Abstract
TGF-β (Transforming Growth Factor-β) cytokines employ the Smad proteins as the intracellular mediator of signaling. Upon TGF-β stimulation, the cytoplasmic Smads become phosphorylated and consequently accumulate in the nucleus to regulate target gene expression. The cytoplasm-to-nucleus redistribution of Smads, as well as the ability of Smads to activate or repress gene transcription, is under multiple layers of regulation by factors not limited to TGF-β. With recent advance in the knowledge of regulatory factors impinged on Smads, we are beginning to understand the complexity in cellular responses to TGF-β.
Keywords: Smad, TGF-β, nucleocytoplasmic trafficking, phosphorylation, ubiquitination, SUMOylation
1. INTRODUCTION
Since its identification in the early 1980s, transforming growth factor β (TGF-β) has been well established as an important regulator of tissue homeostasis and embryonic development in metazoans [1, 2]. The physiological responses to TGF-β and members of this cytokine family are diverse, and dependent on the intrinsic properties of the recipient cells.
The signal transduction of all TGF-β cytokines is mediated by a rather simple and evolutionarily conserved mechanism [3-5]. TGF-β cytokines are bound by their cognate receptors (referred to as the type II receptor) which are constitutively active protein kinases [6, 7]. This ligand binding event induces type II receptor to associate with and consequently phosphorylate a second transmembrane receptor - the type I receptor. Phosphorylation of the type I receptor results in activation of its kinase activity [8]. Downstream of the TGF-β receptor kinases, the family of Smad proteins serves as the essential mediators of signaling [9, 10]. When fused to the heterologous GAL-4 DNA binding domain, Smad was shown to be capable of activating the transcription of a reporter gene with GAL-4-binding sites in the promoter [11]. This, and the later observation that Smads have specific DNA binding activities, led to the conclusion that Smads function as transcription factors [12]. The link between activated TGF-β receptors and Smads was established with the key discovery that Smad is a substrate of the type I TGF-β receptor kinase and as a result the phosphorylated Smad concentrates in the nucleus [13-15]. Thus, being a signal-activated transcription factor, Smad is the central transducer of TGF-β signal.
In mammals, the family of Smad proteins include Smad1, 2, 3, 5 and 8, which all contain an SXS motif at their carboxy-termini that are phosphorylated by ligand-activated TGF-β receptor kinases. These Smads are therefore referred to as R-Smads (Receptor activated Smads). While Smad2 and Smad3 act specifically downstream of only TGF-βs, activin and Nodal, Smad1, 5, and 8 are activated by BMPs (Bone Morphogenetic Proteins) only [9, 10]. Smad4 is not phosphorylated at its carboxy-terminus, but associates with R-Smads and enters the nucleus upon stimulation by all TGF-β cytokines. Thus Smad4 is referred to as the co-Smad [9, 10]. Smad6 and Smad7 belong to the I-Smad (Inhibitory Smad) subgroup, which bind to the type I receptor or Smad4 and block their interaction with R-Smads [16-19]. Hence, Smad6 and Smad7 inhibit TGF-β signaling. The Smads share sequence similarities, most notably in the N-terminal and carboxy-terminal regions, referred to as the MH1 (Mad Homology 1) and MH2 domains respectively. The sequence between the MH1 and MH2 domains is termed the linker region, which is quite variable among Smads.
Knockout mice as well as tissue culture-based studies have implicated R-Smads and Smad4 in regulating growth and differentiation properties of a wide variety of cell types, consistent with the multifaceted physiological functions of TGF-β cytokines [1, 2]. It is becoming increasingly clear that Smads are subject to additional inputs from other extracellular signals. The linker regions of R-Smads are phosphorylated in response to mitogenic signals. In the case of Smad1, such phosphorylation plays important roles in regulating tissue patterning during embryonic development [20, 21]. Through phosphorylating the linker of Smad2 and 3, MAPKs (Mitogen-Activated Protein Kinase) or CDKs (Cyclin-Dependent Kinases) antagonize the growth inhibitory function of TGF-β in epithelial cells [22, 23]. In contrast, the Ras/MAPK pathway cooperates with TGF-β and Smads in regulating EMT (Epithelial-to-Mesenchymal Transition) and extracellular matrix remodeling [24, 25]. Moreover, TGF-β can bypass Smads and directly activate the PI3K (Phosphatidylinositol 3-Kinase) pathway in regulating growth properties of fibroblast [26]. Thus, cross-talks between TGF-β and other signaling pathways are highly dependent on cellular contexts, may take place through Smad-dependent and independent mechanisms, and have profound implications in disease mechanisms of cancer and fibrosis [4, 27, 28].
The complexity in TGF-β responses arises partly from the wide variety of transcriptional cofactors that partner with Smads in promoter specific or cell type specific manners and also non-Smad elements in TGF-β signaling. The subjects on transcriptional regulation by Smads and Smad-independent TGF-β signaling have been discussed extensively in several excellent reviews [4, 5, 29-31]. This present review will instead focus on the signaling activity of Smad, particularly on intracellular trafficking and post-translational modifications of Smads. These two events critically control Smad function as a signal transducer and transcription factor, and serve as the nodes that integrate inputs from TGF-β and other signals.
2. CONTROL OF SUBCELLULAR LOCALIZATION OF SMADS
R-Smads and Smad4 are present mostly in the cytoplasm or evenly throughout the cell at basal state, but become concentrated in the nucleus upon TGF-β stimulation [13, 32, 33]. Such spatial control of Smads ensures conditional access of Smads to their target genes. Moreover, the amount of Smads accumulated in the nucleus and the duration of Smads being engaged with their target promoters are critical determinants of transcriptional outcomes [34-36]. Therefore, nucleocytoplasm translocation of Smads is a crucial regulatory issue in TGF-β signaling.
2.1. Nucleocytoplasmic Shuttling of Smads in Basal State Cells
Although immunofluorescent staining showed that the bulk of R-Smads and Smad4 undergo cytoplasm-to-nucleus movement only upon TGF-β stimulation, two studies using methods designed to capture dynamics of Smads revealed spontaneous bi-directional shuttling of Smads across the nuclear envelope without TGF-β stimulation [37, 38].
This conclusion was further validated recently in live cell analyses [39, 40]. PAGFP (Photo-Activatable Green Fluorescent Protein)-fused Smad2 can be photoactivated in live cells locally by laser, so that a particular pool of PAGFP-Smad2 can be visualized. With this technology, it was shown that Smad2 continuously move into and out of the nucleus in unstimulated cells [40]. Moreover, FLIP (Fluorescence Loss In Photobleaching) analysis of GFP-fused Smad2 and Smad4 showed that when either the cytoplasmic or nuclear pool of GFP-Smads is depleted by photobleaching, the remaining GFP-Smads redistribute and quickly reache a new equilibrium between the two compartments [39]. These real-time analyses strongly suggest that R-Smads and Smad4 do not need TGF-β stimulation to enter the nucleus; but apparently without TGF-β, nuclear import of R-Smads and Smad4 is offset by export forces so these Smads cannot reach a high concentration in the nucleus.
More and more signal transducers have been found to continuously shuttle in and out of the nucleus, thus nucleocytoplasmic trafficking is becoming a common feature of many signaling molecules [41]. However the functional purposes of such Smads shuttling in the absence of TGF-β stimulation remain a question. While Smads need to be in the cytoplasm to sense the ligand-binding state of the TGF-β receptors, it is unclear if the nuclear presence of R-Smads not phosphorylated at their carboxy-termini has any functional consequences.
2.2. Nuclear Import and Export Mechanisms of Smads
The transport of macromolecules across the nuclear envelope is mediated by the nuclear pore complex (NPC), an assembly of multiple copies of over 30 proteins termed nucleoporins [42, 43]. Translocation across the nuclear envelope entails three steps: anchorage to the NPC, migration through the NPC and release into either cyto- or nucleoplasm [44-46]. Many protein cargos are recruited to the NPC by the importin β (also referred to as karyopherin) family of transport receptors. These transport receptors recognize various nuclear-targeting motifs in proteins such as the classic Nuclear Localization Signal (cNLS) and through separate domains all transport receptors also directly interact with nucleoporins. Such properties are essential for transport receptors to carry cargos into or out of the nucleus. Based on limited sequence similarity or shared biochemical properties, over 20 putative transport receptors have been proposed in mammals [47]. Those function in nuclear import are commonly referred to as importins, while those specialize in nuclear export are termed exportins. The association of importins and exportins with their cargos is regulated by Ran GTPase in opposite manners: RanGTP induces dissociation of importins from their substrates, but enhances interaction between exportins and their cargos [44-46]. Since RanGTP is exclusively present in the nucleus, importins and exportins can therefore only deliver their cargos in one direction even though importins and exportins alone can traverse the nuclear envelope in both directions.
2.2.1. R-Smads
Reconstituted nuclear import assay in vitro has been instrumental in deciphering the molecular mechanism of nuclear import by the importin β family of transport receptors [48]. The behavior of Smad2 and Smad3 in this assay suggest that these Smads, without being phosphorylated by TGF-β receptors, can spontaneously enter the nucleus independent of transport receptors [49, 50]. It appears that the Smads can be recruited to NPC by direct protein-protein interactions with nucleoporins Nup214 and Nup153 [37]. This interaction requires a hydrophobic region in the carboxy-terminus of Smad2 and Smad3. Point mutations in the nucleoporin binding domain, or competitive binding to this domain by other proteins all led to much impaired nuclear import of Smad2, supporting the notion that nucleoporin interaction is important for nuclear import of Smad2 [37]. Smad3 is an unique case in that it also binds to importin β1 through an NLS-like motif in the N-terminal region [51-53]. Due to differences in the flanking sequences, Smad2 does not interact with importin β1 [51]. For Smad3, this importin β1 binding is separate from the nucleoporin binding, suggesting the possibility that there may be parallel mechanisms for importing Smad3 into the nucleus. However, in the same reconstituted nuclear import assay, the MH2-domain mediated importin-independent pathway for Smad3 appeared to be more robust than the MH1-domain mediated importin β1-dependent mechanism [50]. A number of other signal transducers such as STATs (Signal Transducer and Activator of Transcription) and ERKs (Extracellular Signal-Regulated Protein Kinases) have been described to employ multiple mechanisms for nuclear import [54, 55]. For Smad3, the question is what determines the choice of alternative nuclear import pathways.
The ability to directly bind nucleoporins could facilitate nuclear export as well, but the participation of exportins may also be needed. Recently, exportin 4 was shown to be capable of binding to the MH2 domain of Smad3, and knockdown of exportin 4 by siRNA inhibited nuclear export of Smad3 [40]. More importantly, TGF-β-induced carboxy-terminus phosphorylation of Smad3 appeared to weaken its interaction with exportin 4, consistent with the idea that by impeding the rate of nuclear export, TGF-β enhances nuclear accumulation of R-Smads [40].
2.2.2. Smad4
Smad4 undergoes cytoplasm-to-nucleus redistribution upon TGF-β stimulation, but unlike most R-Smads it also becomes exclusively localized in the nucleus when the CRM-1-dependent nuclear export is inhibited by Leptomycin B [56, 57]. While the current consensus is that CRM-1 is the export receptor for Smad4, the mechanism underlying TGF-β-independent nuclear import of Smad4 is under debate. Smad4 can directly interact with Nup214 and it enters the nucleus without the assistance of importins in the nuclear import assay [50]. On the other hand, a lysine rich motif is present in the MH1 domain of Smad4. This region in Smad3 binds to importin β1, but in Smad4 it apparently interacts with importin α instead [58]. Mutation of this importin α binding site affected nuclear concentration of Smad4 in response to Leptomycin B treatment, but significant amount of Smad4 was still detected in the nucleus [56]. Moreover, it is unclear if the effect is due to defects in nuclear import or nuclear retention, since the mutations might also interfere with Smad4 binding to DNA as shown in another study [59]. When tested in the reconstituted in vitro assay, nuclear import of Smad4 was robust under the condition that a typical importin α cargo was unable to translocate into the nucleus [50]. Thus, similar to Smad3, Smad4 may also enter the nucleus through multiple pathways.
2.3. Nuclear and Cytoplasmic Retention Factors of Smads
In addition to the nuclear transport machinery, retention factors are another major force in controlling the subcellular localization of Smads. Kinetic analysis of Smad movement in live cells also indicated that the mobility of Smads in the cytoplasm is limited, probably due to certain retention mechanisms [39]. When overexpressed, SARA (Smad Anchor of Receptor Activation), Ski and PKB (Protein Kinase B)/Akt have all been shown to sequester Smad2 or Smad3 in the cytoplasm [49, 60-62]. In the case of SARA, its binding to Smad2 precludes nucleoporin interaction and inhibits nuclear import of unphosphorylated Smad2 [37, 49]. Moreover, the affinity between SARA and Smad2 is substantially decreased upon TGF-β-induced phosphorylation of Smad2, so release from SARA may be part of how TGF-β enhances nuclear accumulation of R-Smads [60]. However, SARA is almost exclusively localized in endosomes, while Smad2/3 are present throughout the cytoplasm, so apparently only a small proportion of Smad2/3 are under the control of SARA [60]. In fact, live cell analysis suggested that the mobility of R-Smads in the cytoplasm is unchanged upon TGF-β signaling, arguing against the idea that release from cytoplasmic retention plays an important role in TGF-β-induced nuclear accumulation of R-Smads [40].
Mirroring the retention factors in the cytoplasm, Smad binding proteins can also sequester Smads in the nucleus. It was first shown that overexpression of FoxH1, itself an exclusively nuclear protein, results in nuclear localization of Smad2 in the absence of TGF-β signal [37, 63]. Similar observations were made with a number of other Smad2 interacting proteins in the nucleus, such as ATF2 [64]. It is conceivable that DNA-binding, characteristic of R-Smads and Smad4, may also help retaining Smads in the nucleus. Retention of Smad in the nucleus is likely a consequence of suppressed Smad nuclear export, perhaps due to competition between the nuclear retention factor and the export factor for association with Smads. Interestingly, when the Smad-binding domain of SARA was artificially targeted to the nucleus by fusion with an NLS, the endogenous Smad2 and Smad3 became sequestered in the nucleus without TGF-β induction [65]. This supports the notion that inhibition of Smad-nucleoporin interaction could block both nuclear export and import of Smad2 and Smad3. Like SARA, Fox H1 and ATF2 both bind to the MH2 region of Smad2, and Fox H1 can compete with the nucleoporin Nup153 for binding to Smad2, which could explain how Fox H1 inhibits Smad2 exit from the nucleus [37]. Given the recently identified role of exportin 4 in Smad3 export to the cytoplasm, it will be interesting to examine if Fox H1 or ATF2 interferes with Smad3 binding to exportin 4 as well.
2.4. How TGF-β targets Smads into the nucleus
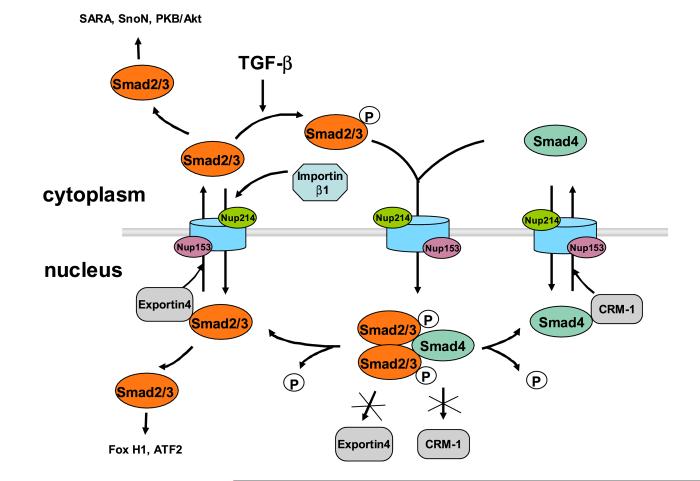
In response to TGF-β, one major event that potentially affects nuclear transport of R-Smads and Smad4 is the association between R-Smad and Smad4, which creates a new demand to import Smads as a complex. This raises the question of how much of the conclusions on nuclear import of monomeric R-Smads and Smad4 can be applied to the R-Smad/Smad4 complex. In addition to nuclear import and export, a host of nuclear and cytoplasmic retention factors must also be at play in concentrating R-Smads and Smad4 in the nucleus upon TGF-β stimulation (Fig. 1).
Figure 1.
Nucleoporins, transport receptors and retention factors regulate the nucleo-cytoplasmic distribution of R-Smads and Smad4. Note the involvement of importin β1 is limited to Smad3 only. The model suggests that Smad2 and Smad3 need to be dephosphorylated and dissociated from Smad4 before they can be exported to the cytoplasm by exportin 4 (for Smad2/3) and CRM-1 (for Smad4).
The case is best illustrated for Smad4 (Fig. 1). Upon association of Smad4 with phosphorylated R-Smads, the interaction between CRM-1 and Smad4 is largely abolished, and consequently Smad4 export is prohibited [66]. On the other hand, there is no indication of increased nuclear import when Smad4 and R-Smads are bound to each other [66]. The complex exhibited similar affinity for nucleoporins, and there is no evidence if Smad4 binding to importin α is enhanced as a consequence to Smad4 association with R-Smads. The in vitro import assay revealed that compared to monomeric Smad4, the complex may contact different nucleoporins when traversing the NPC, but the efficiency of nuclear import remained comparable [66]. Therefore, the failure to export Smad4, due to binding of phospho-R-Smads, may be sufficient to restrict Smad4 to the nucleus. This agrees well with earlier observations that ligand-induced nuclear accumulation of Smad4 depends on the presence of R-Smads in mammalian and Drosophila cells [32, 67].
The scenario is more complex for R-Smads (Fig.1). Live cell analysis did not detect noticeable change in the nuclear import rate of R-Smads in response to TGF-β, consistent with the finding that phosphorylation of R-Smads does not affect its affinity to nucleoporins [37, 40]. Kinetic analysis of live cells also showed no sign of Smads being released from cytoplasmic retention in response to TGF-β stimulation. While it was proposed that phoshorylation of R-Smads may increase its affinity for importin β1, it has not been shown if importin β1 can import the R-Smad/Smad4 complex [51, 52]. The pressing issue is if the R-Smads/Smad4 complex uses different machinery for import compared to unphosphorylated monomeric Smads which are much smaller in size. On the other hand, the mobility of R-Smads in the nucleus seemed to be much decreased upon TGF-β stimulation [51]. This led to the idea that perhaps similar to Smad4, TGF-β keeps R-Smads in the nucleus mostly by inhibiting their nuclear export. Indeed phosphorylation of R-Smads was shown to cause some decrease in binding to exportin 4 [68]. But depletion of exportin 4 by siRNA did not cause much change in R-Smads localization at basal state [68]. So unlike in the case of Smad4, blocking the export is not sufficient to target R-Smads to the nucleus. Apparently, additional elements must be considered to explain how R-Smads are targeted to the nucleus by TGF-β.
3. POST-TRANSLATIONAL MODIFICATION OF SMADS
Like many other signaling molecules, Smads are subject to phosphorylation, ubiquitination, SUMOylation and acetylation (Table 1). These covalent modifications are important in regulating the activity of Smads in TGF-β signaling (Table 1). Some of these modifications are induced by non-TGF-β signals, thus providing ways for other signals to perturb TGF-β signal transduction.
Table 1.
Summary of post-translational modifications of Smads, the effectors and functional consequences.
| Type of Posttranslational Modification | Targeted Smads | Responsible Enzymes | Effects |
|---|---|---|---|
| Carboxy-terminal phosphorylation | R-Smads | Type I TGF-β receptor kinases | Nuclear accumulation, association with Smad4 and various transcription factors etc. |
| N-terminal or linker region phosphorylation | R-Smads | MAPK, CDK, CamK II,GRK2 | Inhibition of nuclear accumulation or transcriptional activity. In the case of GRK2, also failure in C-terminal phosphorylation. |
| Smad4 | MAPK | Enhanced nuclear accumulation in response to TGF-β | |
| Ubiquitination | R-Smads | Smurf 1, 2 | Degradation of R-Smads. |
| Smad2 | Itch | Enhanced Smad2 phosphorylation in response to TGF-β. | |
| Smad4 | Ectodermin/TIF1γ | Smad4 degradation and exclusion from the nucleus | |
| SUMOylation | Smad4 | Ubc9, PIASy | Enhanced stability, nuclear accumulation and transcriptional activity. But also evidence suggesting much suppressed transcriptional activity. |
| Acetylation | Smad2, 3 | CBP and p300 | Increased transcriptional activity. |
| Smad7 | Mostly p300 | Increased stability. |
3.1. Phosphorylation
3.1.1. TGF-β-induced phosphorylation at the carboxy-termini of R-Smads
Phosphorylation of the carboxy-terminal SXS motif is the on-switch that enables R-Smads to interact with Smad4, accumulate in the nucleus, and bind to DNA and transcription cofactors to regulate gene transcription. A long-standing question regarding the control of carboxy-terminal phosphorylation is the phosphatases that act to dephosphorylate and hence inactivate the R-Smads. Conceivably such protein phosphatases can either function in the cytoplasm to limit the amount of phosphorylated R-Smads that can enter the nucleus, or act in the nucleus to terminate the transcriptional regulation by R-Smads. In addition, dephosphorylation of R-Smads is closely linked to and required for nuclear export of R-Smads and Smad4 [37, 38]. Recently, an RNAi (RNA-interference) screening of Ser/Thr phosphatases in cultured Drosophila S2 cells identified PDP (Pyruvate Dehydrogenase Phosphatase) to be required for dephosphorylation of MAD (Smad1 ortholog in Drosophila) [69]. Interestingly, Drosophila strains harboring a defective PDP gene exhibit ectopic carboxy-terminal phosphorylation of MAD at embryonic stages when in wild type animals MAD is not phosphorylated [69]. This provides strong genetic evidence implicating PDP in dephosphorylation of MAD. Moreover, PDP is present in both nucleus and cytoplasm, and in the PDP mutant strain phospho-MAD could be detected in punctate pattern in cytoplasm [69]. These observations suggest the possibility that PDP’s function may be to reverse aberrant phosphorylation of MAD by signals other than DPP, which may take place at discreet loci in cytoplasm during embryonic development. Since PDP is present in mitochondria, it will also be interesting to examine if the punctate distribution of ectopically phosphorylated MAD corresponds to mitochondria. In mammals, two closely related PDPs, PDP1 and PDP2, are found in the genome, and have been characterized as mostly functioning in the mitochondria to regulate the catalytic activity of pyruvate dehydrogenase [70]. siRNA-mediated depletion of PDPs did cause reduced dephosphorylation of Smad1, which is downstream of BMP, but not dephosphorylation of Smad2/3 [69]. This agrees with the general notion that the BMP pathway in vertebrates, rather than the TGF-β pathway, is more related to the DPP pathway in Drosophila. But it is not yet resolved if indeed PDPs distinguish the two subfamilies of Smads, just like the receptor kinases can, or that the observation was just a reflection of more redundancy in phosphatases capable of dephosphorylating Smad2/3.
In the meantime, Lin et al. resorted to an overexpression approach to screen for mammalian phosphatases that target R-Smads, which uncovered PPM1A as a phosphatase for Smad2 and Smad3 [71]. siRNA-based knockdown confirmed the requirement of PPM1A in dephosphorylation of Smad2 and Smad3. PPM1A directly dephosphorylates Smad2 at the carboxy-terminal SXS motif, but not the phospho-Ser/Thr residues in the linker region [71]. Elevated level of PPM1A mitigated transcriptional response to TGF-β, and inhibited the anti-proliferation function of TGF-β. In zebrafish, overexpression of PPM1A perturbed nodal signaling during development, suggesting that the function of PPM1A is evolutionarily conserved [71]. PPM1A is exclusively present in the nucleus, and its activity facilitates nuclear export of Smad2/3 [71]. Therefore, PPM1A’s primary role may be to terminate the TGF-β responses in the nucleus.
Moreover, Knockaert et al. reported that the SCP (Small C-terminal Domain Phosphatases) family of phosphatases function to inhibit BMP signaling and induction of the secondary axis in Xenopus embryonic development [72]. Biochemical evidence indicated that SCPs dephosphorylate Smad1 directly, and depletion of SCPs by RNAi in mammalian cells led to enhanced and prolonged phosphorylation of Smad1 and increased transcriptional activation of target genes in response to BMP [72]. Interestingly, SCPs exhibited much stronger catalytic activity as well as higher interaction affinity towards Smad1 than Smad2/3. Unlike PDPs, SCPs are present entirely in the nucleus, suggesting that these two groups of phosphatases may dephosphorylate Smad1 at different locations and/or different phases of BMP signaling [72].
These studies marked a new direction in understanding how TGF-β signaling is downregulated, and also raised several questions such as whether these phosphatases may also act on the TGF-β receptor kinases. All three studies pointed out the possibility that multiple phosphatases recognize R-Smads as substrates. This may reflect multiple functional purposes expected of such phosphatases, i.e. either to limit the magnitude of signal sent into the nucleus or to terminate the signal once it reaches into the nucleus. It is also conceivable that different cell types, or at different developmental stages, different phosphatases are involved to turn off TGF-β signal. Furthermore, activities of Smad phosphatases may be subject to regulation by various other signaling pathways, thus different phosphatases may serve as conduits through which different signals interfere with the strength of TGF-β signals. In vivo studies of phosphatases against Smads are eagerly awaited to provide more insights on how such negative regulation would affect various physiological processes regulated by TGF-β.
3.1.2. Smad phosphorylation by other pathways
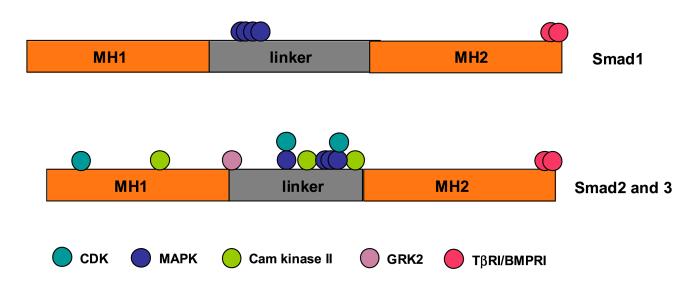
TGF-β-independent phosphorylation of serine or threonine residues was also observed in endogenous R-Smads. These phosphorylation events are triggered by various extracelluar signals, and the targeted sites are mostly in the linker or N-terminal region of Smads that are not always conserved in all R-Smads (Fig. 2). This may provide potential node through which different R-Smads can be differentially regulated.
Figure 2.
Relative positions of phosphorylated residues in R-Smads. The responsible kinases are indicated with color coding. Only phosphorylation sites verified by mutational analyses are highlighted. TβRI: type I TGF-β receptor; BMPRI: type I BMP receptor.
For many of the linker region phosphorylation events, the responsible kinases have been suggested. Initially, MAPKs (Mitogen Activated Protein Kinase) were shown to phosphorylate Smad1 upon EGF (Epidermal Growth Factor) stimulation [73]. Similar, but not identical sites in Smad2 and Smad3 are phosphorylated by MAPKs downstream of activated Ras (Fig. 2) [22]. MAPK is also responsible for phosphorylation of Smad4 on Thr276 [74, 75]. In addition, the linker regions in Smad2 and Smad3 are subject to phosphorylation by Cam Kinase II (Ca- and Calmodulin-dependent kinase II), on three of the atypical SXD Cam Kinase II sites [76]. More recently, the N-terminal and linker regions of Smad3 were shown to be phosphorylated by CDKs, which is regulated in a cell-cycle-dependent manner [23]. GRK-2 (G protein-coupled Receptor Kinase 2) phosphorylates Ser197 in Smad2, a residue in the linker that is not targeted by any of the kinases listed above [77].
The current consensus is that these phosphorylation events all act to repress the function of R-Smads in TGF-β signaling. In Xenopus embryonic development, BMP signaling suppresses neural differentiation induced by FGF (Fibroblast Growth Factor) and IGF (Insulin-like Growth Factor) [20]. FGF and IGF induces linker phosphorylation of Smad1 through MAPK, and elimination of such phosphorylation by mutating the MAPK sites rendered Smad1 much more potent than the wild type Smad1 in inhibiting neural differentiation [20]. Thus, phosphorylation of the Smad1 linker region provides the mechanism by which FGF and IGF attenuate BMP signaling, in order for neural differentiation to proceed. Also in Xenopus embryos, the TGF-β ligand activin can induce mesoderm markers in ectoderm, but the responsiveness is lost at particular embryonic stages. Such loss of competence could be rescued by introducing Smad2 with the MAPK sites mutated, establishing the correlation between Smad2 linker phosphorylation and inactivation of activin signaling [78]. Mice with knock-in point mutations abolishing MAPK phosphorylation sites in Smad1 linker displayed distinct phenotypes compared to mice with Smad1-null mutation or Smad1 with the carboxy-terminal mutation preventing BMP-induced phosphorylation [21]. At the cellular level, mutation of the MAPK sites resulted in distribution of Smad1 to the cytoplasmic membrane, and changes in cell-cell adhesion as well as actin cytoskeleton structures [21]. The observations in mice further validate that the linker region of Smad1 is a point of cross-talk between BMP and MAPKs, and inhibition of BMP signaling may not be the only functional outcome of linker phosphorylation of Smad1. Much less is known about the physiological consequence of Smad4 phosphorylation by MAPK, this phosphorylation event appears to be constitutive and may result in enhanced nuclear accumulation of Smad4 [74, 75].
The underlying mechanism through which N-terminal and linker region phosphorylation inhibits R-Smads functions remains an issue of much debate. MAPK- and Cam Kinase II-mediated phosphorylation did not appear to interfere with the carboxy-terminal phosphorylation of R-Smads by TGF-β or BMP receptor kinases, but was shown to inhibit nuclear accumulation of Smad1, 2, and 3 [73, 76]. The loss of activin responsiveness in Xenopus is largely due to exclusion of Smad2 from the nucleus, and indeed mutating the MAPK sites in the linker restored Smad2’s ability to accumulate in the nucleus [78]. The N-terminal and linker phosphorylation by CDKs resulted in reduced transcriptional activation potency of Smad3, whether this is due to decreased interaction with transcription cofactors remains to be tested, but apparently there is no defect in the carboxy-terminal phosphorylation of Smad3 [23]. In contrast, the GRK-2-mediated phosphorylation of Ser197 in Smad2 inhibited the carboxy-terminal phosphorylation in response to TGF-β, which explains how GRK-2 prevents nuclear accumulation of Smad2 [77].
Many questions remain unanswered. For example, what percentage of endogenous R-Smads and Smad4 are phosphorylated at the linker or N-terminal regions, how such phosphorylations may regulate biochemical properties of R-Smad and result in reduced nuclear accumulation of R-Smads? Structural analysis of Smads with the linker region phosphorylated will provide useful information. Furthermore, protein phosphatases targeting the linker and N-terminal phosphorylation sites are potentially important factors in regulating Smad functions. Identifying such phosphatases will unravel another layer of regulation imposed on Smads.
3.2. Ubiquitination
Ubiquitination of lysine residues was initially recognized as a prelude to proteosome-mediated protein degradation [79]. More recent studies have shown that dependent on the type of ubiquitination (i.e. poly-vs. mono ubiquitination) and the topology of ubiquitin linkage to the protein (i.e. involvement of either Lys48 or Lys63 of the ubiquitin), ubiquitin can serve other functions such as a sorting signal that direct the protein to particular subcellular locales [80, 81].
R-Smads phosphorylated by the TGF-β receptor kinase undergo ubiquitination and subsequent degradation [82]. Together with dephosphorylation, these events lead to termination of the TGF-β signaling. The steady state level of R-Smads is also subject to stability control by ubiquitination, which may limit the extent of TGF-β response. Smurf1 and Smurf2 are HECT-domain containing E3 ubiquitin ligases that recognize R-Smads as substrates through a PPXY motif found in R-Smads [83-85]. In addition to being direct substrates of Smurfs, Smads may serve as a targeting device and direct the associated Smurf to other substrates of ubiquitination. For instance, Smad2 binds to both the proto-oncogene SnoN and Smurf2, and by bringing the two together facilitates SnoN ubiquitination and degradation [86]. Another example is Smad7, which binds to Smurf2 and targets Smurf2 to the TGF-β receptor kinase for degradation [87-89].
Smad4 lacks a PPXY motif to recruit the Smurfs, however, ectodermin/TIF1-γ (also named TIM33, RFG7 and PTC7), a protein harboring a RING domain in addition to a number of other functional domains, was reported recently to suppress Smad4 function and impair Xenopus embryonic development by acting as an E3 ubiquitin ligase of Smad4 [90]. This hypothesis is somewhat controversial, as another recent study in mammals did not find ectodermin/TIF1-γ functions in Smad4 degradation, but rather that ectodermin/TIF1-γ competes with Smad4 as a transcription cofactor of R-Smads and regulates the development of the hematopoietic system [91].
Most of these conclusions were based on in vitro biochemical studies involving overexpressed proteins. More in vivo evidence is needed to verify the proposed model of Smurf function. To this end, dSmurf, the Drosophila Smurf, was shown to restrict DPP signaling during Drosophila embryonic development, supporting the idea that by targeting R-Smads for degradation dSmurf is a negative regulator of TGF-β signaling in vivo [92]. In contrast, knockout of the Smurf1 gene in mice did not manifest phenotypes that implicate Smurf1 function in R-Smads degradation [93]. Rather, Smurf1 appeared to be a critical regulator of the abundance of MEKK2 [93]. This could be simply due to functional redundancy between Smurf1 and Smurf2, and indeed compound Smurf1 and Smurf2 null animals have severe defects and suffer early embryonic lethality [93]. Whether such physiological abnormalities can be attributed to aberrant R-Smads levels and TGF-β signaling is still an issue under investigation.
On the other hand, mice carrying mutations in other E3 ubiquitin ligases have suggested the possibility that ubiquitination may facilitate TGF-β signaling. In embryonic fibroblasts isolated from mice with null deletion of Itch, a gene encoding an E3 ligase, the sensitivity to TGF-β stimulation was much diminished, as evidenced by decrease in R-Smad phosphorylation and nuclear translocation [94]. The most imminent issue is whether the pro-TGF-β function of Itch is direct, perhaps through ubiquitination of Smads or TGF-β receptor kinases. Although in vitro Itch can ubiquitinate Smad2 directly, whether this is the case in vivo and how such ubiquitination benefits Smad2 phosphorylation by TGF-β still needs to be verified [94].
At present, the field lacks critical information regarding which lysine residues in endogenous R-Smad are ubiquitinated by which E3 ligase, the topology of such ubiquitination, and whether the ubiquitination state changes in response to TGF-β and other signals. There are challenging technical difficulties, but primary cell lines with Smurf1 or Itch knock out are available now which will greatly facilitate investigations of these important issues.
3.3. SUMOylation
The small ubiquitin-related modifier (SUMO) proteins (i.e. SUMO-1, 2, 3) can also be covalently linked to lysine residues within the consensus sequence VKDE [95]. Similar to ubiquitination, SUMOylation is catalyzed by a three-tier enzymatic cascade involving E1, E2 and E3 enzymes [95]. The functional consequence of SUMOylation is just beginning to be realized, and so far examples include regulation of transport across the NPC and stabilization of target proteins [96].
Several labs have reported SUMOylation of Smad4, and these studies identified Lys 159, and to a lesser degree Lys 113, as the sites of SUMOylation [97-99]. These lysines lie in the linker region, the most divergent part in Smads. Therefore, it was not surprising that under the same assay conditions, none of the R-Smads were found to be SUMOylated. In these studies, the E2 enzyme Ubc9 and the E3 enzyme PIASy are suggested to be responsible for Smad4 SUMOylation [97-99].
Only a small pool of Smad4 is SUMOylated, and TGF-β may enhance Smad4 SUMOylation through the p38 MAPK [99]. Based largely on mutational analysis of Lys 159 and Lys113, it was concluded that when these residues were mutated to Arg, the stability of Smad4 was slightly improved [97-99]. It is not clear however, if this is due to SUMOylation blocking ubiquitination of the same Lys residues. SUMOylation of Smad4 was proposed to facilitate nuclear accumulation and transcriptional activation by Smad4 [97]. But this was challenged by mutational analyses which showed that mutating Lys 159 and Lys 113 of Smad4 significantly enhanced the transcriptional activation function of Smad4, suggesting that in spite of stabilizing the Smad4 protein, SUMOylation of Smad4 suppresses its function as a transcription factor in TGF-β signaling [98, 100].Interestingly, Smad4 was recruited to punctate structures in the nucleus by the E3 ligase PISAy [98]. Whether and how such sub-nuclear distribution of SUMOylated Smad4 affect Smad4 activity in transcriptional regulation still awaits further investigations.
Most of the conclusions on the effects of SUMOylation on Smad4 activity are based on mutational analyses, and it is hard to determine if the observed functional consequences after mutations are due to the loss of SUMOylation or structural changes introduced by the Lys to Arg switch. However, it is conceivable that SUMOylation of Smad4 may bestow unique properties to Smad4, such as interaction with proteins, which consequently affects the efficiency of Smad4 to regulate TGF-β target gene transcription. It will also be interesting to determine how the p38 MAPK stimulates SUMOylation of Smad4, which is yet another unique cross-talk between TGF-β and other signaling pathways.
3.4. Acetylation
Acetylation is another form of covalent modification that can take place on Lys. The acetylation state of various lysine residues in histones determines chromatin configuration and the accessibility of gene promoters by transcription factors. Thus, histone acetyltransferases and deacetylases are important regulators of gene transcription [101]. Acetylation was also found in a number of transcription factors, which regulates their biochemical properties, including DNA binding, protein-protein interaction and stability [102].
CBP (CREB Binding Protein) and p300 are acetyltransferases that serve as transcriptional coactivators of R-Smads and Smad4 [103, 104]. Indeed, Smad2 and Smad3 were recently found to be acetylated directly by these two acetyltransferases [105]. The site of acetylation was mapped to Lys 378 in Smad3, and mutation of this residue to Arg led to decreased transcriptional activation by Smad3, consistent with the idea that this acetylation event is important for the transcriptional regulatory functions of Smad3.
Smad7 was also found to be subject to acetylation, which is believed to be mediated mostly by p300 but not CBP [106]. Lys 64, and to a less degree, Lys 70 in the MH1 domain of Smad7 were identified as the sites of acetylation [106]. It was shown that overexpression of p300 stabilized Smad7 protein, and such effect depends on the acetyltransferease function of p300. In addition, the carboxy-terminal region of Smad7 binds to the histone deacetylase HDAC1, which can reverse the acetylation of Smad7 by p300 [107]. As expected, acetylation of Lys4 and Lys70 prevented ubiquitination of Smad7, which probably accounted for the observed increase in Smad7 stability [106]. Therefore, acetylation of Smad7 by p300, which is a nuclear protein, may be important in maintaining the stability of Smad7 in the nucleus. Interestingly, Smurf1 has been suggested to ubiquitinate Smad7 and facilitate nuclear export of Smad7 [87]. This raises the question if acetylation also serves as a nuclear retention signal of Smad7, and prevents Smad7 from binding to the TGF-β receptor. Again, it is far from clear what is the percentage of endogenous Smad7 that undergoes acetylation and if this is required for the physiological functions of Smad7.
4. CONCLUDING REMARKS
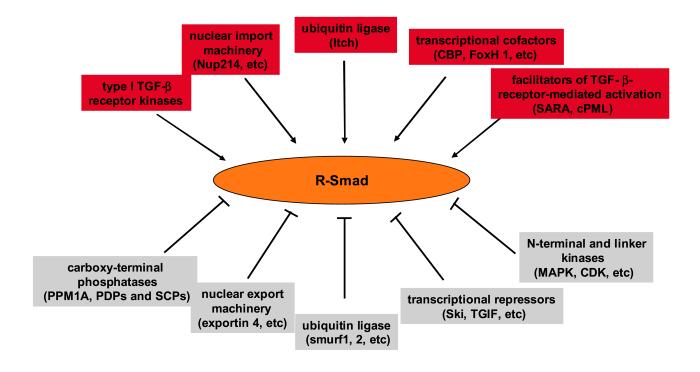
With the discovery of Smads, a rather simple and linear mechanism of TGF-β signal transduction was proposed nearly ten years ago that centers around Smads. Since then, the understanding of Smads has become much more comprehensive (Fig.3). In addition to TGF-β, a wide variety of factors impinge on Smads to either enhance or antagonize TGF-β impacts on cell proliferation, differentiation, angiogenesis, extracellular matrix remodeling, etc.
Figure 3.
Positive and negative regulation of R-Smad functions. Factors that facilitate or inhibit R-Smads functions are categorized and shown in red and grey boxes, respectively.
In TGF-β signaling, selective activation and repression of target gene transcription is critically dependent on three inter-related aspects of Smad regulation: nuclear accumulation, post-translational modification, and interaction with transcriptional cofactors (Fig. 3). Many basic mechanistic questions still remain to be explored on these three issues, such as whether TGF-β and/or other cross-talking signals directly impact on the nuclear transport machinery to modulate nucleocytoplasmic shuttling of Smads; the identity of phosphatases targeting the activated type I TGF-β receptor kinases and the various linker phosphorylation sites in Smads, and how they themselves maybe regulated. Additional challenges lie further ahead in defining the physiological relevance of some of the recent findings. For instance, knock-in mutations of phosphorylation, ubiquitination, SUMOylation or acetylation sites would provide much needed in vivo verification if these post-translational modifications of Smads are important in TGF-β-regulated processes such as embryonic development, wound healing, angiogenesis etc. Such mutants can also be studied in the background of mouse models of various cancers in order to determine the involvement of Smad modification, either by TGF-β or non-TGF-β signals, in cancer development.
All these information will feed into the ever expanding network of TGF-β signaling, which is vital in understanding the complex roles of TGF-β in the pathophysiology of diseases such as cancer. Such knowledge will ultimately help design therapeutic strategies that target TGF-β signaling.
Acknowledgment
The author thanks Dr. Q. Xu for comments on the manuscript, and past and present colleagues for discussions. Work from the author’s laboratory is supported by grants from the National Cancer Institute and the Worcester Foundation for Biomedical Research.
REFERENCES
- [1].Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- [2].Whitman M. Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- [3].Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- [4].Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- [5].ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- [6].Massagué J. Receptors for the TGF-β family. Cell. 1992;69:1067–1070. doi: 10.1016/0092-8674(92)90627-o. [DOI] [PubMed] [Google Scholar]
- [7].ten Dijke P, Ichijo H, Franzén P, Schulz P, Saras J, Toyoshima H, Heldin C-H, Miyazono K. Activin receptor-like kinases: a novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene. 1993;8:2879–2887. [PubMed] [Google Scholar]
- [8].Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- [9].Massagué J, Hata A, Liu F. TGFβ signaling through the Smad pathway. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- [10].Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- [11].Liu F, Hata A, Baker J, Doody J, Cárcamo J, Harland R, Massagué J. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- [12].Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- [13].Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFβ receptor and phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- [14].Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin CH. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- [15].Yingling JM, Das P, Savage C, Zhang M, Padgett RW, Wang XF. Mammalian dwarfins are phosphorylated in response to transforming growth factor beta and are implicated in control of cell growth. Proc Natl Acad Sci U S A. 1996;93:8940–8944. doi: 10.1073/pnas.93.17.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signaling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- [17].Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-β superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- [18].Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- [19].Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Aubin J, Davy A, Soriano P. In vivo convergence of BMP and MAPK signaling pathways: impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev. 2004;18:1482–1494. doi: 10.1101/gad.1202604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFβ/Smad signaling by ongenic ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- [24].Oft M, Akhurst RJ, Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol. 2002;4:487–494. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- [25].Leivonen SK, Chantry A, Hakkinen L, Han J, Kahari VM. Smad3 mediates transforming growth factor-beta-induced collagenase-3 (matrix metalloproteinase-13) expression in human gingival fibroblasts. Evidence for cross-talk between Smad3 and p38 signaling pathways. J Biol Chem. 2002;277:46338–46346. doi: 10.1074/jbc.M206535200. [DOI] [PubMed] [Google Scholar]
- [26].Wilkes MC, Mitchell H, Penheiter SG, Dore JJ, Suzuki K, Edens M, Sharma DK, Pagano RE, Leof EB. Transforming growth factor-beta activation of phosphatidylinositol 3-kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-activated kinase-2. Cancer Res. 2005;65:10431–10440. doi: 10.1158/0008-5472.CAN-05-1522. [DOI] [PubMed] [Google Scholar]
- [27].Javelaud D, Mauviel A. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-beta: implications for carcinogenesis. Oncogene. 2005;24:5742–5750. doi: 10.1038/sj.onc.1208928. [DOI] [PubMed] [Google Scholar]
- [28].Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- [29].Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- [30].Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- [31].Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- [32].Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional responses. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hoodless PA, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signalling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- [34].Wharton SJ, Basu SP, Ashe HL. Smad affinity can direct distinct readouts of the embryonic extracellular Dpp gradient in Drosophila. Curr Biol. 2004;14:1550–1558. doi: 10.1016/j.cub.2004.08.053. [DOI] [PubMed] [Google Scholar]
- [35].Nicolas FJ, Hill CS. Attenuation of the TGF-beta-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-beta-induced growth arrest. Oncogene. 2003;22:3698–3711. doi: 10.1038/sj.onc.1206420. [DOI] [PubMed] [Google Scholar]
- [36].Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- [37].Xu L, Kang Y, Col S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- [38].Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- [39].Nicolas FJ, De Bosscher K, Schmierer B, Hill CS. Analysis of Smad nucleocytoplasmic shuttling in living cells. J Cell Sci. 2004;117:4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- [40].Schmierer B, Hill CS. Kinetic Analysis of Smad Nucleocytoplasmic Shuttling Reveals a Mechanism for Transforming Growth Factor {beta}-Dependent Nuclear Accumulation of Smads. Mol Cell Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xu L, Massague J. Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol. 2004;5:209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- [42].Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- [43].Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- [44].Weis K. Regulating Access to the Genome. Nucleocytoplasmic Transport throughout the Cell Cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- [45].Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- [46].Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- [47].Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Adam SA, Sterne-Marr R, Gerace L. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 1992;219:97–110. doi: 10.1016/0076-6879(92)19013-v. [DOI] [PubMed] [Google Scholar]
- [49].Xu L, Chen YG, Massagué J. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFb-dependent phosphorylation. Nat Cell Biol. 2000;2:559–562. doi: 10.1038/35019649. [DOI] [PubMed] [Google Scholar]
- [50].Xu L, Alarcon C, Col S, Massagué J. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem. 2003;278:42569–42577. doi: 10.1074/jbc.M307601200. [DOI] [PubMed] [Google Scholar]
- [51].Kurisaki A, Kose S, Yoneda Y, Heldin CH, Moustakas A. Transforming growth factor-beta induces nuclear import of Smad3 in an importin-beta1 and Ran-dependent manner. Mol Biol Cell. 2001;12:1079–1091. doi: 10.1091/mbc.12.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of Smad 3. J Biol Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- [53].Xiao Z, Liu X, Henis YI, Lodish HF. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci U S A. 2000;97:7853–7858. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Meyer T, Begitt A, Lodige I, van Rossum M, Vinkemeier U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. Embo J. 2002;21:344–354. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Adachi M, Fukuda M, Nishida E. Two co-existing mechanisms for nuclear import of MAP kinase: passive diffusion of a monomer and active transport of a dimer. Embo J. 1999;18:5347–5358. doi: 10.1093/emboj/18.19.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pierreux CE, Nicolas FJ, Hill CS. Transforming growth factor beta-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol Cell Biol. 2000;20:9041–9054. doi: 10.1128/mcb.20.23.9041-9054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Watanabe M, Masuyama N, Fukuda M, Nishida E. Regulation of intracellular dynamics of Smad4 by its leucine-rich nuclear export signal. EMBO Rep. 2000;1:176–182. doi: 10.1093/embo-reports/kvd029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xiao Z, Latek R, Lodish HF. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene. 2003;22:1057–1069. doi: 10.1038/sj.onc.1206212. [DOI] [PubMed] [Google Scholar]
- [59].Dai JL, Turnacioglu KK, Schutte M, Sugar AY, Kern SE. Dpc4 transcriptional activation and dysfunction in cancer cells. Cancer Res. 1998;58:4592–4597. [PubMed] [Google Scholar]
- [60].Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- [61].Krakowski AR, Laboureau J, Mauviel A, Bissell MJ, Luo K. Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc Natl Acad Sci U S A. 2005;102:12437–12442. doi: 10.1073/pnas.0504107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Conery AR, Cao Y, Thompson EA, Townsend CM, Jr., Ko TC, Luo K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat Cell Biol. 2004;6:366–372. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- [63].Hoodless PA, Tsukazaki T, Nishimatsu S, Attisano L, Wrana JL, Thomsen GH. Dominant-negative Smad2 mutants inhibit activin/Vg1 signaling and disrupt axis formation in Xenopus. Dev Biol. 1999;207:364–379. doi: 10.1006/dbio.1998.9168. [DOI] [PubMed] [Google Scholar]
- [64].Kang Y, Chen CR, Massagué J. A Self-Enabling TGFbeta Response Coupled to Stress Signaling. Smad Engages Stress Response Factor ATF3 for Id1 Repression in Epithelial Cells. Mol Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- [65].Zhao BM, Hoffmann FM. Inhibition of Transforming Growth Factor-{beta}1-induced Signaling and Epithelial to Mesenchymal Transition by the Smad-binding Peptide Aptamer Trx-SARA. Mol Biol Cell. 2006 doi: 10.1091/mbc.E05-10-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chen HB, Rud JG, Lin K, Xu L. Nuclear targeting of transforming growth factor-beta-activated Smad complexes. J Biol Chem. 2005;280:21329–21336. doi: 10.1074/jbc.M500362200. [DOI] [PubMed] [Google Scholar]
- [67].Das P, Maduzia L, Wang H, Finelli A, Cho SH, Smith M, Padgett R. The Drosophila gene medea demontrates the requirement for differentent classes of smads in dpp signaling. Development. 1998;125:1519–1528. doi: 10.1242/dev.125.8.1519. [DOI] [PubMed] [Google Scholar]
- [68].Kurisaki A, Kurisaki K, Kowanetz M, Sugino H, Yoneda Y, Heldin CH, Moustakas A. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol. 2006;26:1318–1332. doi: 10.1128/MCB.26.4.1318-1332.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chen HB, Shen J, Ip YT, Xu L. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes Dev. 2006;20:648–653. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Huang B, Gudi R, Wu P, Harris RA, Hamilton J, Popov KM. Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J Biol Chem. 1998;273:17680–17688. doi: 10.1074/jbc.273.28.17680. [DOI] [PubMed] [Google Scholar]
- [71].Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Knockaert M, Sapkota G, Alarcon C, Massague J, Brivanlou AH. Unique players in the BMP pathway: Small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc Natl Acad Sci U S A. 2006;103:11940–11945. doi: 10.1073/pnas.0605133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGFβ family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- [74].Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Roelen BA, Cohen OS, Raychowdhury MK, Chadee DN, Zhang Y, Kyriakis JM, Alessandrini AA, Lin HY. Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-beta-induced nuclear accumulation. Am J Physiol Cell Physiol. 2003;285:C823–830. doi: 10.1152/ajpcell.00053.2003. [DOI] [PubMed] [Google Scholar]
- [76].Wicks SJ, Lui S, Abdel-Wahab N, Mason RM, Chantry A. Inactivation of smad-transforming growth factor beta signaling by Ca(2+)-calmodulin-dependent protein kinase II. Mol Cell Biol. 2000;20:8103–8111. doi: 10.1128/mcb.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ho J, Cocolakis E, Dumas VM, Posner BI, Laporte SA, Lebrun JJ. The G protein-coupled receptor kinase-2 is a TGFbeta-inducible antagonist of TGFbeta signal transduction. Embo J. 2005 doi: 10.1038/sj.emboj.7600794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Grimm OH, Gurdon JB. Nuclear exclusion of Smad2 is a mechanism leading to loss of competence. Nat Cell Biol. 2002;4:519–522. doi: 10.1038/ncb812. [DOI] [PubMed] [Google Scholar]
- [79].Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- [80].Kerscher O, Felberbaum R, Hochstrasser M. Modification of Proteins by Ubiquitin and Ubiquitin-Like Proteins. Annu Rev Cell Dev Biol. 2006 doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- [81].Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- [82].Lo RS, Massagué J. Ubiquitin-dependent degradation of TGF-beta-activated Smad2. Nat Cell Biolog. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- [83].Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- [84].Feng X-H, Filvaroff EH, Derynck R. Transforming growth factor-β (TGF-β)-induced down-regulation of cyclin A expression requires a functional TGF-β receptor complex. J. Biol. Chem. 1995;270:24237–24245. doi: 10.1074/jbc.270.41.24237. [DOI] [PubMed] [Google Scholar]
- [85].Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- [87].Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- [88].Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- [89].Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- [90].Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- [91].He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- [92].Podos SD, Hanson KK, Wang YC, Ferguson EL. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev Cell. 2001;1:567–578. doi: 10.1016/s1534-5807(01)00057-0. [DOI] [PubMed] [Google Scholar]
- [93].Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Bai Y, Yang C, Hu K, Elly C, Liu YC. Itch E3 ligase-mediated regulation of TGF-beta signaling by modulating smad2 phosphorylation. Mol Cell. 2004;15:825–831. doi: 10.1016/j.molcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- [95].Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- [96].Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- [97].Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J Biol Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- [98].Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem. 2003;278:27853–27863. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- [99].Ohshima T, Shimotohno K. Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4. J Biol Chem. 2003;278:50833–50842. doi: 10.1074/jbc.M307533200. [DOI] [PubMed] [Google Scholar]
- [100].Long J, Wang G, He D, Liu F. Repression of Smad4 transcriptional activity by SUMO modification. Biochem J. 2004;379:23–29. doi: 10.1042/BJ20031867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- [102].Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- [103].Pouponnot C, Jayaraman L, Massagué J. Physical and functional interactions of Smads and p300/CBP. J. Biol. Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- [104].Feng X-H, Zhang Y, Wu R-Y, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Inoue Y, Itoh Y, Abe K, Okamoto T, Daitoku H, Fukamizu A, Onozaki K, Hayashi H. Smad3 is acetylated by p300/CBP to regulate its transactivation activity. Oncogene. 2006 doi: 10.1038/sj.onc.1209826. [DOI] [PubMed] [Google Scholar]
- [106].Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- [107].Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 2005;280:21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]