Abstract
Polo-like kinase (Plk1) is crucial for cell cycle progression through mitosis. Here we present the molecular and structural mechanisms that regulate the substrate recognition of Plk1 and influence its centrosomal localization and activity. Our work shows that Plk1 localization is controlled not only by the polo box domain (PBD); remarkably, the kinase domain is also involved in Plk1 targeting mechanism to the centrosome. The crystal structures of the PBD in complex with Cdc25C and Cdc25C-P target peptides reveal that Trp-414 is fundamental in their recognition regardless of its phosphorylation status. Binding measurements demonstrate that W414F mutation abolishes molecular recognition and diminishes centrosomal localization. Therefore, Plk1 centrosomal localization is not controlled by His-538 and Lys-540, the residues involved in phosphorylated target binding. The different conformations of the loop, which connects the polo boxes in the apo and the PBD-Cdc25C and PBD-Cdc25C-P complex structures, together with changes in the proline adjacent to the phosphothreonine in the target peptide, suggest a regulatory mechanism to detect binding of unphosphorylated or phosphorylated target substrates. Altogether, these data propose a model for the interaction between Plk1 and Cdc25C.
Keywords: cell cycle, enzymes, kinase, protein structure, polo box domain
The genomic integrity of all eukaryotic cells depends on the error-free segregation of chromosomes during mitotic and meiotic cell divisions. During the phase of mitosis, a major reorganization of the cell architecture, including the formation of the mitotic spindle and the actin–myosin contractile ring, is needed to transmit the genetic information to the daughter cells. This tightly regulated space–time process depends on enzyme families that control protein phosphorylation.
Polo-like kinase 1 (Plk1) resides at the centrosome during interphase and is an important regulatory enzyme in cell cycle progression during M phase; it is conserved from yeast to human, and its family is composed of three additional members in mammals (1, 2). Plk1 is involved in important processes such as assembly and dynamics of the mitotic spindle apparatus (3), activation and inactivation of cycle-dependent kinases Cdk/cyclin complex (1), removal of cohesin from chromatin (4), regulation of the anaphase-promoting complex/cyclosome (APC/C; ref. 5), and control of mitotic exit and cytokinesis (6–8). Some Plk1 substrates are Cdc25C phosphatase, several APC/C subunits, cyclin B, SCC1 cohesin, and some kinesin-related motor proteins (2). All these interactions demonstrate the multiple roles of Plk1 during mitosis.
Plk1 is composed of a common N-terminal catalytic domain and a C-terminal regulatory domain with highly conserved sequences named polo boxes (PB). The PB motif is observed only in the Plk and contains a characteristic sequence, which is the hallmark of this protein family. This motif is supposed to be involved in an autoregulatory mechanism or in targeting the kinase to its substrates (9). The intramolecular interactions between the catalytic and noncatalytic domain [PB domain (PBD)], as well as the phosphorylation of Plk1, regulate the activation of its protein kinase activity (10). Phosphorylation represents an important mechanism for Plk1 activation. Highly conserved Ser-137 and Thr-210 residues within the catalytic domain of mammalian Plk1 have been identified as potential activating phosphorylation sites (7, 11).
New evidence indicates that Plk1 forms part of the regulatory circuit that controls mitosis entry by binding to phosphorylated Cdc25C through its PBD. Indeed, Plk1 can phosphorylate and thereby regulate both Cdc25C (12) and the Cdk1 inhibitor Myt1 (13, 14). This would seem consistent with the hypothesis that Plk1 is the “trigger” kinase for the activation of Cdk1. However, an alternative view holds that Plk1 activation depends on the prior activation of Cdk1, in which case Plk1 would function primarily in feedback loops. Whether Plk1 regulates Cdc25C phosphatase at the level of activity or localization (or both) remains to be established.
Recent work has described the structure of the PBD bound to a nonphysiological peptide found in a proteomic screen looking for p-Thr- and p-Ser-binding domains (15–17). This peptide was synthesized as a member of an immobilized library of degenerate phosphopeptides, and it cannot be found in any natural protein. This finding suggested an important role for the PBD in Plk1 centrosomal localization and substrate recognition, especially residues His-538 and Lys-540, which are involved in phosphate binding. In contrast, we show evidence that discards phosphate binding as the driving force for Plk1 centrosomal localization and questions the role of the PBD as the unique determinant of Plk1 localization. Our experiments reveal the importance of the N-terminal domain in centrosomal localization and substrate recognition, indicating that Plk1 centrosomal localization can be achieved independently of substrate binding and of its phosphorylation state. On the other hand, the influence of substrate binding on Plk1 activity shows that Plk1 is not activated upon target binding; an upstream activation is needed before binding to enhance its activity. The structures of the human Plk1-PBD in complex with an unphosphorylated and a phosphorylated target peptide from Cdc25C, a Plk1 natural substrate, reveal the mode of binding of the substrate in both states and the importance of Trp-414 in the PBD-binding pocket. The key role of this residue in substrate recognition and its contribution to Plk1 localization have been confirmed by site-directed mutagenesis.
Results
EGFP-Plk1 and EGFP-Plk1 H538A/K540M Localize at the Centrosome.
Even though the PBD alone can localize to the centrosome, a double mutation in residues H538A and K540M has been shown to hamper its centrosomal localization in permeabilized cells (16). However, when Plk1 and Plk1 H538A/K540M were fused to EGFP, we could not detect any difference in their centrosomal localization. Human PC3 cells were transfected with either EGFP-Plk1 or EGFP-Plk1 H538A/K540M. After 20 h of expression, both the wild-type (WT) Plk1 and the double mutant localized at the centrosome (Fig. 1a Center and Right). Equivalent results were observed in HeLa and NIH 3T3 cells (data not shown).
Fig. 1.
Plk1 and Plk1 H538A/K540M mutant localization at centrosomes and FRAP analysis. (a) PC3 cells were transfected with either EGFP-Plk1 or with EGFP-Plk1 H538A/K540M and stained for γ-tubulin (red) and DNA (blue). (Center) The precise localization of EGFP-Plk1 (green) at the centrosomes (arrowheads) colocalizing with γ-tubulin. (Right) The same pattern is shown when cells express EGFP-Plk1 H538A/K540M. (Left) EGFP expression alone. A detailed view of the centrosomal area is depicted (Insets). (b) Analysis of EGFP- and EGFP-Plk1 H538A/K540M FRAP experiments at the centrosomal region. Each plot represents an average of 10 individually analyzed cells per construct.
Fluorescence Recovery After Photobleaching (FRAP) Shows Similar Dynamic Behavior for EGFP-Plk1 and EGFP-Plk1 H538A/K540M.
FRAP analysis was performed to observe whether the dynamics of Plk1 at the centrosome can be affected by the double mutation in the PBD. The EGFP-Plk1 centrosomal signal was bleached, and the recovery of the signal was monitored in 3- to 5-s lapses. In each case, at least 10 cells were monitored during 3 min. This analysis revealed that Plk1 has a recovery half-time (t1/2) at the centrosome of 9.8 (±0.09) s, and a mobility fraction (Mf) ≈40% (Fig. 1b, green plot). Comparable data were observed with the Plk1 H538A/K540M mutant. The t1/2 is slightly faster, 6.6 (±0.17) s, and the mobility fraction is reduced to 33% (Fig. 1b, red plot). This minor change in diffusion time could be due to the fact that these mutations are very drastic; both polar residues are mutated to hydrophobic ones, and this change could disturb the area, inducing a slightly faster Plk1 diffusion rate. Nevertheless, the Plk1 double mutant recovers its signal at the centrosome following very similar kinetics as the WT Plk1. Thus, Plk1 centrosomal localization does not seem to be ruled by phosphorylated substrate recognition led by the PBD.
Affinity Measurements Imply That the Kinase Domain Is Needed for Recognition of the Phosphorylated Target Peptide.
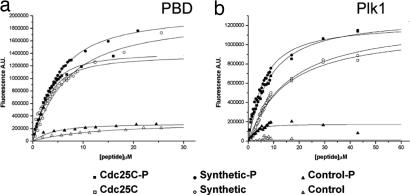
Thereby, we analyzed whether the affinity of Plk1 to its substrates could be influenced by other protein regions outside the PBD. To achieve this aim, we measured the interaction of Plk1 and the PBD to a target peptide from Cdc25C phosphatase. Plk1 recognizes this region of Cdc25C upon Thr-130 phosphorylation (ref. 18; see also Material and Methods for peptides sequences). Surprisingly, the PBD displayed a similar affinity for the unprimed (nonphosphorylated) and the primed (phosphorylated) Cdc25C target peptides (Fig. 2). The Cdc25C peptide affinities were approximately three times higher in both cases when compared with those obtained for an unphosphorylated and a phosphorylated nonphysiological peptide [ref. 15; Fig. 2; supporting information (SI) Table 1]. Nevertheless, the ability of the PBD to discriminate between the primed and unprimed target peptides is low, as deduced from the calculated Kd (SI Table 1). However, when the experiments were performed with the full-length protein, a preference for the primed target peptide could be observed (Fig. 2b; SI Table 1). Plk1 showed ≈7-fold more affinity for the phosphorylated peptide. A control peptide whose sequence corresponds to the T loop of Plk1 with and without pThr was used to show that both the PBD and Plk1 have specificity toward a certain substrate target peptide sequence. In both cases, the proteins depicted negligible binding to the control peptide, notwithstanding its phosphorylation state. Altogether, these measurements suggest that the PBD alone is not able to distinguish between the unprimed and the primed target peptide. The presence of the kinase domain seems essential for this purpose. However, both Plk1 and the PBD recognize a target sequence.
Fig. 2.
The PBD binds Cdc25C and Cdc25C-P with similar affinity, but Plk1 shows preference for the phosphorylated form. PBD (a) and Plk1 (b) affinities using different target peptides were measured by intrinsic Trp fluorescence.
PBD Substrate Binding Promotes Plk1 Activity After Kinase Activation.
Substrate binding to Plk1 is supposed to promote its activity (19). In contrast, we found that the enzyme must be previously activated before the target substrate can enhance kinase activity (SI Fig. 6). His-tagged Plk1 purified from Sf9 cells weakly phosphorylated a common kinase substrate (Histone H1) (SI Fig. 7a, lane 1). When Plk1 was assayed in the presence of increasing amounts of Cdc25C and Cdc25C-P ranging from 0.5 to 1.5 (1:0.5, 1:1.0, and 1:1.5 Plk1/target peptide molar ratios), we did not see any effect on kinase enzymatic activity (SI Fig. 7 b and c Left), indicating that target peptide binding does not promote Plk1 activity. Previous reports have used a phosphatase inhibitor to induce Plk1 activation (12). However, to mimic active Plk1, we incubated the recombinant kinase in the presence of a mitotic HeLa cell extract. After incubation with this extract, pure Plk1 was activated, displaying 5- to 6-fold more activity (SI Fig. 7a, lane 2; see SI Fig. 6 for other control experiments). This method allows the study of Plk1 enzymatic properties in its inactive and active states, which will resemble Plk1 status in G1 and mitosis, respectively. Once the activated Plk1 was assayed with Cdc25C and Cdc25C-P target peptides, by using the same molar ratios as before, a kinase activity enhancement could be observed despite peptide phosphorylation status (SI Fig. 7 b and c Right). When this assay was performed by using both unphosphorylated and phosphorylated nonnatural peptides found in a previous study (15), the kinase activity decreased ≈50% (SI Fig. 7 d and eRight). Thus, although the nonnatural peptide binds to Plk1, it does not elicit the same effect in the enzyme as the Cdc25C one.
Crystal Structures of PBD and Its Complexes with Cdc25C Target Peptides Reveal Different Binding Properties.
The crystal structures of PBD and its complexes with Cdc25C-P and Cdc25C peptides have been solved by molecular replacement and refined to 1.95-, 2.10-, and 2.80-Å resolution (see SI Table 2). The structures show a common scaffold that has been described (16, 17). In each PB, the six β-strands form an antiparallel β-sheet building a shallow cavity where the Cdc25C and Cdc25C-P peptides bind (Fig. 3a; SI Fig. 8). After careful comparison of the three models, the main difference arises from the 20-residue loop, which is disordered in the apo and PBD-Cdc25C structures (residues Ala-493–Arg-507 for apo and Glu-488–Arg-507 for the PBD-Cdc25C complex). This loop [connecting loop (CL)] joins both PB and flanks the binding site of the target peptide (SI Figs. 8 and 10a). However, when the primed peptide was bound, the CL was well defined and thus built into the structure.
Fig. 3.
PBD Cdc25C peptide complex crystal structures. (a) Surface representation of the PBD/Cdc25C and the PBD/Cdc25C-P crystal structures. The coloring scheme represents the contact area between the target peptide and the protein ranging from cyan (no contact) to magenta (strong contact). The peptide is depicted in yellow stick representation. (b) 2Fo−Fc σA-weighted electron-density map contoured at 1 σ showing the residues and the solvent molecules involved in the binding of the Cdc25C-P peptide. A water-mediated interaction of the phosphate moiety with Arg-518 and Lys-556 from two different crystallographically related molecules can be observed.
In both structures where the peptide is bound, only 7 of its 10 residues could be modeled into the electron-density map. A close view reveals the binding differences and common features between the primed and unprimed target peptide. These differences affect the conformation of the PBD-binding cleft amino acids as well as the target peptide conformation in the binding pocket (SI Figs. 9 and 10b). Although the conformation of the central core of the target peptide is similar in both cases (Cys-3–Ser-4–Thr-5), there are conformational changes in the main and side chains of several amino acids, as well as in their protein–peptide hydrogen-bonding networks depending on the peptide phosphorylation state (see detailed description in SI Text). The presence of the phosphate in Thr-5 side chain promotes the interaction of O1P with His538ND1 (2.65 Å) and O2P with Lys-540 NZ (2.69 Å), as was observed in the nonphysiological peptide structures (16, 17). Interestingly, the unprimed Thr-5 also interacts with the side chain of His-538 (Thr5OG1 His538ND1, 3.5 Å), favoring the binding of the unprimed peptide (SI Fig. 9). Moreover, the phosphate moiety is associated with seven water molecules that form an extensive hydrogen-bond network. These water molecules are absent when the unprimed peptide is bound.
Pro-6 in the PBD-Cdc25C complex varies its position, promoting a conformational change in the unprimed peptide (SI Fig. 10b). Consequently, the N terminus of the unprimed peptide occupies the position where the main chain of the CL is located in the PBD-Cdc25-P complex (SI Figs. 9 and 10a). Therefore, none of the interactions observed with the primed peptide could be detected with the unprimed peptide complex.
The Trp-414 builds several interactions between the PBD and the target peptide that are similar despite its phosphorylation status. This residue seems to play an important role in Plk1 subcellular localization; indeed, its mutation (W414F) disrupts Plk1 subcellular localization without disturbing its kinase activity (9, 20). The presence of Trp-414 displaying its indole ring on the bottom of the binding cleft contacts Ser-4, Cys-3, and Leu-2 main chains whose positions are conserved in both PBD-Cdc25C and PDB-Cdc25C-P structures (SI Fig. 9). These data indicate that Trp-414 could be involved in the molecular recognition of the target substrate independently of its phosphorylation state.
A comparison between the structures of the PBD-Cdc25C-P and PBD in complex with the nonnatural peptide reveals differences in the peptide-binding mode (SI Fig. 10c). Even though the p-Thr and the following Ser maintain similar conformations and contact similar residues in the PBD, the position, bond lengths, and number of water molecules that can interact with O1P and O2P in Cdc25C-P are different from those described for the nonnatural peptide (16, 17). It is noteworthy that a close view of the Cdc25C-P phosphate moiety reveals its interaction with the side chain from two basic residues, Arg-518 and Lys-556, which belong to other crystallographically related molecules (Fig. 3b). This shows that the phosphate-binding cleft in the PBD is exposed and thus accessible to residues located outside the PBD. Additional differences are observed in the peptide conformation both in the C- and N-terminal regions. The presence of polar amino acids in the C-terminal region of the nonnatural peptide (Gln-His-Met) contrasts with the hydrophobic ones in these positions in the Cdc25C-P peptide (Cys-Leu-Leu). This distinct chemical character promotes an array of different interactions between the ligand and the protein, which are reflected in the different accommodations that the side chains adopt (SI Fig. 10c). The differences are minor in the residues in the N-terminal side before the pThr (Asn-Pro). So far, all of the PBD structures crystallized with a phosphopeptide conserve this N-terminal region conformation and keep the CL ordered despite all of the crystals belonging to different space groups.
W414F Mutation Abolishes PBD Target Peptide Binding and Reduces EGFP-Plk1 Centrosomal Localization.
To clarify the role of Trp-414 in substrate recognition, we mutated this residue to Phe and analyzed its binding properties to Cdc25C target peptide and its role in Plk1 centrosomal localization. Because of this mutation, the intrinsic fluorescence probe to measure binding was eliminated; consequently, the interaction was measured by using isothermal titration calorimetry. The binding of the target peptide Cdc25C-P with the WT PBD yielded a Kd similar to that calculated by fluorescence (1.8 μM) (Fig. 4a). However, target peptide binding was severely affected by PBD W414F mutation, and no binding could be detected independently of peptide phosphorylation status (Fig. 4a). To examine the effect of this mutation in vivo, the EGFP-Plk1W414F centrosomal localization was studied. The protein was localized at the centrosome, although its fluorescence intensity decreased ≈50% when compared with the WT (Fig. 4 b and c), indicating that Trp-414 is a key residue that links substrate recognition and PBD influence in Plk1 localization, two important properties involved in Plk1 regulatory mechanism.
Fig. 4.
The W414F mutation abolishes PBD target peptide binding and affects Plk1 localization. (a) Isothermal titration calorimetry was used to determine the binding constant of the PBD W414F mutation to the target peptide Cdc25C-P. The interaction of the peptide Cdc25C-P with the PBD was measured as a control (Left). No detectable binding was observed for the PBD W414F mutant (Right) in the presence of the target peptide. (b) PC3 cells were transfected with either Plk1, Plk1-W414F, or the truncated Plk1 Δ400–603 fused to EGFP, and stained for γ-tubulin (red) and DNA (blue). (Left) The precise localization of WT Plk1 (green) at the centrosomes (arrowheads). The same pattern is shown when cells are expressing Plk1-W414F or Plk1 Δ400–603 showing γ-tubulin colocalization (arrowheads). A detailed view of the centrosomal area is depicted (Insets). (c) Centrosomal fluorescence quantitation for the different EGFP-Plk1 constructs and EGFP alone related to γ-tubulin intensity. Arbitrary units were taken considering the EGFP signal as background.
To elucidate whether other regions apart from the PBD can still guide the protein to the centrosome, we analyzed the localization properties of an EGFP-Plk1 Δ400–603 mutant, a trimmed Plk1 lacking the PBD. The truncated Plk1 localized to the centrosome and displayed a similar fluorescence intensity to the W414F mutant (Fig. 4 b and c), indicating that W414F mutation completely abolishes PBD control on Plk1 localization. Moreover, the centrosomal dynamics of this truncated Plk1 are similar to the WT and the H538A/K540M mutant (SI Fig. 11). Even though the interaction of the Plk1 kinase domain with α, β, and γ tubulins has been described (21), our data show in vivo that the kinase and/or the linker of Plk1 contribute to its subcellular localization and could direct Plk1 at certain subcellular areas independently of the presence or the phosphorylation status of its substrates, which are bound by the PBD.
Discussion
Plk1 is one of the major mitotic regulators. Here, we present a combination of experiments to unravel the molecular mechanisms that regulate substrate recognition and Plk1 localization.
Plk1 localization at the centrosome is a well known characteristic of this enzyme that has been described since it was discovered in several organisms (1, 22, 23). Recently, a double mutation in His-538 and Lys-540, the residues involved in phosphate coordination, have been shown to abolish PBD centrosomal localization (16). We have carried out localization experiments (Fig. 1a) with WT Plk1, Plk1 H538A/K540M, and Plk1 Δ400–603 fused to EGFP. In contrast to previous experiments performed with the PBD (16), both the WT Plk1 and Plk1 H538A/K540M and the truncated Plk1 Δ400–603 behave in a similar manner and display centrosomal localization, indicating that the residues responsible for phosphate coordination are not critical for this regulatory feature of Plk1. In addition, FRAP experiments with the WT and the mutants revealed similar kinetics (Fig. 1b and SI Fig. 11). Thus, the full-length Plk1 is not negatively influenced in its centrosomal localization properties or in its dynamic behavior by the mutation in the residues that bind the phosphorylated substrate. In fact, even the depletion of the 200 C-terminal residues does not abolish its centrosomal presence.
The analysis of the interaction of Plk1 and the PBD with their substrates showed that, whereas the PBD displayed a similar affinity for the Cdc25C peptide despite its phosphorylation state in Thr-5, Plk1 had a 7-fold higher affinity for the phosphorylated form (Fig. 2), indicating that the PBD is not a strict pSer/pThr-binding domain, and the recognition of a primed target substrate needs the presence of the kinase and/or the linker. In addition, the PBD-Cdc25C-P structure shows that the phosphate moiety is accessible by residues outside the PBD (Fig. 3b), emphasizing the fact that the PBD-binding cleft is rather exposed, and other residues outside the PBD could be involved in phosphate binding.
In contrast to the proposed working model of Plk1 (19), we observed that the binding of a natural substrate peptide does not promote kinase activity. An upstream activation is required before the target peptide can enhance Plk1 activity. Our assays show that recombinant Plk1 barely phosphorylates histone H1 (SI Figs. 6 and 7). Thus, our recombinant Plk1 is an excellent tool to elucidate whether target peptide binding could elicit kinase activity. Remarkably, neither the Cdc25C target peptides nor the nonphysiological ones were able to switch on the enzyme independently of their phosphorylation status. So far, the only mechanism used to obtain active recombinant Plk1 has been the incubation of the culture with okadaic acid (12), a phosphatase inhibitor that arrests cells at the onset of mitosis. Instead, we activated Plk1 by using extracts from mitotic HeLa cells. A clear activation after incubation with a mitotic extract could be observed (SI Figs 6 and 7a). The effect of the target peptide binding on the activated Plk1 shows a clear increase in the presence of Cdc25C target peptides, independently of their phosphorylation status (SI Fig. 7 b and c Right). This increase doubled the kinase activity at 1.5 target peptide/protein molar ratio.
Previous work has shown that the PBD cannot interact with the kinase domain upon Plk1 Thr-210 phosphorylation (10). Interestingly, upon Plk1 activation in the presence of a mitotic extract, the kinase seems to bind both primed and unprimed peptides in a similar manner as the PBD (Fig. 2a). This behavior suggests that once Plk1 is activated, a conformational change may disrupt contacts between the kinase and the PBD (10). In this conformation, the PBD in Plk1 will resemble the properties of the PBD alone. Therefore, Plk1 loses its selectivity and can bind substrates regardless of their phosphorylation state, and the increase in Plk1 activity is similar despite target peptide phosphorylation.
The molecular basis of substrate recognition by the PBD is understood after a detailed view of the x-ray structures of the PBD and its complexes with a physiological target peptide (Fig. 3 and SI Fig. 9). The central part of the Cdc25C peptide composed of Cys-Ser-Thr is the core of the target peptide, which is recognized by the PBD independently of the phosphorylation status. This observation is supported by the similar conformations and interactions with the PBD amino acids in both structures (SI Fig. 9).
The PBD-Cdc25C and PBD-Cdc25C-P structures highlight the role of Trp-414 in peptide binding. This residue plays a major role in substrate recognition and localization, as shown by using site-directed mutagenesis (Fig. 4). Although there seems to be a link between substrate recognition and localization mediated by the PBD, this mechanism does not depend on the target substrate phosphorylation state; the mutation of the residues responsible for phosphate binding does not abolish Plk1 localization (Fig. 1), and depletion of the PBD does not hamper Plk1 centrosomal localization (Fig. 4 b and c), although its intensity is affected similar to the W414F mutant. Thus, Trp-414 seems to be the molecular link between the substrate recognition and localization functions in the PBD, a key issue in Plk1 regulatory mechanism, which contributes but does not fully control enzyme localization to the centrosome.
Although the structure of the PBD among the three different crystals is well conserved (SI Fig. 10a), the loop that connects PBI and PBII is ordered only in the case of the PBD-Cdc25C-P complex. A similar conformation for this loop was observed in the structures of the PBD with the nonnatural peptide (16, 17), despite the fact that they crystallized in different space groups. It is noteworthy that the N-terminal sequences of Cdc25C and the nonnatural peptide are identical in this region, and a similar interaction is conserved that orders the CL loop. However, in the PBD-Cdc25C complex, this loop is forced to change its conformation due to a rotation in the ring of Pro-6 (SI Fig. 10b), which swaps its conformation, pointing toward the inner face of the pocket. This change in the CL depending on the target peptide threonine phosphorylation could be a regulatory mechanism to distinguish the binding of the unphosphorylated and phosphorylated target substrates, to ensure that Plk1 activity is directed toward its phosphorylated substrates.
The interaction among Cdc25C, Plk1, and the Cdk1/CyclinB complex has clear implications during mitotic onset. The possibility that the subcellular localization of these proteins can be used as a trigger to start mitosis has been considered (2). It is well known that Plk1 is located at the centrosome at the G2/M transition (1, 24), and recent work has shown that the Cdk1/cylinB complex is activated at the centrosome (25). All these data support the activation of mitosis at the centrosome, where several components that trigger the activation of the mitosis-promoting factor seem to be present. Following this line, a hypothetical model for the regulation of Plk1 and Cdc25C interaction during the cell cycle is suggested (Fig. 5). Plk1 could locate at the centrosome at very early stages of the cell cycle, as we and others have observed (Figs. 1a and 4b). During early G2, both Plk1 and Cdc25C are still not primed, although they could interact and form a complex, as has been observed in vitro (12). Interestingly, if this interaction happens, it should not promote any Plk1 kinase activity (SI Fig. 7). However, most probably this interaction cannot occur at this point because of Cdc25C binding to 14-3-3 protein, which likely hides the target area for Plk1 binding (26). On the other hand, in the structures of both the PBD and the PBD-Cdc25C complex, the CL that joins PBI and PBII does not have a stable conformation. When the cell progresses through the G2 stage, upstream factors phosphorylate Cdc25C in the Thr-130 (18). This fact implies an increase in the affinity toward the Plk1-Cdc25C-P complex formation, as we have observed (Fig. 2b); then, Thr-130 phosphorylation in the target peptide could induce a conformational change in the adjacent proline, which favors the stabilization of the CL, as observed in the PBD-Cdc25C-P complex structure. Taking into account Plk1 localization, the model indicates that Cdc25C should be at the centrosome at a certain stage. Although some evidence, such as the centrosomal detection in prophase of an active Cdk1/Cyclin B complex, suggests this possibility (25), it has not yet been shown.
Fig. 5.
Hypothetical model of Plk1 interaction with Cdc25C during the cell cycle.
Finally, the progression toward the end of Gy2 promotes Plk1 activation (27, 28), and complex formation enhances the activation and nuclear translocation of Cdc25C (12, 29). Subsequently, the active Cdc25C triggers mitosis entry. The possibility that Plk1 is activated before substrate recognition is not supported in our Plk1 activity assays (SI Fig. 7). The target peptide elicits the same response independently of its threonine phosphorylation status, and the differences observed in target peptide affinity should have been manifested in our kinase activity measurements.
Our structural, biochemical, and cellular data shed light on the molecular mechanisms of Plk1 substrate recognition and localization, two of the main regulatory mechanisms of this enzyme. Our findings emphasize the possibility to design small molecules that could inhibit normal Plk1 operation in tumor cells targeting the PBD (30). Indeed, it has been shown that Plk1 binds nonphysiological molecules in the PBD (16, 31), which hamper its activity.
Materials and Methods
Full protocols are available in SI Text.
Cell Immunofluorescence, FRAP Analysis, and Fluorescence Centrosomal Quantification.
Transfected PC3 cells were processed for immunofluorescence following standard protocols. Centrosomes were stained with antibodies against γ-tubulin (Sigma, St. Louis, MO; GTU88). Finally, cells were analyzed with a Leica (Deerfield, IL) SP2 confocal microscope. FRAP was performed 20 h after transfection. The 488-nm laser was used in bleaching and imaging experiments. A laser power of 5–7% (5 mW) was used in image acquisitions and 100% (5 mW) in photobleaching. Several images were collected immediately after the photobleach at 3- to 5-s time lapses. The final recovery models were generated as in ref. 32. Leica LAS AF quantification software was used for fluorescence quantification. The EGFP-Plk1 and the γ-tubulin Alexa594 signals were captured in a Leica SP5 confocal microscope. Laser intensity, optical thickness, and photomultiplier acquisition settings were kept identical for all analyzed cells. EGFP-Plk1 centrosomal fluorescence values were related to the γ-tubulin intensity to compensate changes in centrosomal size.
Cloning, Expression, and Purification of the Human Plk1 and PBD.
The Plk1 cDNA sequence was amplified by PCR and cloned into a pFastBac vector (Invitrogen, Carlsbad, CA) to generate a recombinant baculovirus. Infected cells were disrupted by sonication, and the protein was purified in a Ni2+ column and by gel-filtration chromatography. The PBD cDNA sequence (residues 367–603) was amplified by PCR and cloned into vector pGEX-6P-2. PBD expression and purification were performed as described (33).
Recombinant Plk1 Activation and Kinase Assays.
Four micrograms of the recombinant His-Plk1 was mixed with Ni2+ beads and incubated for 1 h to allow binding. Subsequently, His-Plk1 was incubated with either an interphasic or mitotic extract (100 μg) prepared from HeLa cell culture. After incubation, Plk1-Ni2+ beads were washed in lysis buffer. Finally, Plk1-Ni2+ beads were washed in kinase buffer. Pure Plk1 was then eluted by addition of 300 mM imidazole. The kinase reaction was initiated by the addition of 0.2 mM ATP/2 μCi (1 Ci = 37 GBq) [γ-32P]ATP (Amersham, Piscataway, NJ) to the eluted Plk1 and 5 μg of Histone H1 (Roche, Indianapolis, IN) as substrate. After 30 min at 37°C, the reaction was stopped and analyzed by electrophoresis and autoradiography.
Affinity Measurements.
Cdc25C, nonphysiological, and control target peptides (Cdc25C-P sequence LLCS[pT]PNGL, Cdc25C sequence LLCSTPNGL, nonphysiological-P sequence MAGPMQS[pT]PLNGAKK, nonphysiological sequence MAGPMQSTPLNGAKK, control-P sequence CGERKK[pT]LSGTPNY, and control sequence CGERKKTLSGTPNYI) were synthesized by GENOSPHERE Biotechnologies (Paris, France) and checked by mass spectrometry. Fluorescence experiments were performed by using a PTI fluorimeter. Calorimetry was measured by using VP-ITC microcalorimeter (MicroCal, Amherst, MA).
Crystallization, Data Collection, Structure Solution, Model Building, and Refinement.
Crystals of the PBD, PBD-Cdc25C-P, and PBD-Cdc25C were obtained as described in ref. 33. All data were collected by using synchrotron radiation at the European Sychrotron Radiation Facility and Swiss Light Source. Diffraction images were processed as described in ref. 33. Statistics for the crystallographic data are summarized in SI Text. Coordinates have been submitted to the Protein Data Bank (34).
Supplementary Material
Acknowledgments
We thank the staff at the European Synchrotron Radiation Facility and Swiss Light Source for help during data collection. B.G.-A. thanks the European Molecular Biology Organization and Ministerio de Educación y Ciencia (MEC) for postdoctoral fellowships. G.d.C. thanks the MEC for a Ramón y Cajal contract. Financial support was obtained through Grants S-GEN-0166/2006, BFU2005-02403, GEN2003-20642-C09-02, and CSD2006-00023 (to G.M.) and Fondo de Investigaciones Sanitarias PI051186 (to G.d.C.).
Abbreviations
- Plk
polo-like kinase
- FRAP
fluorescence recovery after photobleaching
- PB
polo box
- PBD
PB domain
- CL
connecting loop.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2OGQ, 2OJS, and 2OJX).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609131104/DC1.
References
- 1.Nigg EA. Curr Opin Cell Biol. 1998;10:776–783. doi: 10.1016/s0955-0674(98)80121-x. [DOI] [PubMed] [Google Scholar]
- 2.Barr FA, Sillje HH, Nigg EA. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 3.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 4.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 5.Kotani S, Tugendreich S, Fujii M, Jorgensen PM, Watanabe N, Hoog C, Hieter P, Todokoro K. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- 6.Descombes P, Nigg EA. EMBO J. 1998;17:1328–1335. doi: 10.1093/emboj/17.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KS, Erikson RL. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM. EMBO J. 2001;20:1259–1270. doi: 10.1093/emboj/20.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang YJ, Lin CY, Ma S, Erikson RL. Proc Natl Acad Sci USA. 2002;99:1984–1989. doi: 10.1073/pnas.042689299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian YW, Erikson E, Maller JL. Mol Cell Biol. 1999;19:8625–8632. doi: 10.1128/mcb.19.12.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumagai A, Dunphy WG. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. J Biol Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 14.Okano-Uchida T, Okumura E, Iwashita M, Yoshida H, Tachibana K, Kishimoto T. EMBO J. 2003;22:5633–5642. doi: 10.1093/emboj/cdg535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elia AE, Cantley LC, Yaffe MB. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 16.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 17.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. EMBO J. 2003;22:5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumi T, Maller JL. Mol Biol Cell. 1995;6:215–226. doi: 10.1091/mbc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Weerdt BC, Medema RH. Cell Cycle. 2006;5:853–864. doi: 10.4161/cc.5.8.2692. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Lewellyn AL, Chen LG, Maller JL. J Biol Chem. 2004;279:21367–21373. doi: 10.1074/jbc.M400482200. [DOI] [PubMed] [Google Scholar]
- 21.Feng Y, Hodge DR, Palmieri G, Chase DL, Longo DL, Ferris DK. Biochem J. 1999;339:435–442. [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton B, Glover DM. Nature. 1993;363:637–640. doi: 10.1038/363637a0. [DOI] [PubMed] [Google Scholar]
- 23.Glover DM, Ohkura H, Tavares A. J Cell Biol. 1996;135:1681–1684. doi: 10.1083/jcb.135.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover DM, Hagan IM, Tavares AAM. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- 25.Jackman M, Lindon C, Nigg EA, Pines J. Nat Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 26.Kumagai A, Yakowec PS, Dunphy WG. Mol Biol Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian YW, Erikson E, Li C, Maller JL. Mol Cell Biol. 1998;18:4262–4271. doi: 10.1128/mcb.18.7.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian YW, Erikson E, Maller JL. Science. 1998;282:1701–1704. doi: 10.1126/science.282.5394.1701. [DOI] [PubMed] [Google Scholar]
- 29.Toyoshima-Morimoto F, Taniguchi E, Nishida E. EMBO Rep. 2002;3:341–348. doi: 10.1093/embo-reports/kvf069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strebhardt K, Ullrich A. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 31.Gumireddy K, Reddy MV, Cosenza SC, Boominathan R, Baker SJ, Papathi N, Jiang J, Holland J, Reddy EP. Cancer Cell. 2005;7:275–286. doi: 10.1016/j.ccr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García-Álvarez B, Ibañez S, Montoya G. Acta Crystallogr F. 2006;62:372–375. doi: 10.1107/S1744309106007494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.