Abstract
The double-ring chaperonin GroEL mediates protein folding, in conjunction with its helper protein GroES, by undergoing ATP-induced conformational changes that are concerted within each heptameric ring. Here we have examined whether the concerted nature of these transitions is responsible for protein substrate release in an all-or-none manner. Two chimeric substrates were designed, each with two different reporter activities that were recovered after denaturation in GroES-dependent and independent fashions, respectively. The refolding of the chimeras was monitored in the presence of GroEL variants that undergo ATP-induced intraring conformational changes that are either sequential (F44W/D155A) or concerted (F44W). Our results show that release of a protein substrate from GroEL in a domain-by-domain fashion is favored when the intraring allosteric transitions of GroEL are sequential and not concerted.
Keywords: allostery, chaperonins, cooperativity, protein folding
The Escherichia coli chaperonin GroEL is a molecular machine that assists protein folding by undergoing allosteric transitions between protein substrate binding and release states (for reviews, see refs. 1–3). It is made up of two homoheptameric rings, stacked back-to-back, with a cavity at each end (4) in which protein folding can take place in a confining and protective environment. GroEL functions in conjunction with a heptameric ring-shaped cochaperonin, GroES (5), that caps the cavity of the so-called cis ring (6), thereby triggering dissociation of bound protein substrates into the cavity. The allosteric transitions of GroEL are induced by ATP binding that occurs with positive cooperativity within rings and negative cooperativity between rings (7, 8). Experimental data (refs. 9 and 10 and G. Curien, J. Grason, and G. Lorimer, personal communication) and simulations (11) have shown that the intraring allosteric transitions of GroEL are concerted, in accordance with the Monod–Wyman–Changeux model of cooperativity (12), as proposed in the nested model (7). In contrast, the intraring allosteric transitions of the eukaryotic heterooligomeric chaperonin CCT were recently shown to occur in a sequential fashion (13) in accordance with the Koshland–Némethy–Filmer model of cooperativity (14). The implications of these different allosteric mechanisms for the folding function of chaperonins have not yet been explored.
The work described here was undertaken to establish whether the concerted intraring ATP-induced allosteric transitions of GroEL facilitate simultaneous release of different parts of a bound protein substrate, as previously speculated (13). By contrast, it was suggested (13) that ATP-induced sequential conformational changes in CCT may facilitate sequential release and, as a result, domain-by-domain substrate folding. A powerful tool available for investigating this question is a wild-type variant of GroEL that contains a fluorescence reporter introduced by the F44W mutation and has the D155A substitution that converts its ATP-induced intraring allosteric transitions from concerted to sequential (15). In other words, this substitution converts the concerted t7→r7 allosteric transition of a wild-type GroEL ring into a sequential allosteric transition such as t7→t4r3→r7. [t and r stand for the respective conformations of a subunit in the low (T) and high (R) affinity states of a ring for ATP, and tnr7-n stands for a ring with n adjacent subunits in the t state and 7 − n adjacent subunits in the r state.] Other single sequential pathways or combinations of pathways are also possible. Hence, demonstration that protein substrate release from the F44W/D155A mutant, but not from the F44W wild-type variant, occurs domain-by-domain would indicate that release of a protein substrate in an all-or-none manner requires a concerted ATP-induced allosteric transition. Two chimeric protein substrates that are appropriate for addressing this question were designed and constructed as described below.
Results
Two fusion proteins each composed of two different GroEL substrates with distinct reporter activities were constructed so that release and folding of each part of these chimera substrates could be monitored separately. A crucial feature of their design is that the two proteins forming each chimera are such that release from GroEL and folding of one of them requires ATP only, whereas release and folding of the other requires both ATP and GroES. Bovine mitochondrial rhodanese was fused to either EGFP or mouse dihydrofolate reductase (mDHFR) because it has been shown that release from GroEL and folding of rhodanese requires ATP and GroES (16), whereas release and folding of EGFP (17) and mDHFR (18) requires ATP only. In the first chimera constructed, the C terminus of EGFP was joined to the N terminus of rhodanese via a KAAATLEFEAS linker peptide (Fig. 1A). The actual linker peptide in the chimera is probably longer because the C-terminal end of EGFP appears to be unstructured (19). This chimera has a molecular mass of ≈66 kDa, and it may, therefore, be too large to be encapsulated in the cis cavity underneath GroES (20), although it has been reported that an 86-kDa protein complex can still be encapsulated (21). The chimera's folding can, however, also be assisted by GroES binding to the trans ring (opposite the polypeptide) of GroEL, as shown for other GroES-dependent substrates (22). Moreover, the size of the substrate is not relevant to this study because the effects of the concerted or sequential ATP-induced conformational changes on substrate release were examined in the absence of GroES. The second chimera, which has a smaller mass of ≈57 kDa and can, therefore, be encapsulated in the cis cavity underneath GroES (20) consists of mDHFR fused to the N terminus of rhodanese with a GSDLRSHHHHHHQAS linker peptide (Fig. 1B). Both chimeras were expressed in E. coli and purified. The first chimera was found to emit 510-nm fluorescent light upon excitation at 465 nm and to have rhodanese activity, whereas the second chimera was found to have both DHFR and rhodanese activities (data not shown). The respective activities of the chimeras are completely lost when they are denatured in 6 M GuHCl (data not shown).
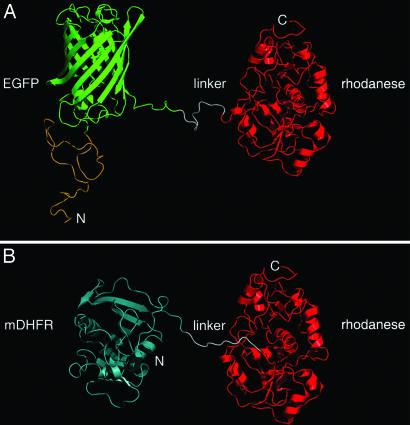
Fig. 1.
Models of the EGFP–rhodanese and mDHFR–rhodanese chimeras. (A) In the EGFP–rhodanese chimera, the C terminus of EGFP (green) is connected to the N terminus of rhodanese (red) by a linker peptide of 11 residues (white). The N terminus of EGFP is fused to a peptide of 39 aa with an N-terminal His6-tag (yellow). (B) In the mDHFR–rhodanese chimera, the C terminus of mDHFR (cyan) is joined to the N terminus of rhodanese (red) via a linker peptide (white) of 15 residues that contains His6-tag. Rhodanese (29), EGFP (19), and mDHFR (30) are represented by ribbon diagrams of their crystal structures (Protein Data Bank codes 1RHD, 1EMA, and 1U70, respectively). In the case of mDHFR, the only available crystal structure corresponds to a mutant ternary complex. The N and C termini of the chimeras are indicated.
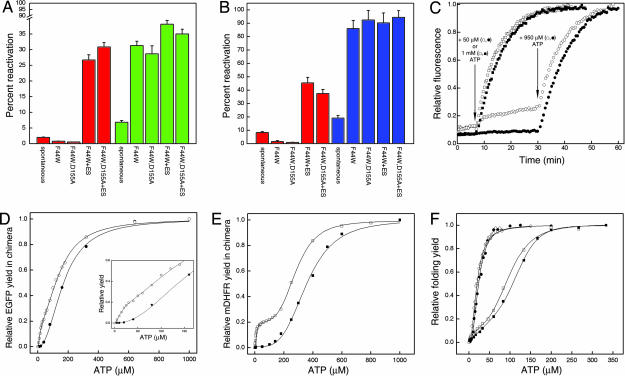
Spontaneous (unassisted) reactivation of the 0.5 μM denatured EGFP–rhodanese chimera resulted in respective yields of ≈7.5% and 2.5% (relative to the same concentration of chimera that was not denatured) in the extent of regain of EGFP fluorescence and rhodanese activity (Fig. 2A). In the case of the mDHFR–rhodanese chimera, spontaneous reactivation resulted in yields of ≈19% and 8% in the respective regains of DHFR and rhodanese activities (Fig. 2B). These differences in yields indicate that the two parts of the chimera can fold independently of each other. The yields are low but consistent with the higher yield of spontaneous folding of EGFP (≈90%) (17) and mDHFR (≈100%) (18) compared with rhodanese (≈25%) (16) under similar conditions. Recovery of the EGFP fluorescence of the chimera (0.5 μM) was found to increase ≈4-fold relative to spontaneous folding in the presence of 1 μM GroEL (F44W or F44W/D155A) and 1 mM ATP, and ≈5-fold when 2 μM GroES was also present (Fig. 2A). In contrast, recovery of the rhodanese activity of the chimera increased ≈12-fold when 1 μM GroEL (F44W or F44W/D155A), 2 μM GroES, and 1 mM ATP were added but was lower than spontaneous folding when GroES was absent (Fig. 2A). Hence, recovery of the rhodanese activity and EGFP fluorescence of the chimera is GroE-dependent in a manner that parallels that of rhodanese and EGFP when they are not fused to each other. Qualitatively similar results were obtained for the mDHFR–rhodanese chimera, although the yields were substantially higher (Fig. 2B). It is important to note that in the presence of 3 μM GroEL and 6 μM GroES (i.e., 6- and 12-fold molar excess of GroEL and GroES relative to the chimera), the respective yields of recovery of rhodanese activity and EGFP fluorescence of the chimera were found to be 70 ± 2.0% and 78 ± 2.5%. This result, which shows that at least 48 ± 3.0% of the individual EGFP–rhodanese chimera molecules regained both activities, indicates that the activities of the two parts of the chimera are not mutually exclusive. This conclusion is also true for the mDHFR–rhodanese chimera, as may be seen from inspection of the data in Fig. 2B.
Fig. 2.
Reactivation of the denatured EGFP–rhodanese and mDHFR–rhodanese chimeras under different conditions. (A) Yields of reactivation of the EGFP–rhodanese chimera in the absence of GroE (spontaneous) and in the presence of GroEL (F44W or F44W/D155A) and 1 mM ATP with or without GroES. The extent of reactivation was determined 4 h after initiation of folding by measuring rhodanese activity (red bars) and EGFP fluorescence (green bars). Each experiment was repeated four times, and standard deviations were determined. The final concentration of the chimera was 0.5 μM. (B) Yields of reactivation of the mDHFR–rhodanese chimera in the absence of GroE (spontaneous) and in the presence of GroEL (F44W or F44W/D155A) and 1 mM ATP with or without GroES. The extent of reactivation was determined 3 h after initiation of folding by measuring rhodanese (red bars) and mDHFR (blue bars) activities. Each experiment was repeated four times, and standard deviations were determined. The final concentration of the chimera was 0.5 μM. (C) Time-resolved regain of the EGFP fluorescence of the chimera in the presence of the F44W and F44W/D155A GroEL mutants and different concentrations of ATP. Denatured chimera and the F44W (filled symbols) or F44W/D155A (open symbols) GroEL mutants were mixed and incubated for 7 min before addition of 50 μM (circles) or 1 mM (squares) ATP. The addition of 50 μM ATP was followed by a second addition of 950 μM ATP to these samples at the time indicated by the arrow. (D) Relative yields of the EGFP fluorescence of the chimera in the presence of the F44W or F44W/D155A GroEL mutants at different concentrations of ATP. The denatured chimera and the F44W (filled circles) or F44W/D155A (open circles) GroEL mutants were mixed, and refolding was then initiated by addition of ATP and allowed to proceed until maximum reactivation was reached. The yields of refolded EGFP were normalized relative to the maximum yield that was obtained at 1 mM ATP and corrected for spontaneous refolding, which is reflected in the slow phase in C and is seen before addition of ATP. The data for the F44W and F44W/D155A mutants were fitted to Hill equations for one or two (31) allosteric transitions, respectively (the data for the F44W/D155A mutant cannot be fitted well to the Hill equation for one allosteric transition). (Inset) A magnification of the plot at low ATP concentrations. (E) Relative yields of refolding of mDHFR in the chimera in the presence of the F44W (filled squares) or F44W/D155A (open squares) GroEL mutants at different concentrations of ATP. The yields were normalized, and the data were fitted as described in the legend to D. Spontaneous refolding before addition of ATP as in C was not observed. (F) Relative yields of refolded EGFP (circles) and mDHFR (squares) in the presence of the F44W (filled symbols) or F44W/D155A (open symbols) GroEL mutants at different concentrations of ATP. Denatured EGFP or mDHFR (not fused to rhodanese) and the F44W or F44W/D155A GroEL mutants were mixed, and refolding was then initiated by addition of ATP and allowed to proceed until maximum reactivation was reached. The yields of refolded substrate are normalized relative to the maximum yields obtained at high ATP concentrations. The data were fitted to the Hill equation for one allosteric transition. Note that maximum yields are reached at ATP concentrations that are lower than those at which maximum yields of the chimeras are reached (D and E).
In the presence of low concentrations of ATP (and no GroES), a striking difference was seen between the extent of the regain of EGFP fluorescence when the EGFP–rhodanese chimera was in the presence of the GroEL variants F44W and F44W/D155A (Fig. 2C). For example, upon addition of 50 μM ATP, there was almost no regain in EGFP fluorescence when refolding took place in the presence of the F44W wild-type variant, whereas a considerable regain in EGFP fluorescence was observed when the F44W/D155A mutant was present. A second addition of ATP (so that the total ATP added was 1 mM) led to a further regain in EGFP fluorescence when refolding took place in the presence of the F44W/D155A mutant. This second addition of ATP also led to a regain in EGFP fluorescence when refolding took place in the presence of the F44W variant (similar to the final regain in the presence of the F44W/D155A mutant). Similar final recoveries of the EGFP fluorescence of the chimera were also observed when 1 mM ATP was added in one step to either the F44W or the F44W/D155A variants. A plot of the recoveries of the EGFP fluorescence of the chimera at different ATP concentrations (Fig. 2D) shows that the difference between the two GroEL variants at 50 μM ATP is also seen at other ATP concentrations but is most pronounced at low ATP concentrations. Importantly, the final recovery of the EGFP fluorescence of the chimera, in the presence of the F44W/D155A mutant, was found (Fig. 2D) to have a bisigmoidal dependence on ATP concentration that mirrors the break in symmetry previously observed in its ATPase activity (15). By contrast, the final recovery of the EGFP fluorescence of the chimera in the presence of the F44W mutant was found to have a simple monosigmoidal dependence on ATP concentration (Fig. 2D). Similar results were obtained for the mDHFR–rhodanese chimera (Fig. 2E). The extent of regain of the DHFR activity of the chimera in the presence of the F44W/D155A mutant was found to have a pronounced bisigmoidal dependence on ATP concentration, whereas in the presence of the F44W mutant it was found to have a monosigmoidal dependence on ATP concentration.
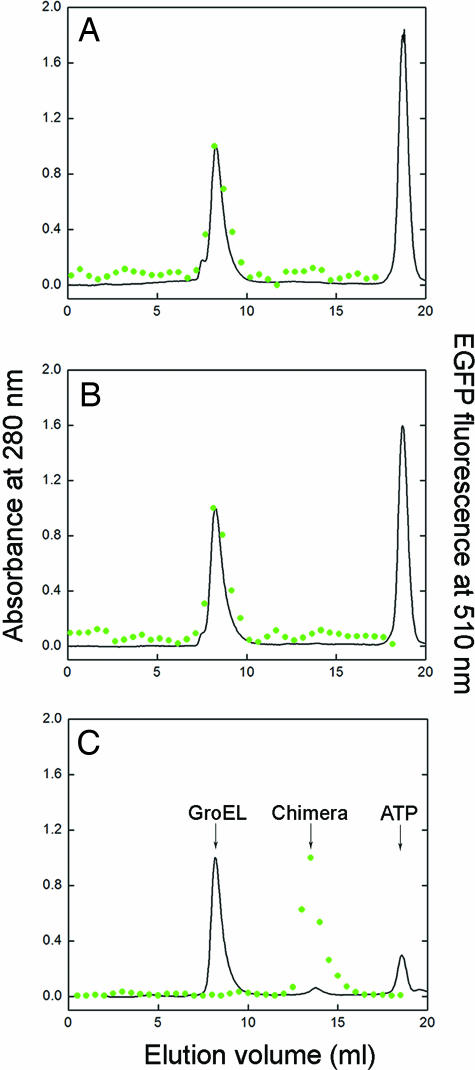
Importantly, little difference was found between the two mutants in assisted refolding of EGFP or mDHFR (Fig. 2F) by themselves (i.e., not fused to rhodanese) at different concentrations of ATP. These findings indicate that the difference between the two GroEL variants in refolding of the EGFP and mDHFR components of the respective chimeras is not the result of differences in their catalytic rate constants of ATP hydrolysis or affinities for ATP. This conclusion is consistent with previous work (15) in which no such differences were found. The results in Fig. 2F also indicate that the difference between the two mutants in reactivation of the chimeras cannot be accounted for by a lower affinity for EGFP or mDHFR of the F44W/D155A mutant compared with the F44W mutant. (The affinity for EGFP of the F44W/D155A mutant might indeed be lower as suggested by a slow regain in the EGFP fluorescence of the chimera before the first addition of ATP) (Fig. 2C). Hence, addition of a low concentration of ATP to the F44W/D155A mutant in complex with the chimera, in the absence of GroES, leads to dissociation and folding of the EGFP or mDHFR components of the respective chimeras, whereas the rhodanese component remains unfolded and GroEL-bound. The conclusion that folding of the EGFP or mDHFR components occurs while the rhodanese component remains unfolded and GroEL-bound is supported by gel-filtration experiments (Fig. 3) showing that folded EGFP coelutes with GroEL after addition of 1 mM ATP to the complex of GroEL (F44W or F44W/D155A) with the chimera. It is also supported by surface plasmon resonance spectroscopy experiments in which the preformed GroEL–substrate complex was immobilized on a chip by using the substrate's His tag that showed ATP dose-dependent dissociation of GroEL from immobilized EGFP but not from immobilized rhodanese or the immobilized EGFP–rhodanese chimera, even at 5 mM ATP (data not shown). Although our data indicate that unfolded rhodanese remains GroEL-bound in the presence of ATP under the conditions in our study, it is possible that it actually undergoes cycles of binding and release (23). Assuming such a scenario cannot account for the biphasic kinetics observed in the case of the F44W/D155A mutant and would, thus, not affect the conclusions of this work. The gel-filtration experiments (Fig. 3) also rule out the possibility that the folded chimera can bind to GroEL. Interestingly, in the case of both mutants, a higher concentration of ATP is required to release the EGFP (Fig. 2D) and mDHDR (Fig. 2E) components of the respective chimeras relative to EGFP and mDHFR (Fig. 2F) alone, thereby indicating that the chimeras have a larger retarding effect on the ATP-induced allosteric transitions of GroEL.
Fig. 3.
Gel filtration of the products of GroEL-assisted reactivation of the denatured EGFP–rhodanese chimera. (A and B) Elution profiles of the products of the chimera refolding reaction in the presence of 1 mM ATP and the respective GroEL mutants F44W or F44W/D155A. (C) The column was calibrated by separating the native chimera that was mixed with 1 μM F44W GroEL and 0.2 mM ATP in G10K buffer. The peaks from left to right correspond to GroEL, the chimera, and ATP, respectively. The elution profiles were determined by measuring the absorbance at 280 nm (black line) and the fluorescence of EGFP (green circles).
Discussion
It has been shown that nonfolded protein substrates bind to GroEL at multiple attachment points located in different and usually adjacent subunits (24). Given that an ATP-induced conformational change in GroEL is a necessary condition for release and folding of (authentic) protein substrates (1–3), it is reasonable to assume that such a change should occur in all of the subunits to which the substrate is bound. Otherwise, complete release of the protein chain might not take place, thereby leading to inefficient (or even mis-) folding. A concerted t7→r7 allosteric switch provides a mechanism for ensuring that the entire protein chain is released. Such a mechanism would also guarantee that different parts of a protein substrate that is attached to nonadjacent subunits (as in the case of rhodanese; see ref. 24) are released together. The purpose of the work described here was to test this idea by monitoring the refolding of two chimeric substrates, each with two different reporter activities, in the presence of GroEL variants that undergo ATP-induced intraring sequential (F44W/D155A) or concerted (F44W) conformational changes. A key aspect of the design of the chimeras was to fuse a GroES-dependent substrate (rhodanese) with a GroES-independent one (EGFP or mDHFR) to test whether addition of ATP releases the GroES-independent part of the chimera when symmetry in GroEL is broken (i.e., when only part of the ring has undergone a conformational switch).
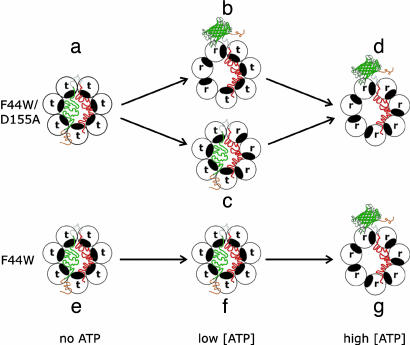
In the absence of ATP, the GroEL rings in complex with the chimera are in the T (t7) state (Fig. 4, species a and e). Upon addition of a small concentration of ATP to the F44W/D155A mutant, in the absence of GroES, there is a break in symmetry because only some (but not all) of the subunits in a ring undergo a t→r transition (Fig. 4, species b and c). This break in symmetry in the F44W/D155A mutant at low ATP concentrations, which does not occur in the F44W wild-type variant, is reflected in the data shown in Fig. 2 D and E. In some rings, the t→r transition takes place in the subunits bound to the EGFP (or mDHFR) part of the chimera, thereby leading to EGFP (or mDHFR) release and folding (Fig. 4, species b). In other rings, the subunits bound to the EGFP (or mDHFR) part of the chimera do not undergo the t→r transition, and as a result, EGFP (or mDHFR) is not released (Fig. 4, species c). The rhodanese part of the chimeras may remain bound to these rings (Fig. 3) or may be released and rebound (23). Hence, at low concentrations of ATP, such as 50 μM, EGFP (or mDHFR) is released only from a certain fraction of rings (Fig. 2 D and E). In the case of the F44W variant, there is no break in symmetry in the presence of low concentrations of ATP (Fig. 2 D and E), and therefore, almost no EGFP or mDHFR is released at 50 μM ATP (Fig. 4, species f). In the presence of a high concentration of ATP, such as 1 mM, the maximum yield of reactivation of EGFP (or mDHFR) is reached because both variants are then predominantly in the R (r7) state (Fig. 4, species d and g). In the absence of GroES, rhodanese is not released and refolded [or is released and then rebound (23)]; therefore, the folded EGFP or mDHFR parts of the respective chimeras remain attached to GroEL (Fig. 3). This result, therefore, also indicates that GroEL-bound nonfolded rhodanese does not block the T→R transition and that the R state retains high affinity for this substrate. The rhodanese component of the chimera is required, however, for the break in symmetry to be reflected in the folding activity.
Fig. 4.
Scheme showing coupling between ATP-induced conformational changes in GroEL and release and folding of EGFP in the EGFP–rhodanese chimera. The ATP-induced conformational change of a subunit from the t state (with low affinity for ATP and high affinity for nonfolded substrates) to the r state (with high affinity for ATP and low affinity for nonfolded substrates) is represented by a counterclockwise rotation that causes the protein substrate-binding site (black) to face away from the cavity. The wild-type variant (F44W) of GroEL and the F44W/D155A mutant undergo ATP-induced concerted (t7→r7) and sequential (e.g., t7→t4r3→r7) allosteric transitions, respectively. The color-coding of the unfolded and folded (ribbon diagram of EGFP) parts of the chimera are according to Fig. 1A. For simplicity, only the substrate-bound ring of GroEL is shown in this scheme.
In conclusion, our findings show that ATP-induced sequential allosteric transitions in the F44W/D155A mutant result in biphasic release and folding of the EGFP or mDHFR components of the chimeras. In contrast, ATP-induced concerted transitions in the F44W wild-type variant result in monophasic release and folding. Hence, the extent of EGFP (or mDHFR) folding mirrors the fraction of subunits of these GroEL variants that is in the r state. This fraction has either a sigmoidal (F44W) or bisigmoidal (F44W/D155A) dependence on ATP concentration. In other words, EGFP (or mDHFR) folding does not depend, in the case of the wild-type variant, on whether the ATP is bound to subunits associated with the rhodanese or EGFP (or mDHFR) parts of the chimera. In the case of the F44W/D155A mutant, however, the effect of ATP on EGFP (or mDHFR) folding does depend on whether it is bound to the subunits associated with the rhodanese or EGFP (or mDHFR) parts of the chimera. Our results therefore indicate that domain-by-domain folding of a multidomain protein can occur when the intraring allosteric transitions of GroEL are sequential. The results also support the proposal that the concerted nature of the intraring allosteric transitions of GroEL is responsible for release of protein substrates in an all-or-none manner. By contrast, sequential intraring allosteric transitions in CCT (13) are likely to facilitate a domain-by-domain release mechanism that may be required for the efficient folding of larger eukaryotic proteins. This model for CCT-assisted folding can be tested by using the approach described here with appropriately designed substrates.
Materials and Methods
Construction of the EGFP–Rhodanese and mDHFR–Rhodanese Chimeras.
The gene for bovine mitochondrial rhodanese (EC 2.8.1.1) subcloned into the NcoI/HindIII sites of the pPROEX HT plasmid (Invitrogen, Carlsbad, CA) was amplified with the oligonucleotides 5′-GCAAGCAAGCTTccATGGTTCATCAGGTGCTC-3′ (forward) and 5′-GCAACGAAGCTTTCAGGCCTTCCCACCCTTCC-3′ (reverse). These oligonucleotides introduced HindIII restriction sites (italic) that flanked the 5′ and 3′ ends of the rhodanese gene and two nucleotides (lowercase) that corrected the reading frame with respect to the EGFP or mDHFR genes. The EGFP–rhodanese chimera was generated by digesting the PCR product with HindIII and inserting it into the pBAD/HisA(EGFP) vector that was previously digested by the same enzyme, treated with calf intestinal alkaline phosphatase, and purified. The pBAD/HisA(EGFP) vector was previously constructed by removing the gene for EGFP(Q185H) from pEGFP (Clontech, Saint-Germain-en-Laye, France) into the PstI/EcoRI sites of pBAD/HisA (Invitrogen) as described (25), thereby adding to the protein an N-terminal tail that contains a His6-tag. A stop codon between EGFP and rhodanese was removed by using site-directed mutagenesis performed with a QuikChange kit (Stratagene, La Jolla, CA) and the oligonu-cleotides 5′-CGGCATGG A C G A G C T G T A CAAG*AAA- G C G GCCGCGACTCTAGAATTCG-3′ (forward) and 5′-CGAATTCTAGAGTCGCGGCCGCTTT*CTTGTACAG- CTCGTCCATGCCG-3′ (reverse). The asterisk indicates the position of the stop codon nucleotide that was deleted. The mDHFR–rhodanese chimera was generated by inserting the above-mentioned digested PCR product containing the gene for rhodanese into the vector pDS56 (26) containing the gene for mDHFR with a His6-tag at its C terminus that was previously digested by HindIII, treated with calf intestinal alkaline phosphatase, and purified. A stop codon between the mDHFR and rhodanese genes was replaced with the codon for Gln as described above by using the oligonucleotides 5′-CTCATCACCATCACCATCACC*AAGCTTCCATGGTTC- ATCAG-3′ (forward) and 5′-CTGATGAACCATGGAAGC- TTG*GTGATGGTGATGGTGATGAG-3′ (reverse). The asterisk was inserted at the right of the mutated base. The complete constructs were verified through DNA sequencing.
Protein Expression and Purification.
The EGFP–rhodanese chimera protein was obtained as follows. E. coli BL21(DE3) cells bearing the pBAD/HisA (chimera) vector were grown overnight in Luria broth (LB) containing 50 μg/ml ampicillin at 37°C. The overnight culture was diluted 1:100 in LB containing 50 μg/ml ampicillin, and the cells were then grown at 37°C until an OD of 0.5 was reached. The temperature was then lowered to 20°C and 0.2% (wt/vol) l-arabinose was added to initiate expression. The cells were grown for an additional 7 h (after 3.5 h, 0.2% l-arabinose was added again) and then centrifuged at 3,000 × g for 15 min. The pellet was resuspended in 20 mM sodium phosphate buffer (pH 7.5) containing 0.5 M NaCl, 10 mM imidazole, and 10 mM 2-mercaptoethanol (buffer A) and then centrifuged as before. The pellet was stored at −80°C until use and then resuspended in buffer A containing 1 mM PMSF. The cells were disrupted by using sonication, and the lysate was clarified by centrifugation at 30,000 × g for 30 min. The supernatant was mixed with Ni-NTA agarose beads, and the slurry was then agitated for 1 h by inverting to allow protein binding. The slurry was then poured into a column and washed with buffer A containing increasing concentrations of 10, 50, 100, and 500 mM imidazole. Green-colored fractions that eluted at 100 mM imidazole were analyzed with SDS/PAGE. The appropriate fractions were combined, transferred into 50 mM Tris·HCl buffer (pH 7.5) containing 10 mM KCl, 10 mM MgCl2 (G10K buffer), and 1 mM DTT by using a PD-10 desalting column (Amersham Pharmacia, Uppsala, Sweden), and then concentrated. Aliquots of protein were snap-frozen and stored at −80°C. The mDHFR–rhodanese chimera was obtained by growing overnight E. coli TG1 cells bearing the pDS56 (chimera) vector in LB containing 50 μg/ml ampicillin at 37°C. The overnight culture was diluted 1:100 in LB containing 50 μg/ml ampicillin, and the cells were then grown at 37°C until an OD of 0.5 was reached when 1 mM isopropyl-β-d-thiogalactopyranoside was added to initiate expression. The cells were grown for an additional 7 h (1 mM isopropyl-β-d-thiogalactopyranoside was added again after 3.5 h) and then centrifuged at 3,000 × g for 15 min. The pellet was resuspended in buffer A, centrifuged as before, and stored at −80°C until use. The cells were disrupted by sonicating the resuspended pellet in buffer A containing 1 mM PMSF, and the lysate was clarified by centrifugation at 30,000 × g for 30 min. Then 35 ml of supernatant was loaded on a 1-ml HisTrap HP column (Amersham Pharmacia) and eluted with a 40-ml gradient (0.5 ml/min) of 0–2 M imidazole in buffer A. Rhodanese-containing fractions were combined, concentrated, and then loaded on a Superdex 200 gel-filtration column preequilibrated by using G10K buffer containing 1 mM Na2S2O3 and 1 mM DTT. Rhodanese-containing fractions were analyzed with SDS/PAGE. The appropriate fractions were combined and concentrated and then aliquots of protein were snap-frozen and stored at −80°C. Protein concentrations were determined by using the Bradford assay with BSA as a standard. The GroEL mutants F44W and F44W/D155A and GroES were constructed (15) and purified (27) as described.
Refolding Assays.
The EGFP–rhodanese chimera (25 μM, unless stated otherwise, in 20 μl of G10K buffer containing 1 mM DTT) was lyophilized in a siliconized test tube, dissolved in 20 μl of 6 M GuHCl and 5 mM DTT, and then incubated with vortex mixing for at least 1 h at room temperature to ensure complete unfolding. Refolding was initiated by diluting the denatured chimera 1:100 in G10K buffer containing 5 mM DTT, 20 mM Na2S2O3 (refolding buffer), and other components when appropriate at 25°C in a siliconized test tube. In cases of unassisted (spontaneous) reactivation, 15 μM BSA was added to the refolding buffer. In cases of GroE-assisted reactivation, 1 μM GroEL, ATP (at the indicated concentration), and 2 μM GroES (when appropriate) were added to the refolding buffer. Reactivation of EGFP in the chimera was monitored by measuring the fluorescence emission of the reactivation mix at 510 nm upon excitation at 465 nm. Recovery of the rhodanese activity of the chimera was monitored by removing 100-μl aliquots of the reactivation mix at different time points after initiation of folding and incubating them for 5–10 min at room temperature with 500 μl of an assay mix containing 50 mM KH2PO4, 50 mM Na2S2O3, and 50 mM KCN. The reaction was stopped by adding 100 μl of 37% formaldehyde, and then 600 μl of ferric nitrate reagent [0.1 g/liter Fe(NO3)3 and 13% HNO3] was added. The solution was clarified by centrifugation, and absorbance at 460 nm was measured (28). Denaturation and refolding of EGFP by itself (i.e., not in the chimera) was carried out as described (17).
The mDHFR–rhodanese chimera (10 μM unless stated otherwise) was dissolved in 6 M GuHCl and 5 mM DTT and then incubated with vortex mixing for 1 h at room temperature to ensure complete unfolding. Refolding was initiated by diluting the denatured chimera 1:50 in G10K buffer containing 50 μM dihydrofolic acid, 20 mM Na2S2O3, 5 mM DTT, and other components when appropriate at 25°C in a siliconized test tube. In cases of unassisted (spontaneous) reactivation, 15 μM BSA was added to the refolding buffer. In cases of GroE-assisted reactivation, 0.8 μM GroEL, ATP (at the indicated concentration), and 1.6 μM GroES (when appropriate) were added to the refolding buffer. The extent of reactivation of mDHFR in the chimera was measured by adding final concentrations of 60 μM NADPH and 100 μM dihydrofolic acid to the refolding mixture and determining the initial reaction velocity from the decrease in the absorbance at 340 nm as a function of time. Denaturation and refolding of mDHFR by itself (i.e., not in the chimera) were carried out as described above.
Gel Filtration of Reactivation Products.
Reactivation of denatured EGFP–rhodanese chimera was initiated by diluting it 1:100 in 1 ml of refolding buffer containing 1 mM ATP and the F44W or F44W/D155A GroEL mutants at 1 μM. The fluorescence emission of EGFP was monitored, and when it reached a maximum the sample was concentrated to <100 μl by using a Microcon ultrafiltration device (Millipore, Billerica, MA) with a 100-kDa cutoff (the flow-through during concentration had no fluorescence) and then loaded onto a Superdex 200 gel-filtration column. The absorbance at 280 nm was monitored, fractions of 500 μl were collected, and the fluorescence of EGFP was measured.
Acknowledgments
We thank Dr. M. Panda (University of Texas, San Antonio, TX) for the pPROEX HT vector bearing the rhodanese gene and for helpful advice and Drs. Debbie Fass and Tammy Horovitz for comments on the manuscript. This work was supported by Israel Science Foundation Grants 67/05 (to A.H.) and 325/03 (to G. H.). A.H. is an incumbent of the Carl and Dorothy Bennett Professorial Chair in Biochemistry.
Abbreviation
- mDHFR
mouse dihydrofolate reductase.
Footnotes
Author contributions: Y.K. and N.P. contributed equally to this work; A.H. designed research; Y.K. and N.P. performed research; Y.K., N.P., G.H., and A.H. analyzed data; and A.H. wrote the paper.
The authors declare no conflict of interest.
References
- 1.Thirumalai D, Lorimer GH. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 2.Saibil HR, Horwich AL, Fenton WA. Adv Protein Chem. 2002;59:45–72. doi: 10.1016/s0065-3233(01)59002-6. [DOI] [PubMed] [Google Scholar]
- 3.Horovitz A, Willison KR. Curr Opin Struct Biol. 2005;15:646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 5.Hunt JF, Weaver AJ, Landry SJ, Gierasch L, Diesenhofer J. Nature. 1996;379:37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 6.Xu Z, Horwich AL, Sigler PB. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 7.Yifrach O, Horovitz A. Biochemistry. 1995;34:5303–5308. doi: 10.1021/bi00016a001. [DOI] [PubMed] [Google Scholar]
- 8.Yifrach O, Horovitz A. J Mol Biol. 1996;255:356–361. doi: 10.1006/jmbi.1996.0028. [DOI] [PubMed] [Google Scholar]
- 9.Yifrach O, Horovitz A. J Am Chem Soc. 1998;120:13262–13263. [Google Scholar]
- 10.Shiseki K, Murai N, Motojima F, Hisabori T, Yoshida M, Taguchi H. J Biol Chem. 2001;276:11335–11338. doi: 10.1074/jbc.M010348200. [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Sigler PB, Xu Z, Karplus M. J Mol Biol. 2000;302:303–313. doi: 10.1006/jmbi.2000.4014. [DOI] [PubMed] [Google Scholar]
- 12.Monod J, Wyman J, Changeux J-P. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 13.Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Nat Struct Mol Biol. 2005;12:233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- 14.Koshland DE, Jr, Némethy G, Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 15.Danziger O, Rivenzon-Segal D, Wolf SG, Horovitz A. Proc Natl Acad Sci USA. 2003;100:13797–13802. doi: 10.1073/pnas.2333925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza JA, Rogers E, Lorimer GH, Horowitz PM. J Biol Chem. 1991;266:13044–13049. [PubMed] [Google Scholar]
- 17.Makino Y, Amada K, Taguchi H, Yoshida M. J Biol Chem. 1997;272:12468–12474. doi: 10.1074/jbc.272.19.12468. [DOI] [PubMed] [Google Scholar]
- 18.Viitanen PV, Donaldson GK, Lorimer GH, Lubben TH, Gatenby AA. Biochemistry. 1991;30:9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- 19.Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 20.Sakikawa C, Taguchi H, Makino Y, Yoshida M. J Biol Chem. 1999;274:21251–21256. doi: 10.1074/jbc.274.30.21251. [DOI] [PubMed] [Google Scholar]
- 21.Song J-L, Li J, Huang Y-S, Chuang DT. J Biol Chem. 2003;278:2515–2521. doi: 10.1074/jbc.M209705200. [DOI] [PubMed] [Google Scholar]
- 22.Farr GW, Fenton WA, Chaudhuri TK, Clare DK, Saibil HR, Horwich AL. EMBO J. 2003;22:3220–3230. doi: 10.1093/emboj/cdg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissman JS, Kashi Y, Fenton WA, Horwich AL. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 24.Farr GW, Furtak K, Rowland MB, Ranson NA, Saibil HR, Kirchhausen T, Horwich AL. Cell. 2000;100:561–573. doi: 10.1016/s0092-8674(00)80692-3. [DOI] [PubMed] [Google Scholar]
- 25.Habuchi S, Cotlet M, Gronheid R, Dirix G, Michiels J, Vanderleyden J, De Schryver FC, Hofkens J. J Am Chem Soc. 2003;125:8446–8447. doi: 10.1021/ja0353311. [DOI] [PubMed] [Google Scholar]
- 26.Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle M-T, Dobberstein B. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 27.Danziger O, Shimon L, Horovitz A. Protein Sci. 2006;15:1270–1276. doi: 10.1110/ps.062100606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sörbo BH. Methods Enzymol. 1955;2:334–337. [Google Scholar]
- 29.Ploegman JH, Drent G, Kalk KH, Hol WGJ. J Mol Biol. 1978;123:557–594. [PubMed] [Google Scholar]
- 30.Cody V, Luft JR, Pangborn W. Acta Crystallogr D. 2005;61:147–155. doi: 10.1107/S0907444904030422. [DOI] [PubMed] [Google Scholar]
- 31.Kafri G, Willison KR, Horovitz A. Protein Sci. 2001;10:445–449. doi: 10.1110/ps.44401. [DOI] [PMC free article] [PubMed] [Google Scholar]