Abstract
The striking feature of carbohydrates is their constitutional, conformational and configurational diversity. Biology has harnessed this diversity and manipulates carbohydrate residues in a variety of ways, one of which is epimerization. RmlC catalyzes the epimerization of the C3′ and C5′ positions of dTDP-6-deoxy-D-xylo-4-hexulose, forming dTDP-6-deoxy-L-lyxo-4-hexulose. RmlC is the third enzyme of the rhamnose pathway, and represents a validated anti-bacterial drug target. Although several structures of the enzyme have been reported, the mechanism and the nature of the intermediates have remained obscure. Despite its relatively small size (22 kDa), RmlC catalyses four stereospecific proton transfers and the substrate undergoes a major conformational change during the course of the transformation. Here we report the structure of RmlC from several organisms in complex with product and product mimics. We have probed site-directed mutants by assay and by deuterium exchange. The combination of structural and biochemical data has allowed us to assign key residues and identify the conformation of the carbohydrate during turnover. Clear knowledge of the chemical structure of RmlC reaction intermediates may offer new opportunities for rational drug design.
Keywords: site directed mutagenesis, X-ray crystallography, drug design, epimerization, enzyme
Introduction
Carbohydrates serve not only as energy stores, but are important structural elements and informational molecules in many organisms 1. They are distinguished by their density of asymmetric centers and functional groups. The inversion of stereochemistry at the asymmetric centers (epimerization) of carbohydrates is a simple means of creating molecular diversity; the resulting diastereoisomers are chemically distinct. Biology has developed a number of different strategies to accomplish such epimerization reactions and uses several different classes of enzymes to catalyze this transformation 2; 3; 4.
L-Rhamnose is a 6-deoxyhexose that is commonly found in bacterial glycoconjugates. Neither the sugar, nor the enzymes required for its synthesis are found in humans and the pathway has been validated as a potential therapeutic target in several clinically important systems, including Mycobacterium tuberculosis (M. tuberculosis)5; 6; 7. In Pseudomonas aeruginosa (P. aeruginosa), L-rhamnose is integral to both the lipopolysaccharide (LPS) core oligosaccharide and O-antigen polysaccharides; chromosomal mutations in the rmlC gene of serotype O5 and serotype O6 of P. aeruginosa resulted in LPS core truncation and loss of pathogenicity 8. In Streptococcus mutans, rhamnose-containing polysaccharide antigens mediate colonization of tooth surfaces 9 and genetic disruption of the rhamnose pathway prevents bacteria initiating or sustaining an infection 10. In M. tuberculosis, L-rhamnose links the peptidoglycan and arabinogalactan in a unique and complex cell wall structure that is essential for viability 11.
RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3′, 5′-epimerase, EC 5.1.3.13) is the third enzyme of the dTDP-L-rhamnose biosynthetic pathway that converts glucose-1-phosphate to dTDP-L-rhamnose12. The pathway requires four enzymes, RmlA, RmlB, RmlC and RmlD, the structures of which have been determined13; 14; 15; 16; 17. RmlC epimerizes the C3′ and C5′ positions of dTDP-6-deoxy-D-xylo-4 hexulose making dTDP-6-deoxy-L lyxo-4-hexulose (Figure 1a). The 22-kDa RmlC protein does not require a cofactor. There are now four reports of RmlC structures15; 18; 19; 20 and co-complexes have been obtained with dTDP-phenol15, dTDP18, dTDP-D-glucose19 and dTDP-D-xylose19. The co-complexes of RmlC from Streptococcus suis (S. suis) with dTDP-D-glucose and dTDP-D-xylose respectively provided experimental evidence for the location of the active site19. This study identified a conserved His residue as the base for both epimerization reactions, a conserved Lys residue which stabilizes the intermediate enolate anion and a conserved Tyr which acts as an acid 19.
Figure 1.
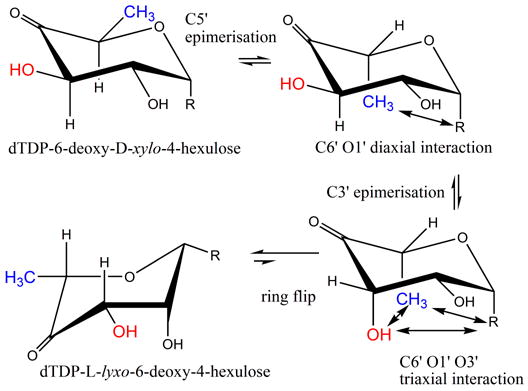
(a) R=OdTDP. The RmlC reaction converts dTDP-6-deoxy-D-xylo-4-hexulose to dTDP-6-deoxy-L-lyxo-4-hexulose (thick arrow). This process involves a ring flip as well as epimerization. The steps are shown according to the current convention, however, this route goes through some very high energy intermediates (notably the C1′, C3′, C5′ triaxial product).
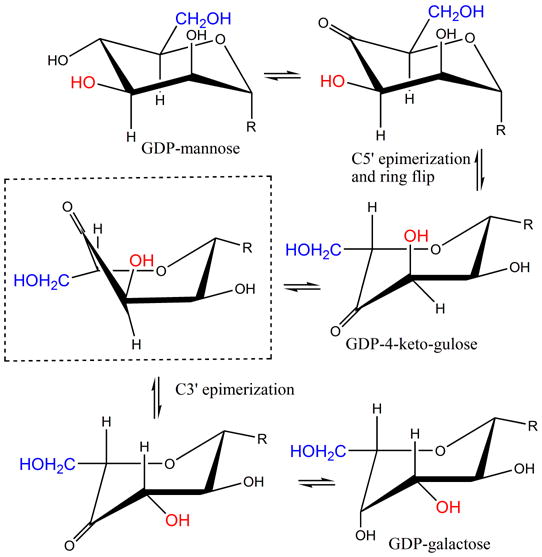
(b) R=OGPD. The GME reaction converts GDP-D-mannose to GDP-L-galactose. Shown boxed is the predicted twist boat intermediate. The dominant order of the epimerization (C5′ first) and the ring flipped form of GDP-L-ribo-4-hexulose were determined experimentally 24.
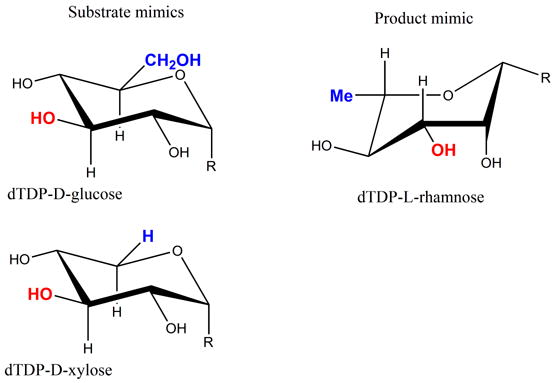
(c) R = OdTDP. Substrate and product mimics employed in this study.
GDP-mannose epimerase (GME) and GDP-6-deoxy-4-keto-D-mannose epimerase/reductase (GMER) also catalyze C3′, C5′ epimerization. As part of this epimerization, both enzymes catalyze NAD-dependent redox chemistry at the C4′ position 21; 22; 23; 24, the reactions catalyzed by GME are shown in Figure 1b. Both GME and GMER are members of the short-chain dehydrogenase (SDR) superfamily of enzymes and share no sequence or structural similarity to RmlC, despite the obvious chemical similarity in the epimerization. In the GME structure a single pair of residues has been identified to function as the acid and base for both epimerizations24. A structure with the mono-epimerized product revealed that in GME the carbohydrate ring flip occurs during the first epimerization at C5′24. The chemical requirements for proton abstraction and consideration of conformational energy of carbohydrates suggested that GME and by analogy GMER operate on a twist boat structure for the second epimerization. In light of this we have re-examined RmlC. Since RmlC also carries out a double epimerization, it too faces the same “problem” in the second epimerization. As conventionally written, (Figure 1a), the enzyme operates on a very high energy 1, 3, 5 tri-axial intermediate. An alternative would be for the ring to “flip” to relieve the strain. However, the ring flip disrupts the stereochemical requirements for enolate stabilization. For this stabilization, the C-H bond must be orthogonal to the plane of the carbonyl group such that the orbital rehybridization occurs during deprotonation to create an extended (over 3 atoms) conjugated π system. We have now determined structures of RmlC from P. aeruginosa, S. suis and M. tuberculosis with product mimics (Figure 1c). RmlC from Salmonella enterica (S. enterica) serovar Typhimurium was the first dTDP-6-deoxy-D-xylo-4-hexulose 3′, 5′-epimerase to be structurally characterized and serves as a prototype. When compared to the S. enterica enzyme, RmlC enzymes from P. aeruginosa (65% identity), S. suis (25% identity) and M. tuberculosis (40% identity) show considerable diversity in sequence. The structures of various co-complexes of RmlC from a wide range of organisms suggest that RmlC favors the binding of sugars with an equatorial configuration at the anomeric position. We have measured rates of deuterium incorporation and our data indicate C5′ (in the enzyme) is more acidic than C3′. These data suggest that despite the complete lack of similarity in structure and sequence, common epimerization mechanisms are shared between RmlC and GME. This is an example where the common chemical requirements of a transformation drive the convergent evolution of the enzyme mechanisms.
Results
P. aeruginosa RmlC structure with dTDP-xylose
The structure of RmlC from P. aeruginosa contains 184 amino acids, consisting of the whole RmlC (residues Met1 to Pro181) plus part of the His-tag linker (Ser-Met-Ser); the remainder of the N-terminal His-tag is disordered. The structure is essentially identical to those described previously, containing thirteen β-strands that form a sandwich. One β sheet is crucial for forming a dimer interface (Figure 2a). The changes in the proteins resulting from binding of nucleotide have been described in detail previously15; 19. In the dTDP-D-xylose complex the carbohydrate ring is not found at the active site and only a portion of the ligand is observed (Figure 3a). We obtained a similar result with dTDP-D-glucose (data not shown); however the crystals were of lower resolution. The α-phosphate of the sugar nucleotide is anchored to the protein by salt contacts and by bridging water molecules to the protein via the same interactions observed for dTDP-D-glucose, dTDP-D-xylose complexes of S. suis19 and for the dTDP complex of RmlC from Methanobacterium thermoautotrophicum18 (Figure 2b). However, in both monomers the β-phosphate is twisted out of its normal binding site (Figure 2b) and the β-phosphate has a significantly higher B-factor (>10 Å2) than the α-phosphate, consistent with disorder of the carbohydrate. This twisting out of the normal binding site suggests the sugar ring is not recognized by the protein/ HPLC analysis of the nucleotide and sugar nucleotide content of P. aeruginosa RmlC:dTDP-xylose crystals reveals only a very small amount of dTDP compared to dTDP-xylose. This establishes dTDP-xylose is indeed present in the crystal structure and hence must be disordered. At the active site in one monomer there is some weak density which is consistent with a tartrate molecule (Figure 3a); in the other monomer the tartrate density is less distinct. Tartrate does not appear to inhibit the enzymes in our assays. In the S. suis19 enzyme E78 makes a bidentate hydrogen bond with dTDP-glucose and dTDP-xylose. The Glu residue is not conserved, nor is the structure of the loop containing E78 19 conserved in other RmlC enzymes.
Figure 2.
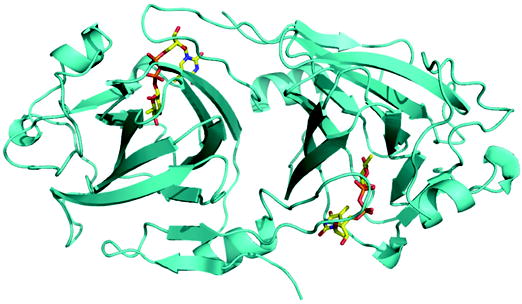
(a) Ribbon diagram of RmlC dimer from P. aeruginosa in complex with product (dTDP-6-deoxy-L-lyxo-4-hexulose). The product (an equatorially configured sugar nucleotide) is drawn in stick representation. The ligand atoms are shown in stick representation, with carbon yellow, nitrogen blue, oxygen red and phosphorus orange.
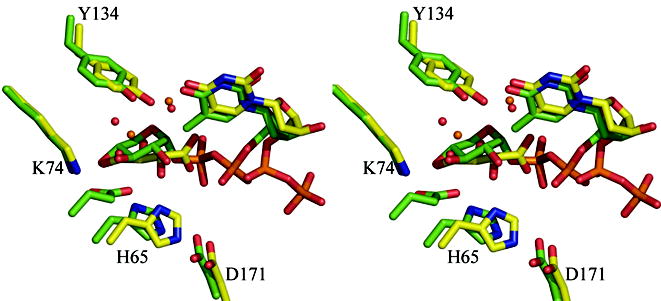
(b) Superposition of the dTDP-D-xylose complex with RmlC from P. aeruginosa and from S. suis 19. The β phosphate of dTDP-D-xylose in the P. aeruginosa is twisted out of the active site and a tartrate molecule occupies the active site in one monomer. In the S. suis structure the carbohydrate occupies the active site but is bound to E78 (shown but not labeled), a non-conserved residue on a loop not found in any other RmlC structure. The color scheme for P. aeruginosa complex is as Figure 2(a). For S. suis carbon atoms are colored green, other atoms are colored as P. aeruginosa. Water molecules in the P. aeruginosa complex are shown as orange spheres, in the S. suis they are shown as red spheres. Amino acid numbering is for the P. aeruginosa sequence.
(c) Superposition of the dTDP-6-deoxy-L-lyxo-4-hexulose (product) complex with RmlC from P. aeruginosa and the dTDP-glucose complex from S. suis 19. Although the carbohydrate is bound at the active site in both complexes, the equatorial linkage between sugar and nucleotide in the P. aeruginosa complex means the sugar ring is positioned differently with respect to the key catalytic residues. The P. aeruginosa complex aligns better to the proposed chemical role of the active site residues. The color scheme is as Figure 2(b). The O6′ of dTDP-D-glucose from S. suis replaces one of the conserved structural water molecules and is bound by E78 (shown but not labeled). Amino acid numbering is for the P. aeruginosa sequence.
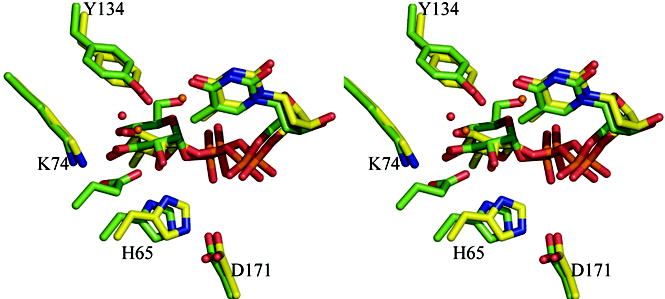
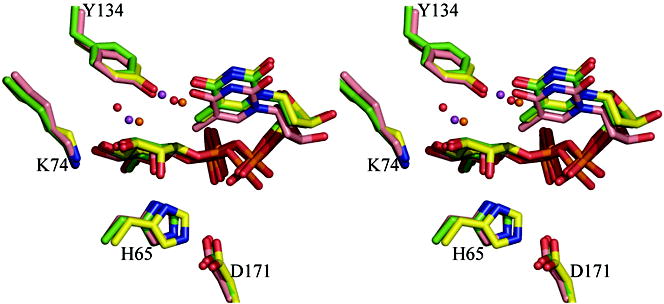
(d) Superposition of the dTDP-L-rhamnose complexes for three enzymes M. tuberculosis, S. suis and P. aeruginosa. The match between the structures is particularly striking especially given the different crystallization conditions and low sequence identity between the structures (25%). Two water molecules one close to C3′ and one close to C5′ are structurally conserved. The S. suis and P. aeruginosa structure are colored as Figure 2(c). The carbon atoms in M. tuberculosis RmlC complex are colored pink; other atoms are colored as P. aeruginosa. The two water molecules in the M. tuberculosis structure are shown as magenta colored spheres. Amino acid numbering is for the P. aeruginosa sequence.
Figure 3.
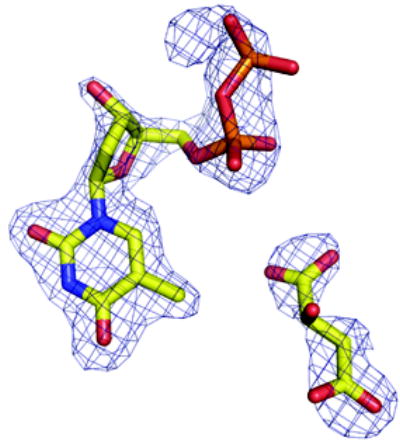
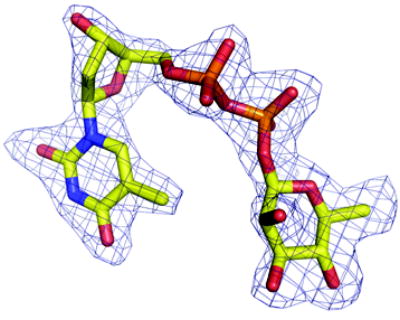
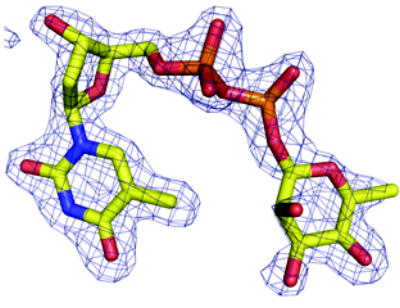
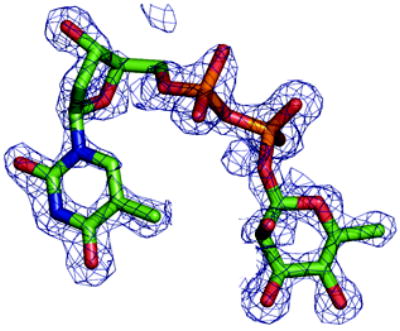
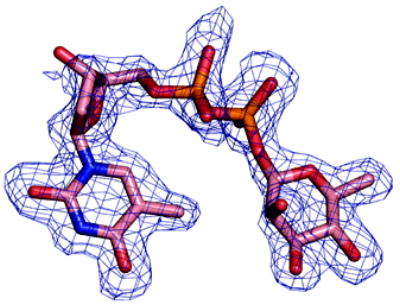
(a) Density from initial Fo-Fc map contoured at 2.5σ for the P. aeruginosa RmlC complex with dTDP-D-xylose. The sugar is disordered and tartrate is seen at the active site of one subunit (shown here). Atoms are colored as Figure 2(a)
(b) Density from initial Fo-Fc map contoured at 2.5σ for the P. aeruginosa RmlC complex with dTDP-6-deoxy-L-lyxo-4-hexulose. Atoms are colored as Figure 2(a).
(c) Density from initial Fo-Fc map contoured at 2.5σ for the P. aeruginosa RmlC complex with dTDP-L-rhamnose. Atoms are colored as Figure 2(a).
(d) Density from initial Fo-Fc map contoured at 2.5σ for the S. suis RmlC complex with dTDP-L-rhamnose. Carbon atoms are colored green, other atoms are colored as Figure 2(a). Two subunits have clear density, the other two the sugar component is significantly more disordered.
(e) Density from initial Fo-Fc map contoured at 2.5σ for the M. tuberculosis RmlC complex with dTDP-L-rhamnose. Carbon atoms are colored pink; other atoms are colored as Figure 2(a). The quality of the density varies between subunits, with two being excellent, one weak and the fourth poor. In the subunit with weak density there is evidence for a second conformation of the sugar but we were unable to satisfactorily model it.
P. aeruginosa RmlC structure with product
To probe the nature of RmlC substrate binding we incubated the P. aeruginosa enzyme with the authentic substrate dTDP-6-deoxy-D-xylo-4-hexulose. Since the substrate is chemically labile, we chose the P. aeruginosa enzyme for these experiments as it crystallizes closer to neutral pH, where the keto sugar is more likely to remain intact. Although the complex was crystallized in tartrate, the sugar ring is bound at the enzyme active site. This observation is in contrast to both the dTDP-D-glucose and dTDP-D-xylose complexes of the P. aeruginosa enzyme. The density clearly shows that the sugar has an equatorial linkage to the nucleotide and indicates a distorted chair consistent with a keto function (Figure 3b). The density is consistent with the product dTDP-6-deoxy-L-lyxo-4-hexulose rather than substrate or monoepimerized product bound at the active site. The positions of the thymidine and α and β phosphates with respect to the proteins are identical to the dTDP-D-glucose and dTDP-D-xylose complexes of S. suis. However, the carbohydrate rings are placed differently with respect to the protein because in the P. aeruginosa RmlC-product complex the nucleotide is equatorial to the carbohydrate ring not axial as in the S. suis complexes (Figure 2c). The C6′ of the product is located in a hydrophobic region formed by F123 and G122. NZ of K74 and ND1 of H121 are coplanar with the sugar ring, and NZ of K74 hydrogen bonds (2.9 Å) to the O4′, the location of the negative charge in the enolate. Unlike the S. suis RmlC complexes, this interaction of O4′ with the conserved Lys is now optimal for enolate stabilization (Figure 2c). H65 and Y134 of P. aeruginosa RmlC are located on opposite sides of the plane of the carbohydrate ring, entirely consistent with their identification as the acid-base pair. The distances from OH of Y134 to C3′ and C5′ are 2.2 Å and 3.7 Å, respectively. Despite the close contact between the Y134 and C3′, the electron density does not suggest a full covalent link and may result from our failure to model any isomerization of keto group from the C4′ to C3′ position of the ring. In solution there is an equilibrium between dTDP-keto-sugar C4′ and C3′ keto groups 25. However, the resolution of the data is insufficient to accurately model such an isomerization. The distances between the sugar C3′/C5′ and ND1 of H65 are 3.4/3.6 Å (Figure 2c) and are consistent with H65 acting as the base for both C3′ and C5′ epimerization. This complex suggests that Y134 could act as the acid for both C3′ and C5′. However, on the same face of the carbohydrate as the Tyr residue there are two water molecules which could also be involved in this process. One water molecule is located close to C5′ and the other close to C3′ (Figure 2c). In the dTDP-D-glucose complex with the S. suis protein, the water molecule close to C3′ is preserved but the water close to C5′ is displaced by the O6′ of glucose (Figure 2c).
RmlC with dTDP-L-rhamnose
To further probe the recognition of an equatorial linked sugar, we employed dTDP-L-rhamnose as a product mimic. It differs from the authentic product in the reduction of the C4′-keto group to a hydroxyl group. It was straightforward to obtain dTDP-L-rhamnose complexes with P. aeruginosa, M. tuberculosis and S. suis RmlC (Figure 3c, d, e). In all three complexes, the ligand and the key catalytic residues superimpose (Figure 2d). Comparison of the dTDP-L-rhamnose complex with the product complex from RmlC of P. aeruginosa revealed that the Tyr residue and the sugar ring have moved apart, resulting in a more normal 3.3 Å separation between C3′ and the OH of Y134 and a 4.5 Å separation between C5′ and the OH of Y134. Otherwise, the product and dTDP-L-rhamnose structures are essentially identical. The structural data suggest Tyr as the protein residue acting as the acid in both epimerizations. However, all three dTDP-L-rhamnose complexes also have the two water molecules at the same positions as seen in the product complex (Figure 2d). The consistency between these complexes is convincing, particularly given the differences in protein sequence and crystallization conditions. It also stands in marked contrast to our difficulties in obtaining dTDP-D-glucose or dTDP-D-xylose complexes with any RmlC protein except that from S. suis.
Biochemical characterization
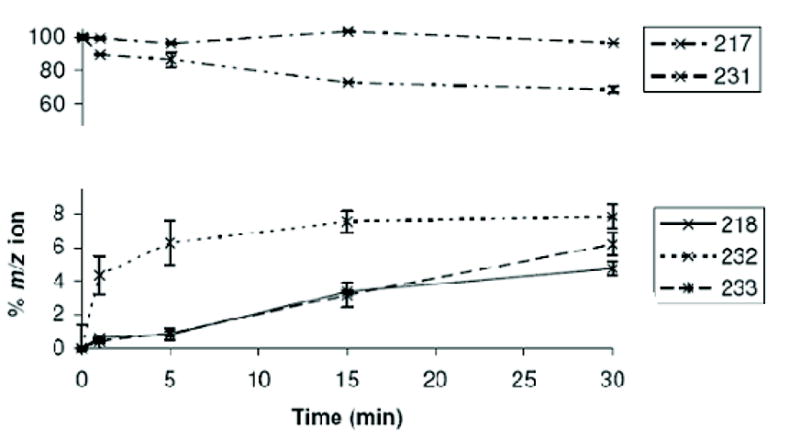
We have already reported assays of RmlC from S. suis 19 in which the key catalytic residues have been mutated. The conventional biochemical assay measures the amount of NADH consumed by RmlD in a coupled system and, therefore, only detects complete product formation by RmlC. This assay cannot be used to probe the individual steps in the reaction. In contrast, deuterium isotope incorporation directly measures the exchange of the protons at C3′ and C5′. This exchange occurs when protons are abstracted from the substrate by a base and replaced by deuterons. While epimerization usually involves exchange, exchange can occur without epimerization26. This is the case here as in D2O, whilst there is a measurable background (uncatalyzed) rate of proton exchange at C3′ and C5′ of dTDP-6-deoxy-D-xylo-4-hexulose in D2O, there is no observable uncatalyzed epimerization. The position and extent of deuterium incorporation from solvent can be assessed by analysis of GC-MS fragmentation patterns of alditol peracetates following reduction-hydrolysis-reduction-acetylation of dTDP-6-deoxy-D-xylo-4-hexulose and products of RmlC action27 (Figure 4). In order to correct for non-enzymatic deuterium incorporation, two no-enzyme controls were performed at pH 5.0 and pH 9.0 respectively. The point mutants H63A, K73A and Y133F of RmlC from S. enterica serovar Typhimurium (corresponding to H65, K74, and Y134 from P. aeruginosa RmlC) were analyzed both for activity and for their ability to catalyze deuterium incorporation into the C3′ and C5′ positions of dTDP-6-deoxy-D-xylo-4-hexulose (Table 2 and Figure 4). The mutants were examined by circular dichroism which confirmed no detectable structural changes that would influence the native fold. H63A is catalytically inactive and shows no deuterium incorporation above background at either C3′ or C5′. K73A is reduced in activity by over 100 fold and a small amount of enzyme catalyzed deuterium incorporation was observed at C5′, while only background levels were seen at C3′. This suggests that for the K73A mutant, C5′ exchange is more rapid than at C3′. The catalytic activity of the Y133F mutant is reduced 1000-fold but shows some deuterium incorporation at C3′ but none at C5′ (above background). This indicates that RmlC can catalyze exchange of the proton at C3′ without Tyr133 but not at C5′. While this assay measures proton/deuterium exchange rather than epimerization, it is possible that a small amount of C3′ mono-epimerized product is produced by Y133F. In order to probe the different rates of exchange at the two positions, deuterium incorporation catalyzed by wild-type S. enterica serovar Typhimurium RmlC was monitored over a 30 min period at 10°C. The rate of C5′ incorporation is more rapid than at C3′ (Figure 4). These data support a hypothesis that the dominant order of enzyme catalyzed exchange is generally C5′ followed by C3′. The data do not exclude the possibility that exchange also occurs first at C3′ followed by C5′, but the data indicate this is a minor component. Although exchange and epimerization are distinct, the first step proton abstraction is common to both. Based on our exchange data in which proton abstraction from C5′ is more rapid, we suggest that the enzyme may favor an order of epimerization with C5′ first. Isothermal titration calorimetry shows quite clearly that dTDP-rhamnose (Figure 5) and dTDP are bound with a significantly higher affinity than dTDP-glucose and dTDP-xylose (Table 3). Whether there is a real difference between dTDP and dTDP-L-rhamnose binding cannot be reliably derived from the data we have. The polar nature of sugars means the hydrogen bonds they make with proteins are often cancelled by the ones they break with water. It seems likely that binding will be dominated by the nucleotide portion of the ligand. There are many examples of thermodynamically weak sugar protein interactions which have multiple hydrogen bonds28; 29.
Figure 4.
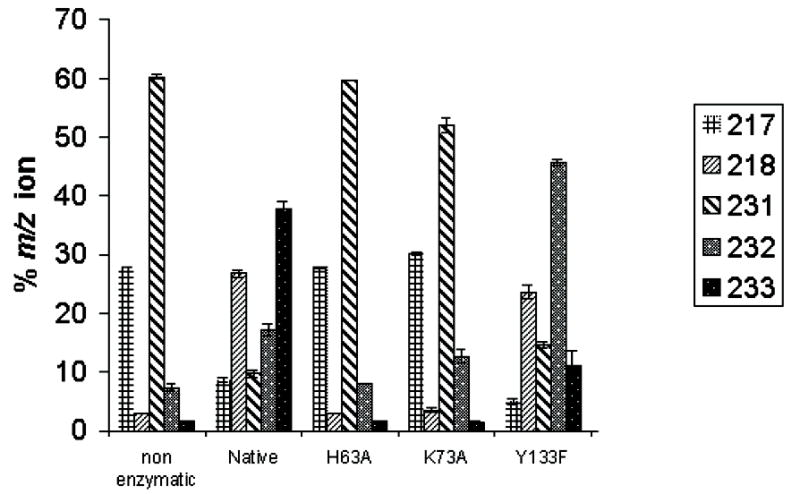
(a) Deuterium incorporation data for mutants of S. enterica serovar Typhimurium RmlC. The masses reflect the fragments of alditol per-acetates associated with deuterium incorporation into substrate. Cleavage between C3′ and C4′ with retention of charge at C3′ yielded an ion at m/z 217 (no deuterium incorporation) or m/z 218 (deuterium incorporation at C3′). Cleavage between C2′ and C3′ with retention of charge at C-3 yielded an ion at m/z 231 (no deuterium incorporation), m/z 232 (deuterium at C3′ or C5′) and m/z 233 (deuterium at C3′ and C5′). Starting material (no D incorporation) has two peaks at 217/231. D incorporation at C5′ only has peaks at 217 and 232. D incorporation at C3′ has two peaks at 218 and 232. D incorporation at both C3′ and C5′ has peaks at 218 and 233. Only the Y133F mutant shows any specific C3′ incorporation. The H63A shows no incorporation, whilst K79A shows some incorporation at C5′ but none at C3′. Y133F mutant shows some C3′ incorporation but no C5′ incorporation.
(b) A time course at 10°C measuring C3′ and C5′ deuterium incorporation into substrate, showing loss of signals from non-deuterated substrate (217, 231) and the evolution of deuterium incorporation. The C5′-specific peak at 232 is rapidly established and complete within 10 minutes, the same time frame as loss of the substrate 231 peak. The 218 peak appears more slowly and at approximately the same rate as the 233 peak. This suggests that there is no significant amount of specific C3′ incorporation on its own, rather that C3′ incorporation occurs (within error) after C5′ incorporation.
Table 2.
Apparent kinetic constants for S. enterica serovar Typhimurium RmlC mutants with dTDP-6-deoxy-D-xylo-4-hexulose.
| Enzyme | Km (mM) | kcat (1/s) |
|---|---|---|
| Native | 0.71 ± 0.17 | 39 ± 6.6 |
| RmlC K73A | 0.35 ± 0.051 | 0.095 ± 0.0083 |
| RmlC Y133F | 0.48 ± 0.21 | 0.016 ± 0.0017 |
| RmlC H63A | N.D | 0 |
Figure 5.
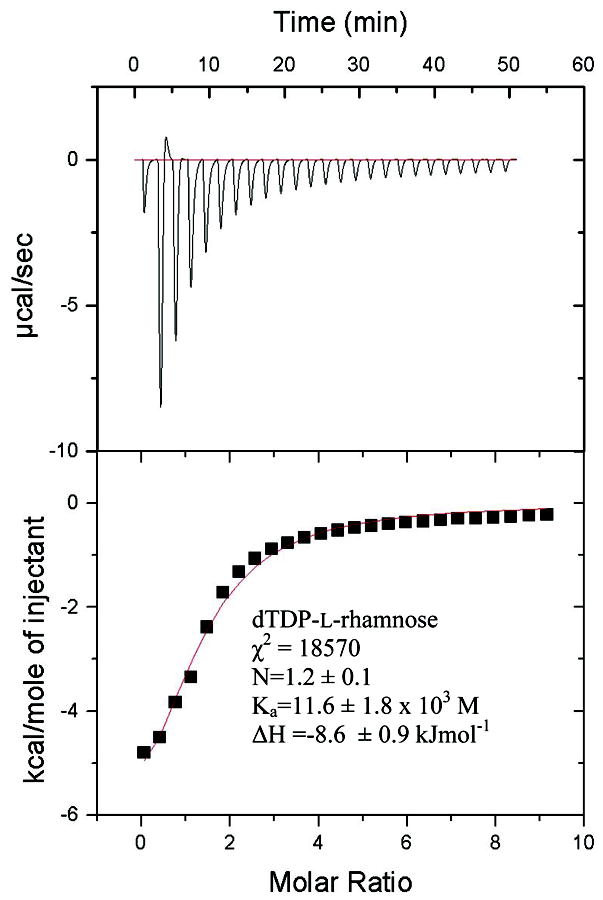
Raw isothermal titration calorimetry data for dTDP-L-rhamnose binding to P. aeruginosa RmlC. The small endothermic contribution in the second injection appears to be a consistent feature in three different experiments. χ2 is a measure of the goodness of fit, N is the stoichiometry, Ka is the association constant and ΔH is the enthalpy change. The concentration of the sugar was estimated by extinction coefficient. Fixing the stoichiometry to be 1: 1 gives a Ka value of 9.0 × 103M. Eliminating the endothermic point from the fitting routine gives a stoichiometry of 1.04 and Ka of 10 × 103M. Eliminating the endothermic point and setting stoichiometry to 1 gives a Ka 9.7 × 103M. Thus the value of Ka is robustly estimated from the data.
Table 3.
Isothermal titration calorimetry data for P. aeruginosa RmlC with sugar nucleotides. The uncertainties are estimated from the fitting and underestimate the true error.
| Ka (× 103M) | ΔH (kJmol−1) | |
|---|---|---|
| dTDP-L-rhamnose | 11.6 ± 1.8 | −8.6 ± 0.9 |
| dTDPa | 13.2 ± 1.7 | −25 ± 2 |
| dTDP-xylose | 1.32 ± 0.05 | −11.8 ± 0.2 |
| dTDP-glucose | 0.96 ± 0.26 | −9.3 ± 6.2 |
The values for dTDP did vary depending on experiment and on the processing parameters. As is seen for dTDP-L-rhamnose (Fig 5) there is an endothermic event at the start of titration that we cannot interpret. If the endothermic points are omitted, N is set to 1 and the values recalculated the Ka drops to 4.5 × 103M. Our interpretation is that dTDP binds more tightly than dTDP-xylose/glucose, we do not think our data robust enough to compare dTDP and dTDP-L-rhamnose.
Discussion
The RmlC reaction must proceed from substrate to a mono-epimerized intermediate, and subsequently to a doubly epimerized product (Figure 1a). Our data suggest that the dominant order of epimerization may be C5′ followed by C3′ and our discussion focuses on this pathway. Our data do not exclude the possibility that there is no obligate order and that C3′ epimerization can occur first. GME does exhibit a preference for C5′ followed by C3′ (Figure 1b). Structural and biochemical data are both consistent with an RmlC mechanism in which both the C5′ and C3′ protons are abstracted by the absolutely conserved His65 residue (P. aeruginosa numbering), which is part of a His-Asp diad. Y134 has been shown to be essential for epimerization and for deuterium incorporation at C5′. This supports our proposal that Y134 has a key role in proton donation to C5′ on the opposite face of the sugar to H65. Y134 is not essential for deuterium incorporation at C3′ and water may be able to compensate for its absence during deprotonation but not during epimerization. In our structural analysis we noted a conserved water molecule close to the position appropriate for donation of a proton to C3′, this may partly compensate for the Tyr. NovW, a validated C3′ mono-epimerase involved in novobiocin biosynthesis also acts on dTDP-6-deoxy-D-xylo-4-hexulose, retains the key active site Tyr residue despite conducting no chemistry at C530. The use of a His-Tyr couple is quite unusual in acid base enzyme chemistry. Histidine is, of course, widely used as both acid and base in many enzymes; GMER is thought to use a Cys-His23 couple to carry out two epimerizations on sugar nucleotide. In catalyzing the dehydration of the C6′ position, RmlB abstracts the same C5′ proton as RmlC, but it uses a glutamic acid to accomplish this31. In amino acid racemases, which also remove a proton α to a carbonyl group, a Cys-Cys couple is very commonly used32. Tyrosine is less widely used as base, although it is well known as the base in the short chain dehydrogenase superfamily33 where it abstracts a hydroxyl proton. A tyrosine-water combination acts as one of the acid base groups during the racemisation of alanine by Treponema denticola cystalysin; lysine acts as the other group34. Most recently and perhaps the most relevant to RmlC mechanism, the enzyme iminodisuccinate epimerase which has a novel fold, has been predicted to use the a His-Tyr acid base couple and Lys to stabilize the enolate anion35. Proton abstraction from carbon by tyrosine is also seen in Chondroitin AC lyase36.
Structural data show RmlC enzymes from P. aeruginosa, M. tuberculosis and S. suis recognize dTDP-L-rhamnose in an identical manner. This is in contrast to the situation with dTDP-D-glucose and dTDP-D-xylose. A product (dTDP-6-deoxy-L-lyxo-4-hexulose) complex obtained with P. aeruginosa RmlC confirms that dTDP-L-rhamnose is a very good product mimic. Isothermal titration calorimetry data establishes that RmlC does bind dTDP-L-rhamnose more tightly than dTDP-glucose or dTDP-xylose (Table 3). We interpret these data to indicate that dTDP-L-rhamnose is a more akin to the transition state of the enzyme at the rate determining step than either dTDP-xylose or dTDP-glucose. We suggest that the transition state has an equatorial linkage at the C1 position (dTDP-L-rhamnose) rather than an axial one (dTDP-xylose/glucose). This would tend to favor a mechanism in which the ring flip occurs as an integral part of catalysis rather than at the end (as has conventionally been written Figure 1a). A ring flip at the end of epimerization would seem unlikely because the O1′, C6′ and O3′ tri-axial clash would make the product very high in energy and presumably create a correspondingly high activation energy (Figure 6a). Structural studies on GME suggested that during the first epimerization at C5′, the carbohydrate ring flips moving from an equatorial to axial linked carbohydrate24. This study experimentally established that a chair conformer with a C6′ O1′ diaxial clash is higher in energy than the ring flipped form which relieves such a strain. A similar process seems likely to occur in RmlC. We propose that as the proton is transferred to C5′ by the Tyr, the carbohydrate ring flip occurs to give the equatorially linked sugar. If not, an axial C6′ would be created by epimerization of the C5′, not only would this have the high energy C6′ O1′ diaxial interaction, but in addition C6′ would severely clash with His. A major re-organization of the active site would be necessary to accommodate a non-ring-flipped mono-epimerized intermediate and in all the structure of RmlC, there is no evidence for such flexibility. By combining the ring flip with proton transfer the C6′ remains fixed in space and avoids these very unfavorable interactions.
Figure 6.
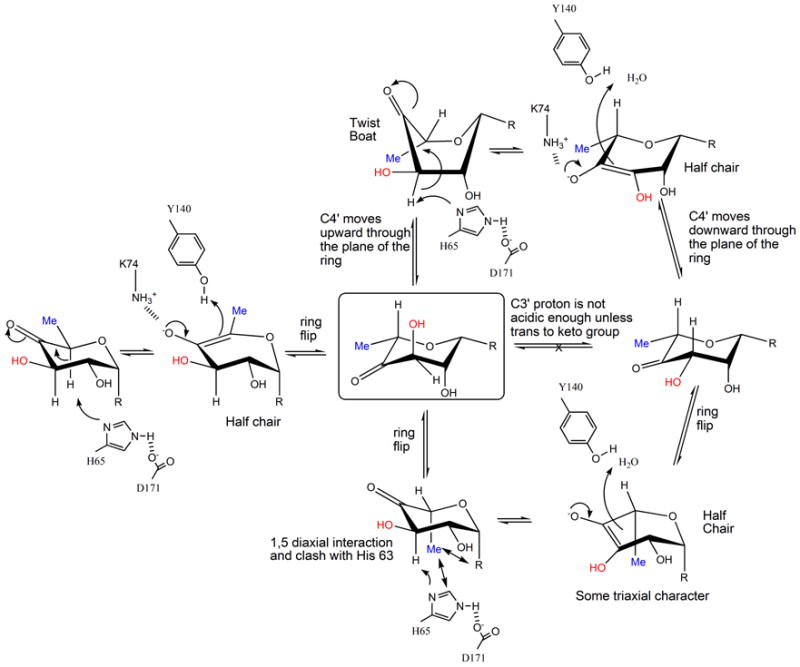
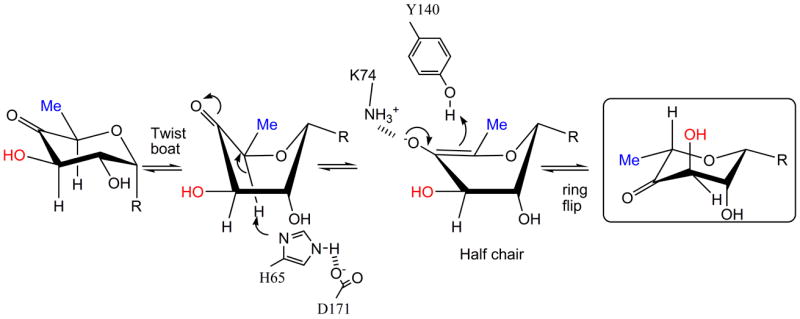
(a) R=OdTDP. A possible mechanism for RmlC based on structural and biochemical data. The key active site residues are shown, the H65 is the catalytic base for both epimerizations, K73 stabilizes the enolate and Y134 acts as the acid for the first epimerization. The mono-epimerized intermediate is shown boxed and has the equatorial linkage between carbohydrate ring and nucleotide. It cannot proceed directly to product because the C3′ proton is only sufficiently acidic when it is orthogonal to the plane of the carbonyl function.
(b) R=OdTDP. An alternative route for the first epimerization using a twist boat form of substrate, the mono epimerized intermediate is shown boxed. The apparent preference of RmlC for the equatorial linked sugar nucleotide suggests this is a possibility.
The ring-flipped intermediate poses a challenge for the enzyme for the second epimerization. The proton attached to C3′ is not orthogonal to the plane of the carbonyl, as a result its pKa is too high for removal by His37. As with GME24, there are two routes to create the required orthogonal arrangement of the π* anti-bonding orbital of the carbonyl and σ-bonding orbital of the C3′. The ring could flip back to the high energy diaxial chair conformer that is conventionally written (Figure 1a). Alternatively, the keto group could move through the plane of the ring to give a twist boat (Figure 6a). We strongly favor the twist boat for three reasons. The diaxial chair conformer would seem to be very energetically unfavorable and it clashes with protein. In contrast there is good precedent from a variety of enzymes that twist boat structures are energetically accessible during enzyme catalysis38. Secondly, epimerization of the diaxial intermediate would create a transition state with some triaxial character. This is likely to be extremely unfavorable and have prohibitively high activation energy. Thirdly, the structural data and thermodynamic strongly support an optimized interaction between an equatorial-linked sugar nucleotide. The equatorial configuration at C1′ is seen in the twist boat but not in the diaxial chair conformer.
It is possible that the substrate adopts a twist boat structure (Figure 6b). This could explain our apparent inability to obtain P. aeruginosa dTDP-xylose complex in which the carbohydrate portion is located at the active site. The structure shows the β-phosphate interaction with the protein is actually changed from that normally seen and thermodynamics establishes the sugar binds much more weakly than dTDP-L-rhamnose. We suggest that the enzyme does not favor binding of the axially configured dTDP-xylose and a result displaces the β phosphate from its normal binding site to position the sugar out of the active site. This would be consistent with the weaker binding of dTDP-glucose and dTDP-xylose relative to dTDP-L-rhamnose and dTDP (Table 3). As dTDP-D-xylose is smaller than the substrate (Figure 1c), non-binding cannot be explained by additional unfavorable steric clashes. Significantly, the dTDP-D-xylose and dTDP-D-glucose complexes of S. suis which do locate an axially linked carbohydrate at the active site do not superimpose with each other39. They also involve a key interaction with a non conserved residue and are not optimally aligned for enolate stabilization. It may be these complexes are artifacts of the S. suis enzyme. Despite differences in protein sequence, the three dTDP-L-rhamnose complexes, which have the equatorial linkage, superimpose with each other very well indeed and are properly aligned to the catalytic residues (Figure 4d). The substrate for RmlC has a C4′ carbonyl group that will reduce the energy penalty for a twist boat structure by decreasing the C1′, C4′ diaxial interaction (Figure 6b). However, the twist boat, even for the keto sugar, will be higher in energy compared to the normal chair form of the substrate. This would require significant stabilization by the enzyme. The binding of the higher energy forms of carbohydrate substrate is well known from studies of other enzymes40. Further investigation will require sophisticated calculations which are beyond the scope of this work.
Despite having no similarity in sequence or structure, the chemical mechanisms of RmlC and GME are very similar. Both use a single amino acid as the catalytic base for both epimerizations, His in RmlC and Cys in GME. Both enzymes also seem to use a single amino acid as the proton source for both epimerizations, Lys in GME, Tyr in RmlC. In RmlC, water may be involved in C3′ epimerization. Our data has provided the experimental evidence that supports a dominant order of C5′ epimerization followed by C3′ for both enzymes. The mono-epimerized intermediate in GME has been shown to be ring flipped (equatorial position for the nucleotide) and the structural data obtained in this study strongly suggest this is also the case for RmlC. In both enzymes, the chemical requirements of the transformation to avoid unfavorable steric clashes, whilst activating the C3′ proton for removal, indicates the mono-epimerized intermediate adopts a twist boat character during catalysis. This convergence in mechanism from two quite different classes of enzyme appears to be driven by the unique chemistry of carbohydrate epimerization. The likely twist boat nature of the intermediate in the RmlC mechanism will help guide inhibitor design against this validated drug target.
Materials and methods
Protein expression and purification
The rmlC gene of M. tuberculosis strain H37Rv was cloned into pET23b using the Ligation Independent Clone system (Stratagene). The bacteria were grown in tryptone-phosphate medium until the OD600 reached 1.0, at which time rmlC expression was induced by adding 1 mM IPTG and incubation was continued for 4 h. The protein was purified by elution from an anion exchange HQ column (Applied Biosystems) with an increasing NaCl gradient from 50 mM to 400 mM at pH 8.5 buffered with 20 mM Tris-HCl buffer. The second step was an elution from a hydrophobic exchange ET column (Applied Biosystems) with a decreasing 50% to 0% saturated ammonium sulfate gradient buffered at pH 7.3 by 20 mM phosphate. The purified protein was dialyzed against several changes of a solution containing 25 mM Tris-HCl buffer at pH 7.8 and concentrated to 7 mg·ml−1. The rmlC gene of P. aeruginosa was cloned into pET23a(+) with an N-terminal 6x His tag and a linker consisting of Gly-Ser-Met-Ala. The protein was overexpressed in E. coli BL21 (DE3) cells grown in Luria broth medium. Once the OD600 reached 0.8, 1 mM IPTG was added and incubation was continued for 6 h. The protein was purified in a similar manner to M. tuberculosis RmlC; the principal difference was the use of a hydrophobic exchange HP column with a decreasing 25% to 0% saturated ammonium sulfate gradient. The protein was dialyzed against 25 mM Tris-HCl buffer at pH 7.8 and concentrated to 8 mg·ml−1. The expression and purification of RmlC from S. suis has been described in detail previously19 as has the protocol for RmlC from S. enterica serovar Typhimurium15. The purity of the proteins was determined by SDS-PAGE and their integrity by N-terminal sequencing, bioassays and MALDI-TOF mass spectrometry.
Protein crystallization
The sugar nucleotides dTDP-rhamnose and dTDP-6-deoxy-D-xylo-4-hexulose were made enzymatically according to published procedures41. Crystals of M. tuberculosis RmlC (7 mg·ml−1) complexed with dTDP-L-rhamnose were obtained in 23% PEG 8000, 0.2 M calcium acetate hydrate, and 0.1 M sodium cacodylate at pH 5.8. The protein was incubated with 10 mM dTDP-L-rhamnose overnight before crystallization; crystals took 5 to 7 days to appear. Crystals of apo P. aeruginosa RmlC (7 mg·ml−1) were grown in 10% PEG 8000, 0.2 M sodium tartrate, 0.1 M MOPS pH 6.5 and took 2 to 3 days to appear. Crystals of P. aeruginosa RmlC complexed with dTDP-L-rhamnose crystals were obtained in 20% PEG 8000, 0.2 M sodium tartrate, 0.1 M MES at pH 5.8 with 10 mM dTDP-rhamnose after 10 days. Crystals of P. aeruginosa RmlC complexed with product were obtained by incubating RmlC with 20 mM dTDP-6-deoxy-D-xylo-4-hexulose, for 2 h at room temperature, prior to setting up crystal plates with 25% PEG 8000, 0.2 M sodium tartrate and 0.1 M MES at pH 6.2. With dTDP-D-xylose P. aeruginosa RmlC crystal conditions were changed to 25% PEG 8000, 0.2 M sodium tartrate, 0.1 M MES at pH 6.4, and 20 mM dTDP-xylose. All of the P. aeruginosa RmlC complex crystals grew to full size over 14 days. Crystals of S. suis RmlC in complex with dTDP-L-rhamnose (10 mM) were obtained from 25% PEG 2000, 0.1 M Tris-HCl at pH 7.6 and 4 mM NiCl2 after 14 days.
Data collection and structural determination
Data to 1.7 Å resolution of M. tuberculosis RmlC in complex with dTDP-L-rhamnose were collected on SRS station 14.1 with an x-ray wavelength of 1.49 Å on an ADSC detector. P. aeruginosa apo RmlC data (2.5 Å) were collected in-house on a DIP2000 imaging plate system mounted on a Nonius rotating anode generator using Cu Kα radiation. Data for P. aeruginosa RmlC in complex with dTDP-rhamnose (2.0 Å), complexed with dTDP-xylose (1.8 Å), and for the S. suis RmlC dTDP-rhamnose complex (1.6 Å) were all collected at SRS station 9.6 with an x-ray wavelength of 0.86 Å on a MAR CCD detector. Data to 1.7 Å resolution were collected from crystals of P. aeruginosa RmlC in complex with product on ID14.2 at the ESRF. All data were processed and scaled using MOLSFLM42 and SCALA43. Full details of data and refinement statistics are listed in Table 1. Structures were determined by molecular replacement using aMoRe44 or MOLREP45, using RmlC from S. enterica serovar Typhimurium as a model. Manual intervention was carried out in O46. Automated refinement of the structures used REFMAC547. All structures were checked using PROCHECK48. Ligands were included in the models when the Fo-Fc electron density was clear at least one subunit (Figure 3).
Table 1.
X-ray crystallographic data
| PAb apo | PAb product | PAb dTDPR | PAb dTDPX | MTb dTDPR | SSb dTDPR | |
|---|---|---|---|---|---|---|
| Wavelength (Å) | 1.54 | 0.933 | 0.87 | 0.87 | 1.488 | 0.87 |
| Resolution (Å) Highest Shell | 40.41-2.53 (2.67-2.53) | 20.59-1.7 (1.79-1.70) | 39.68-2.0 (2.11-2.0) | 43.44-1.8 (1.9-1.8) | 36.2-1.79 (1.89-1.79) | 43-1.6 (1.69-1.60) |
| Cell dimension |
a=b=57.8
c=161.6 |
a =64.4
b =146.2 c =44.9 |
a =64.9
b =125.5 c =109.4 |
a =64.8
b =148.1 c =45.4 |
a =44.8
b =56.8 c =90.7 α=89.9° β=82.3° γ=78.3° |
a =51.4
b =141.5 c =53.6 |
| Space group | P41212 | P21212 | C2221 | P21212 | P1 | P21 |
| Unique reflections | 9710 | 40067 | 30581 | 35190 | 84823 | 99569 |
| Average redundancy | 7.3 (6.3) | 6.1(4.7) | 4.0(4.0) | 3.0(3.1) | 2.2(2.2) | 4.9(4.1) |
| I/σ | 6.8(1.8) | 5.6(1.8) | 6.3(2.1) | 5.5(2.2) | 7.5(1.8) | 6.6(2.8) |
| Completeness (%) | 99.2(99.4) | 84.3(44.1) | 99.9(99.9) | 80.1(66.1) | 92.6(89.3) | 99.1(94.7) |
| Rmergea (%) | 8.8(41.9) | 8.6(38.4) | 8.4(35.6) | 9.6(34.9) | 6.5(29.9) | 7.0(24.8) |
| No of molecules in asymmetric unit | 1 | 2 | 2 | 2 | 4 | 4 |
| Refinement | ||||||
| R (%) | 20.0(31.9) | 15.0(17.4) | 19.0(24.5) | 18.6(21.7) | 15.2(18.3) | 16.7(17.8) |
| Rfree (5% of data) (%) | 27.8(44.7) | 19.8(25.3) | 25.6(29.2) | 23.7(28.8) | 19.6(25.9) | 21.2(27.4) |
| rmsd bonds (Å) / angles (º) | 0.014/1.4 | 0.011/1.4 | 0.008/1.12 | 0.009/1.6 | 0.012/1.3 | 0.012/1.3 |
| B-factor deviation bonds / angles (Å2): | ||||||
| main chain | 0.6/1.0 | 1.3/1.5 | 0.5/0.6 | 0.7/0.7 | 0.9/1.2 | 0.9/1.2 |
| side chains | 1.5/2.3 | 2.2/3.1 | 0.9/1.3 | 1.2/1.9 | 1.9/2.6 | 1.8/2.5 |
| Residues in Ramachandran Core (%) | 89 | 94 | 93 | 93 | 91 | 91 |
| Protein atoms | 1490 | 2982 | 2980 | 2992 | 6164 | 6335 |
| Water atoms | 92 | 531 | 590 | 455 | 1168 | 1383 |
| dTDP ligand atoms | 10 | 70 | 70 | 50 c | 140 | 140 |
| Protein average B-factor (Å2) | 26 | 15 | 35 | 20 | 26 | 12 |
| Average B-factor (Å2) of dTDP-ligand | N/A | 21 | 27 | 36c | 35 | 15 |
| Average B-factor (Å2) waters | 40 | 37 | 55 | 37 | 47 | 30 |
| PDB accession code | 2ixj | 2ixk | 2ixh | 2ixi | 2ixc | 2ixl |
Rmerge = ∑hkl ∑l|Ii − <I>|/∑hkl ∑l <I>, where Ii is an intensity for the ith measurement of a reflection with indices hkl and <I> is the weighted mean of the reflection intensity
PA P. aeruginosa, MT M. tuberculosis, SS S. suis. dTDPR:dTDP-L-rhamnose, dTDPX: dTDP-D-xylose, dTDPG: dTDP-D-glucose.
The carbohydrate portion of this ligand is partly disordered.
Biochemical characterization
Apparent Km and kcat measurements were made via a three-enzyme coupled assay described in detail previously41 (Table 2). The assay monitors the consumption of NADH by RmlD under conditions where the concentration of RmlC is rate limiting and thus the rate constants are apparent. Deuterium incorporation at C5′ and/or C3′ was also measured by an established protocol27; 49. RmlC was mixed with ca 1 nmol dTDP-6-deoxy-D-xylo-4-hexulose in 30 μl of HEPES buffer (50 mM, pH 7.6) made up in D2O for 2 hours at 37°C (Fig. 4a) or at 10°C for varying times for the time course experiment (Fig. 4b). The reaction was quenched by the addition of 50 μl of ethanol to precipitate protein, which removed by centrifugation. NaBH4 (2 mg) was then added to reduce both keto substrate and keto product at C4′ to the hydroxyl group. This generates a mixture of four compounds as their dTDP adducts: 6-deoxy-D-glucose (quinovose) and 6-deoxy-D-galactose (fucose) from substrate, 6-deoxy-L-talose and 6-deoxy-L-mannose (rhamnose) from double epimerized product (to date, mono-epimerized products have not been detected in RmlC catalyzed reactions). Since the equilibrium position for the reactions lies very heavily in favor of substrate, the two former compounds dominate. Further, the borohydride reduction is stereoselective, giving the 6-deoxy-D-gluco-configured product as the major isomer on steric grounds50. The samples were then hydrolyzed with trifluoroacetic acid, reduced with NaBH4 (at C1′) and acetylated to yielded alditol acetates which were analyzed by GC/MS. The m/z 217, 218, 231, 232, and 232 ions of the dominant per-O-acetylated quinovositol peak (which was derived from the substrate) were quantitated by GC-MS against an internal myo-inositol standard (Figure 4). The interpretation of the fragmentation pattern has been described elsewhere27; 49. Briefly, cleavage between C3′ and C4′ with retention of charge at C3′ yielded an ion at m/z 217 (no deuterium incorporation at C3′) or m/z 218 (deuterium incorporation at C3′). Thus the presence of the 218 ion is definitive for C3′ exchange and the presence of 217 for non exchange at C3′. Cleavage between C2′ and C3′ with retention of charge at C3′ yielded an ion at m/z 231 (no deuterium incorporation at either position), m/z 232 (deuterium at either C3′ or C5′) and m/z 233 (deuterium at C3′ and C′5). Thus the presence of the ion at 233 is definitive for C3′ and C5′ exchange, the ion at 231 is definitive for no exchange at all. An ion at 232 accompanied by one at 217 but not 218 indicates C5′ exchange in the absence of any C3′ exchange. Whereas ions at 218 and 232 but not 233, indicate C3′ exchange in the absence of any C5′ exchange.
The Promega GeneEditor site-directed mutagenesis kit was used to generate mutants of RmlC from S. enterica serovar Typhimurium with the following primers:
H63A : CTCAGAGGGCTAGCTTTTCAGAGAGGAG,
K73A : GAAAATGCACAGGGGGCGTTAGTTCGTTGTGC and
Y133F : GAGTTTCTGTTCAAAGCAAC.
Isothermal titration calorimetry
Titrations were performed in triplicate on a VP-ITC system (MicroCal) in 20mM Tris-HCl base, pH 7.45. In a typical titration, twenty five 10μl injections of ligands (5mM dTDP-L-rhamnose, dTDP-glucose and dTDP-xylose and 2mM dTDP) were made into a solution P. aeruginosa RmlC from (0.1mM) in the ITC cell at 25°C. Nucleotide concentrations were estimated by extinction coefficient. Control experiments were performed to assess the heat of dilution of each ligand by injecting this into buffer. There was no evidence of any self association of the ligands (would be shown by a trend in the heats of dilution). Heats of dilution were subtracted from the original heats of interaction prior to data analysis. All solutions were degassed prior to use. The resulting isotherms were fit using the ORIGIN software package (MicroCal). The data were fitted to the simplest model based on independent binding sites. The Ka values derived in Table 3 come from unrestrained refinement; fixing the stoichiometry (N) to 1 does not change the value of Ka within error, except for dTDP. For dTDP fixing N = 1, drops Ka to 103M. This does not change the interpretation of our data. No matter how Ka is calculated for dTDP, it is always significantly higher than dTDP-xylose/glucose. The same is true for dTDP-L-rhamnose it binds significantly more tightly than dTDP-xylose/glucose. Whether dTDP-L-rhamnose binds significantly more tightly than dTDP (or within error equally tightly) does not affect our analysis and is a secondary issue. We do not believe the data we have obtained reliably establish this. The errors in Table 3 are those derived from the fit, the impurity inherent in all sugar nucleotide preparations will increase the actual error of the experiment. For calorimetry we synthesized fresh dTDP-L-rhamnose enzymatically using 1.1 mM dTDP-D-glucose, 1.1 μM S. suis RmlB, 0.4 μM S. enterica serovar Typhimurium RmlC, 0.5 μM M. tuberculosis RmlD, in 180 mM Tris-HCl pH 7.5, 18 mM MgCl2 and 1.8 mM NADPH. Reaction mixtures were incubated for 90 min at 37°C, then protein was removed from the mixture by centrifugation through an Amicon microcon 10 kDa molecular cut-off regenerated cellulose column. Compounds were separated on a Phenomenex Kingsorb 5μ C-18 column (250 × 21.2 mm) at a flow rate of 8 ml min−1. The column was equilibrated with 20 mM triethylammonium acetate (TEAA) pH 6.0 containing 3% acetonitrile prior to sample loading. After 5 min isocratic elution with 20 mM TEAA pH 6.0 and 3% acetonitrile, compounds were eluted with a gradient of 3.0 – 3.7% acetonitrile over 30 minutes. Eluant was monitored by a UV detector at 267 nm. This procedure is based on a published method for purifying nucleotide sugars51. The dTDP-L-rhamnose peak eluted at approximately 3.4% acetonitrile and was collected, the mass of the major component confirmed as 547 by LCT-MS in negative ion mode and the compound freeze dried. dTDP-glucose and dTDP were purchased from FLUKA. dTDP-xylose was synthesized chemically according by established methods52.
Supplementary Material
Acknowledgments
JHN is a Career Development Fellow of Biotechnology and Biology Research Council (BBSRC). The work is supported by grants from the Wellcome Trust and NIH. CW and JSL are Canada Research Chairs and acknowledge funding from the Natural Sciences Engineering Research Council and the Canadian Cystic Fibrosis Foundation respectively. RAF thanks the BBSRC and the EPSRC for financial support. We thank Wulf Blankenfeldt and Uli Schwartz-Linek for assistance and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharon N, Lis H. Lectins as cell recognition molecules. Science. 1989;246:227–233. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- 2.Allard ST, Giraud MF, Naismith JH. Epimerases: structure, function and mechanism. Cell Mol Life Sci. 2001;58:1650–65. doi: 10.1007/PL00000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field RA, Naismith JH. Structural and mechanistic basis of bacterial sugar nucleotide-modifying enzymes. Biochemistry. 2003;42:7637–7647. doi: 10.1021/bi0345079. [DOI] [PubMed] [Google Scholar]
- 4.Tanner ME. Sugar nucleotide-modifying enzymes. Curr Org Chem. 2001;5:169–192. [Google Scholar]
- 5.Ma Y, Stern RJ, Scherman MS, Vissa VD, Yan W, Jones VC, Zhang F, Franzblau SG, Lewis WH, McNeil MR. Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob Agents Chemother. 2001;45:1407–16. doi: 10.1128/AAC.45.5.1407-1416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babaoglu K, Page MA, Jones VC, McNeil MR, Dong CJ, Naismith JH, Lee RE. Novel inhibitors of an emerging target in Mycobacterium tuberculosis; Substituted thiazolidinones as inhibitors of dTDP-rhamnose synthesis. Bioorganic & Medicinal Chemistry Letters. 2003;13:3227–3230. doi: 10.1016/s0960-894x(03)00673-5. [DOI] [PubMed] [Google Scholar]
- 7.Mills JA, Motichka K, Jucker M, Wu HP, Uhlik BC, Stern RJ, Scherman MS, Vissa VD, Pan F, Kundu M, Ma YF, McNeil M. Inactivation of the mycobacterial rhamnosyltransferase, which is needed for the formation of the arabinogalactan-peptidoglycan linker, leads to irreversible loss of viability. J Biol Chem. 2004;279:43540–43546. doi: 10.1074/jbc.M407782200. [DOI] [PubMed] [Google Scholar]
- 8.Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology. 2000;146:2803–14. doi: 10.1099/00221287-146-11-2803. [DOI] [PubMed] [Google Scholar]
- 9.Tsukioka Y, Yamashita Y, Oho T, NaKano Y, Koga T. Biological function of the dTDP-Rhamnose synthesis pathway in Streptococcus mutans. Journal of Bacteriology. 1997;179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita Y, Tomihisa K, Nakano Y, Shimazaki Y, Oho T, Koga T. Recombination between gtfB and gtfC is required for survival of a dTDP-rhamnose synthesis-deficient mutant of Streptococcus mutans in the presence of sucrose. Infect Immun. 1999;67:3693–7. doi: 10.1128/iai.67.7.3693-3697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Pan F, McNeil M. Formation of dTDP-rhamnose is essential for growth of mycobacteria. J Bacteriol. 2002;184:3392–5. doi: 10.1128/JB.184.12.3392-3395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melo A, Glaser L. The mechanism of 6-deoxyhexose synthesis. II. Conversion of deoxythymidine diphosphate 4-keto-6-deoxy-D-glucose to deoxythymidine diphosphate L-rhamnose. J Biol Chem. 1968;243:1475–1478. [PubMed] [Google Scholar]
- 13.Blankenfeldt W, Asuncion M, Lam JS, Naismith JH. The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA) EMBO J. 2000;19:6652–63. doi: 10.1093/emboj/19.24.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allard ST, Giraud MF, Whitfield C, Messner P, Naismith JH. The purification, crystallization and structural elucidation of dTDP-D-glucose 4,6-dehydratase (RmlB), the second enzyme of the dTDP-L-rhamnose synthesis pathway from Salmonella enterica serovar typhimurium. Acta Crystallogr D Biol Crystallogr. 2000;56 :222–5. doi: 10.1107/s0907444999016200. [DOI] [PubMed] [Google Scholar]
- 15.Giraud MF, Leonard GA, Field RA, Bernlind C, Naismith JH. RmlC, the third enzyme of dTDP-L-rhamnose pathway, is a new class of epimerase. Nat Struct Biol. 2000;7:398–402. doi: 10.1038/75178. [DOI] [PubMed] [Google Scholar]
- 16.Blankenfeldt W, Kerr ID, Giraud MF, McMiken HJ, Leonard G, Whitfield C, Messner P, Graninger M, Naismith JH. Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure. 2002;10:773–86. doi: 10.1016/s0969-2126(02)00770-0. [DOI] [PubMed] [Google Scholar]
- 17.Beis K, Allard ST, Hegeman AD, Murshudov G, Philp D, Naismith JH. The structure of NADH in the enzyme dTDP-d-glucose dehydratase (RmlB) J Am Chem Soc. 2003;125:11872–8. doi: 10.1021/ja035796r. [DOI] [PubMed] [Google Scholar]
- 18.Christendat D, Saridakis V, Dharamsi A, Bochkarev A, Pai EF, Arrowsmith CH, Edwards AM. Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP. J Biol Chem. 2000;275:24608–12. doi: 10.1074/jbc.C000238200. [DOI] [PubMed] [Google Scholar]
- 19.Dong CJ, Major LL, Allen A, Blankenfeldt W, Maskell D, Naismith JH. High-resolution structures of RmIC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure. 2003;11:715–723. doi: 10.1016/s0969-2126(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 20.Kantardjieff KA, Kim CY, Naranjo C, Waldo GS, Lekin T, Segelke BW, Zemla A, Park MS, Terwilliger TC, Rupp B. Mycobacterium tuberculosis RmlC epimerase (Rv3465): a promising drug-target structure in the rhamnose pathway. Acta Crystallogr D Biol Crystallog. 2004;60:895–902. doi: 10.1107/S0907444904005323. [DOI] [PubMed] [Google Scholar]
- 21.Rizzi M, Tonetti M, Vigevani P, Sturla L, Bisso A, De Flora A, Bordo D, Bolognesi M. GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily. Structure. 1998;6:1453–1465. doi: 10.1016/s0969-2126(98)00144-0. [DOI] [PubMed] [Google Scholar]
- 22.Menon S, Stahl M, Kumar R, Xu GY, Sullivan F. Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from Escherichia coli. J Biol Chem. 1999;274:26743–50. doi: 10.1074/jbc.274.38.26743. [DOI] [PubMed] [Google Scholar]
- 23.Somers WS, Stahl ML, Sullivan FX. GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site. Structure. 1998;6:1601–12. doi: 10.1016/s0969-2126(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 24.Major LL, Wolucka BA, Naismith JH. Structure and function of GDP-mannose-3′,5′-epimerase: An enzyme which performs three chemical reactions at the same active site. J Am Chem Soc. 2005;127:18309–18320. doi: 10.1021/ja056490i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naundorf A, Klaffke W. Substrate specificity of native dTDP-D-glucose-4,6-dehydratase: Chemo-enzymatic syntheses of artificial and naturally occurring deoxy sugars. Carbohydr Res. 1996;285:141–150. doi: 10.1016/s0008-6215(96)90180-9. [DOI] [PubMed] [Google Scholar]
- 26.Merkel AB, Major LL, Errey JC, Burkart MD, Field RA, Walsh CT, Naismith JN. The position of a key tyrosine in dTDP-4-Keto-6-deoxy-D-glucose-5-epimerase (EvaD) alters the substrate profile for this RmlC-like enzyme. J Biol Chem. 2004;279:32684–32691. doi: 10.1074/jbc.M404091200. [DOI] [PubMed] [Google Scholar]
- 27.York WS, Darvill AG, Mcneil M, Stevenson TT, Albersheim P. Isolation and Characterization of Plant-Cell Walls and Cell-Wall Components. Methods in Enzymology. 1986;118:3–40. [Google Scholar]
- 28.Bryce RA, Hillier IH, Naismith JH. Carbohydrate-protein recognition: molecular dynamics simulations and free energy analysis of oligosaccharide binding to concanavalin A. Biophys J. 2001;81:1373–88. doi: 10.1016/S0006-3495(01)75793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moothoo DN, Canan B, Field RA, Naismith JH. Man alpha1-2 Man alpha-OMe-concanavalin A complex reveals a balance of forces involved in carbohydrate recognition. Glycobiology. 1999;9:539–45. doi: 10.1093/glycob/9.6.539. [DOI] [PubMed] [Google Scholar]
- 30.Tello M, Jakimowicz P, Errey JC, Meyers CLF, Walsh CT, Buttner MJ, Lawson DM, Field RA. Characterisation of Streptomyces spheroides NovW and revision of its functional assignment to a dTDP-6-deoxy-D-xylo-4-hexulose 3-epimerase. Chem Commun. 2006:1079–1081. doi: 10.1039/b515763c. [DOI] [PubMed] [Google Scholar]
- 31.Allard ST, Beis K, Giraud MF, Hegeman AD, Gross JW, Wilmouth RC, Whitfield C, Graninger M, Messner P, Allen AG, Maskell DJ, Naismith JH. Toward a structural understanding of the dehydratase mechanism. Structure. 2002;10:81–92. doi: 10.1016/s0969-2126(01)00694-3. [DOI] [PubMed] [Google Scholar]
- 32.Williams G, Maziarz EP, Amyes TL, Wood TD, Richard JP. Formation and stability of the enolates of N-protonated proline methyl ester and proline zwitterion in aqueous solution: A nonenzymatic model for the first step in the racemization of proline catalyzed by proline racemase. Biochemistry. 2003;42:8354–8361. doi: 10.1021/bi0345992. [DOI] [PubMed] [Google Scholar]
- 33.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–13. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 34.Cellini B, Bertoldi M, Paiardini A, D’Aguanno S, Voltattorni CB. Site-directed mutagenesis provides insight into racemization and transamination of alanine catalyzed by Treponema denticola cystalysin. J Biol Chem. 2004;279:36898–36905. doi: 10.1074/jbc.M404449200. [DOI] [PubMed] [Google Scholar]
- 35.Lohkamp B, Bäuerle B, Rieger P-G, Schneider G. Three-dimensional structure of iminodisuccinate epimerase defines the fold of the MmgE/PrpD protein family. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.07.051. Available on line 29th July 2006. [DOI] [PubMed] [Google Scholar]
- 36.Rye CS, Matte A, Cygler M, Withers SG. An atypical approach identifies TYR234 as the key base catalyst in chondroitin AC lyase. Chembiochem. 2006;7:631–637. doi: 10.1002/cbic.200500428. [DOI] [PubMed] [Google Scholar]
- 37.Corey EJ. The stereochemistry of -haloketones V. Prediction of the stereochemistry of -brominated ketosteroids. J Am Chem Soc. 1954;76:175–179. [Google Scholar]
- 38.Vasella A, Davies GJ, Matthias B. Glycosidase mechanisms. Trends in Chemical Biology. 2002;6:619–629. doi: 10.1016/s1367-5931(02)00380-0. [DOI] [PubMed] [Google Scholar]
- 39.Dong C, Major LL, Allen A, Blankenfeldt W, Maskell D, Naismith JH. High-resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure. 2003;11:715–723. doi: 10.1016/s0969-2126(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 40.Sinnott ML, Guo XM, Li SC, Li YT. Leech Sialidase-L Cleaves the Glycon Aglycon Bond with the Substrate in a Normally Disfavored Conformation. J Am Chem Soc. 1993;115:3334–3335. [Google Scholar]
- 41.Graninger M, Nidetzky B, Heinrichs DE, Whitfield C, Messner P. Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-L-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J Biol Chem. 1999;274:25069–77. doi: 10.1074/jbc.274.35.25069. [DOI] [PubMed] [Google Scholar]
- 42.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB newsletter on protein crystallography No. 1992;26:1–10. [Google Scholar]
- 43.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D Biol Crystallog. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Navaza J. AMoRe: An Automated Package for Molecular Replacement. Acta Crystallogr, Sect A: Found Crystallogr. 1994;50:157–163. [Google Scholar]
- 45.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 46.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron Density Maps and the Location of Errors in These Models. Acta Crystallogr, Sect A: Found Crystallog. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallog. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J Appl Cryst. 1993;26:548–558. [Google Scholar]
- 49.Kirkpatrick PN, Scaife W, Hallis TM, Liu H, Spencer JB, Williams DH. Characterisation of a sugar epimerase enzyme involved in the biosynthesis of a vancomycin-group antibiotic. Chem Commun. 2000:1565–1566. [Google Scholar]
- 50.Stern RJ, Lee TY, Lee TJ, Yan W, Scherman MS, Vissa VD, Kim SK, Wanner BL, McNeil MR. Conversion of dTDP-4-keto-6-deoxyglucose to free dTDP-4-keto-rhamnose by the rmIC gene products of Escherichia coli and Mycobacterium tuberculosis. Microbiology. 1999;145:663–71. doi: 10.1099/13500872-145-3-663. [DOI] [PubMed] [Google Scholar]
- 51.Järvinen N, Mäki M, Räbinä J, Roos C, Mattila P, Renkonen R. Cloning and expression of Helicobacter pylori GDP-L-fucose synthesizing enzymes (GMD and GMER) in Saccharomyces cerevisiae. Eur J Biochem. 2001;268:6458–6464. doi: 10.1046/j.0014-2956.2001.02601.x. [DOI] [PubMed] [Google Scholar]
- 52.Ballell L, Young RJ, Field RA. Synthesis and evaluation of mimetics of UDP and UDP-alpha-D-galactose, dTDP and dTDP-alpha-D-glucose with monosaccharides replacing the key pyrophosphate unit. Org Biomol Chem. 2005;3:1109–1115. doi: 10.1039/b500418g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


