavage even with a good coding flank. W956 appears to be the strongest candidate among Trp residues for a role in interacting with C1b, consistent with a role in stabilizing an extrahelical base. Alternatively, W956 (which is near the catalytic glutamate E962 in the primary sequence) could act equivalently to the second Trp at the Tn5 active site (W323) that lies between the bases of the transposon end (19). A second Trp (W182) is also present at the Hermes active site (22), and because the equivalent Trp is conserved among the hAT transposases, it may represent another feature of hairpinning transposases.
Materials and Methods
DNA Substrates.
Previously described oligonucleotides were synthesized and gel purified to construct intact 12RSS and 23RSS (DAR39/40 and DAR61/62) or prenicked 12RSS and 23RSS (DAR42/DG10/DAR40; DAR42/DG4/DAR61) (7). Prenicked bad flank oligonucleotides ending TTC and TTT (5′ to 3′) differed from DAR42 and DAR40 only at the three coding-flank positions nearest the RSS. Oligonucleotides containing dSpacer substitutions (Integrated DNA Technologies, Coralville, IA) in positions −2, −1, +1, and +2 from the coding signal border on top and bottom strands were used to produce the abasic substrates (Fig. 2a). Oligonucleotides were radiolabeled with polynucleotide kinase or terminal transferase before annealing.
Proteins.
The plasmids pFB-MR1 and pFB-MR2 encode the maltose binding protein fusions of RAG1 and RAG2 and MR1 and MR2 for expression with baculovirus (7). Singly or coexpressed, RAG1 and RAG2 were purified by polyhistidine and maltose-binding protein affinity tags as described in ref. 7. Vaccinia viruses containing T7 RNA polymerase and FLAG-tagged full-length RAG2 were kind gifts from M. Oettinger (Massachusetts General Hospital, Boston, MA) (42). Purification of RAG2 expressed in HeLa S3 cells by using anti-FLAG M2-agarose (Sigma, St. Louis, MO) has been described in ref. 7. A truncated form of HMGB1 (1–163) was provided by Mary H. O'Dea (National Institute of Diabetes and Digestive and Kidney Diseases). Site-specific mutations of the tryptophan residues in murine core RAG1 were produced by using a QuikChange kit (Stratagene, La Jolla, CA) within the baculovirus expression vector pFB-MR1. Mutations were checked by DNA sequencing, and for W893A the amino acid substitution was confirmed by mass spectrometry.
Sf9 insect cells in serum-free medium were coinfected with MR1 mutant and MR2 baculovirus. Batch purifications were performed in parallel to WT protein for comparison of activities. Cells were lysed in 10 ml of purification buffer [20 mM Tris·HCl (pH 8)/500 mM NaCl/10% glycerol/1 mM DTT] plus 1 mM PMSF, 0.1% Triton X-100. One milliliter of amylose resin (washed with purification buffer) was added to the supernatants and mixed for 90 min at 4°C. The resin was transferred to 10-ml columns, washed with 30 ml of purification buffer, then eluted with buffer containing 10 mM maltose. Proteins were dialyzed into 20 mM Tris·HCl (pH 8), 150 mM KCl, 10% glycerol, and 1 mM DTT and stored at −80°C.
Cleavage Assay and EMSA.
Reactions containing 2 nM 12RSS, 20 ng/μl RAG1/2, and 2 ng/μl HMGB1 protein (with or without 12.5 nM 23RSS) were catalyzed by 4 mM MgCl2 or 1 mM MnCl2 for 60 min at 37°C as described in ref. 4. Alternatively, 2 nM labeled 23RSS and then 2 nM unlabeled 12RSS was used for certain coupled-cleavage reactions. Cleavage products were separated on 15% TBE/urea-polyacrylamide gels, quantified by using a Molecular Dynamics PhosphorImager (Piscataway, NJ), and expressed as a percentage of the total signal in each lane by using ImageQuant software. Complexes analyzed by EMSA were formed and loaded onto a native 6% acrylamide gel (80:1 acrylamide:bisacrylamide) as in earlier reports (4).
In Vivo Recombination Assays.
Tryptophan to alanine (or phenylalanine) mutations were introduced into pJH548 (encoding RAG1) by site-directed mutagenesis. These constructs were transfected into NIH 3T3 cells together with pJH549 (encoding RAG2) and the recombination substrate pJH200, as described in ref. 43.
Transposition Assays.
Intact substrates were used for transposition into 100 ng of pBR322 by using 5 mM MgCl2 to catalyze the reaction (44).
Supplementary Material
Acknowledgments
We thank Mary H. O'Dea for providing purified HMGB1, MR1, and MR2; Eric Anderson (National Institute of Diabetes and Digestive and Kidney Diseases) for mass spectrometry analysis of W893A; Nancy Craig, Alison Hickman, and Wei Yang for helpful discussion; and Santiago Ramon-Maiques and Andrew Jobson for critical reading. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
Abbreviations
- Ab
abasic
- D
diversity
- dsb
double-strand break
- J
joining
- RSS
recombination signal sequence
- SC
single complex
- V
variable.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611293104/DC1.
References
- 1.Tonegawa S. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 3.Curry JD, Geier JK, Schlissel MS. Nat Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 4.Jones JM, Gellert M. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagawa F, Kodama M, Nishihara T, Ishiguro K, Sakano H. Mol Cell Biol. 2002;22:7217–7225. doi: 10.1128/MCB.22.20.7217-7225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson PC, Desiderio S. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 7.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 8.Eastman QM, Leu TM, Schatz DG. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 9.van Gent DC, Ramsden DA, Gellert M. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 10.Sawchuk DJ, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig MC, Cortes P. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Gent DC, Hiom K, Paull TT, Gellert M. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal A, Eastman QM, Schatz DG. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 13.Hiom K, Melek M, Gellert M. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 14.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landree MA, Wibbenmeyer JA, Roth DB. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice PA, Baker TA. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 19.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 20.Ason B, Reznikoff WS. J Biol Chem. 2002;277:11284–11291. doi: 10.1074/jbc.M111119200. [DOI] [PubMed] [Google Scholar]
- 21.Allingham JS, Wardle SJ, Haniford DB. EMBO J. 2001;20:2931–2942. doi: 10.1093/emboj/20.11.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F. Nat Struct Mol Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 24.Gerstein RM, Lieber MR. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 25.Cuomo CA, Mundy CL, Oettinger MA. Mol Cell Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsden DA, McBlane JF, van Gent DC, Gellert M. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 27.Kale SB, Landree MA, Roth DB. Mol Cell Biol. 2001;21:459–466. doi: 10.1128/MCB.21.2.459-466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mundy CL, Patenge N, Matthews AG, Oettinger MA. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swanson PC. Mol Cell Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankhead T, Chaconas G. Proc Natl Acad Sci USA. 2004;101:13768–13773. doi: 10.1073/pnas.0405762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu K, Lieber MR. Mol Cell Biol. 1999;19:8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DR, Oettinger MA. Mol Cell Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts RJ, Cheng X. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 34.Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F. Mol Cell. 2005;20:143–154. doi: 10.1016/j.molcel.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Eastman QM, Villey IJ, Schatz DG. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo X, Bailin T, Sadofsky MJ. Mol Cell Biol. 2001;21:2038–2047. doi: 10.1128/MCB.21.6.2038-2047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin E, Lithwick G, Levy AA. Genetics. 2001;158:949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu D, Crawford NM. Genetics. 1998;149:693–701. doi: 10.1093/genetics/149.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mol Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadofsky MJ, Hesse JE, van Gent DC, Gellert M. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 41.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Nat Struct Mol Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 42.Elkin SK, Matthews AG, Oettinger MA. EMBO J. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melek M, Gellert M. Cell. 2000;101:625–633. doi: 10.1016/s0092-8674(00)80874-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.