Summary
PAS domains sense oxygen, redox potential and light, and are implicated in behavior, circadian rhythmicity, development and metabolic regulation. Although PAS domains are widespread in archaea, bacteria and eukaryota, the mechanism of signal transduction has been elucidated only for the bacterial photo sensor PYP and oxygen sensor FixL. We investigated the signaling mechanism in the PAS domain of Aer, the redox potential sensor and aerotaxis transducer in Escherichia coli. Forty-two residues in Aer were substituted using cysteine-replacement mutagenesis. Eight mutations resulted in a null phenotype for aerotaxis, the behavioral response to oxygen. Four of them also led to the loss of the non-covalently bound FAD cofactor. Three mutant Aer proteins, N34C, F66C and N85C, transmitted a constant signal-on bias. One mutation, Y111C, inverted signaling by the transducer so that positive stimuli produced negative signals and vice versa. Residues critical for signaling were mapped onto a three-dimensional model of the Aer PAS domain, and an FAD-binding site and “active site” for signal transduction are proposed.
Keywords: aerotaxis, bacterial chemotaxis, electron transport system, oxygen sensing
Introduction
PAS domains comprise a sensing module superfamily that spans the three divisions of cellular life: archaea, bacteria and eukaryota (Zhulin et al., 1997b; Ponting and Aravind, 1997; Taylor and Zhulin, 1999). Stimuli recognized by PAS domains include, but are not limited to, light (Borgstahl et al., 1995; Yeh and Lagarias, 1998; Jiang et al., 1999), oxygen (Gong et al., 1998; Pellequer et al., 1999), and redox potential (Rebbapragada et al., 1997; Soderback et al., 1998). Biological processes in which PAS domains have been implicated include global regulation of metabolism (Taylor and Zhulin, 1999), behavior (Rebbapragada et al., 1997) and development (Fabret et al., 1999) in prokaryotes, and circadian clocks (Dunlap, 1999), hypoxia responses (Semenza, 1999), ion channel function (Morais Cabral et al., 1998) and development (Sonnenfield, 1997) in eukaryotes (for a review, see Taylor and Zhulin, 1999).
The specificity in sensing arises from cofactors that are associated with PAS domains: p-hydroxycinnamic acid in the bacterial photoactive yellow protein, PYP (Borgstahl et al., 1995); a heme in the bacterial oxygen sensor FixL (Gong et al., 1998); and flavin adenine dinucleotide (FAD) in a bacterial redox potential sensor NifL (Soderback et al., 1998). A plant photoreceptor, NPH1 that also senses redox potential has a flavin mononucleotide (FMN) cofactor in its PAS domain (Christie et al., 1999). Protein sequence analysis (Zhulin et al., 1997b; Ponting and Aravind, 1997) and molecular modeling (Pellequer et al., 1998) suggested a common fold for putative PAS domains from a wide variety of prokaryote and eukaryote sensor proteins. Recent studies revealed a striking similarity in the three-dimensional structure of PAS domains in proteins that are functionally and taxonomically distant (Pellequer et al., 1999). PAS domains from PYP (Borgstahl et al., 1995), FixL (Gong et al., 1998) and the human HERG potassium channel (Morais Cabral et al., 1998) have a distinctive αβ fold with a five-stranded antiparallel β-barrel core. It is expected that the more than 300 known PAS domains will have a similar three-dimensional structure. At this time, relatively little is known about the mechanisms of signal transduction by PAS domains other than PYP, HERG and FixL. The common structure of PAS domains suggests that there might be similar strategies for signaling, but, different cofactors associated with the fold may introduce significant variation in the signaling pathways.
In Escherichia coli, the PAS-domain-containing Aer protein regulates the motile behavior of bacteria in gradients of oxygen (aerotaxis), redox potential and certain nutrients (Bibikov et al., 1997; Rebbapragada et al., 1997; Taylor and Zhulin, 1998). The PAS domain comprises the N-terminal sensing module, and indirect evidence suggests the presence of an FAD cofactor in the protein (Bibikov et al., 1997). Oxidation and reduction of FAD within the PAS domain is postulated to produce a signal that is transmitted to the C-terminal signaling module (Taylor and Zhulin, 1998), which is homologous to chemoreceptor signaling domains (Le Moual and Koshland, 1996; Kim et al., 1999). In this study, we confirm the predicted sensory role of the PAS domain, demonstrate FAD association with the domain and its involvement in signal transduction, and identify individual amino acid residues that are critical for folding and signaling by a FAD-binding PAS domain.
Results
The Aer PAS domain is the sensory input module
The hypothesis that the PAS domain in the Aer aerotaxis transducer is the site of sensory specificity arose from a comparison of similar domains in other oxygen/redox-sensing proteins (Rebbapragada et al., 1997; Bibikov et al., 1997; Zhulin et al., 1997b). In this study evidence supporting the role of the PAS domain in sensing was provided by constructing a chimera (Aesr) that fused the N-terminus of Aer to the C-terminus of the serine chemoreceptor Tsr (Figure 1). The Tsr protein has a ligand-binding domain in the periplasm and a signaling domain in the cytoplasm. In Aer, the PAS and signaling domains are both in the cytoplasm, connected to the membrane by a 38–40 residue hydrophobic sequence. The topology of the Aesr chimera is predicted to be similar to Aer. Expressing the Aesr chimera from the pAVR4 plasmid in an aer tsr strain (BT3354) fully restored aerotaxis (Figure 2), but the cells were insensitive to serine (data not shown). This suggested that the specificity for oxygen/redox sensing in Aer is determined by residues 2 to 260, which include the PAS domain (residues 1 to 119). Bibikov et al. 2000 reached a similar conclusion with a slightly different Aesr construct. We used site directed mutagenesis to confirm that the PAS domain is essential for sensing by the Aer protein.
Fig. 1.
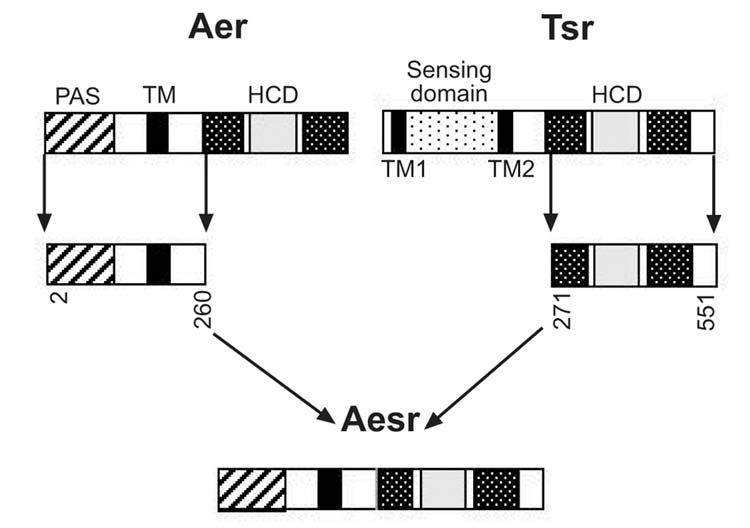
Construction of the Aesr chimera by fusion of residues 2–260 from Aer to residues 271–550 from Tsr (Experimental procedures). TM, transmembrane domain; K1, R1, receptor methylation domains; HCD, highly conserved domain for signaling.
Fig. 2.
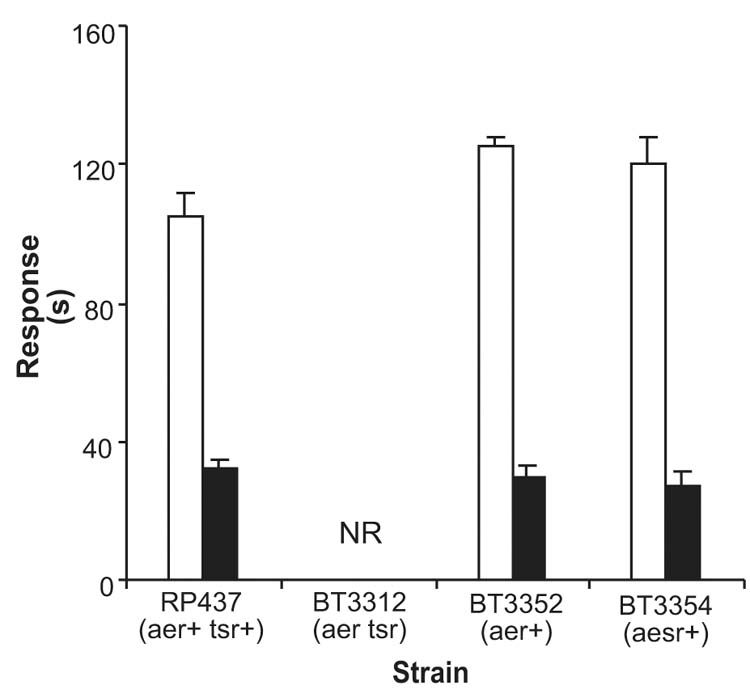
Aesr chimera restores aerotaxis in E.coli BT3354. BT3312 (aer tsr) cells were transformed with pAVR4 (aesr+) to create BT3354. The duration of responses to a 0 to 21% oxygen increase (
 ) and a 21% to 0 oxygen decrease (
) and a 21% to 0 oxygen decrease (
 ) were measured by the temporal oxygen gradient assay (Experimental procedures). BT3352 (BT3312 pAVR2); RP437 (wild type); NR, no response.
) were measured by the temporal oxygen gradient assay (Experimental procedures). BT3352 (BT3312 pAVR2); RP437 (wild type); NR, no response.
Indirect evidence that the Aer protein contains an FAD cofactor was reported, based on the increase in the total membrane FAD content when Aer was overexpressed (Bibikov et al., 1997). To confirm the presence of FAD in Aer, we isolated the His6x-tagged Aer protein as described in Experimental procedures. Purified Aer protein was denatured in urea and the released cofactor was analyzed by high performance liquid chromatography (HPLC). A peak with a retention time and spectrum identical to FAD was detected (data not shown). The highest FAD ratio observed was approximately 1 FAD molecule per Aer monomer, but the non-covalently bound FAD was labile and easily dissociated from the protein. No FMN, riboflavin or heme was detected in the purified protein.
Previous studies indicated that oxygen does not bind directly to Aer (Taylor and Zhulin, 1998). The aerotaxis signal originates in the electron transport system and a protein-protein interaction is postulated between the PAS domain of Aer and a component in the electron transport system. A signal is transmitted via Aer to the central chemotaxis pathway. Residues 2 to 166 of Aer comprise the PAS domain and the adjacent α-helix. Overexpressing Aer2-166 from pAVR1 in a tsr aer+ strain (BT3356) decreased the Aer-mediated smooth-swimming response to an oxygen increase (0 to 21%) from 102 ± 9 s to 57 ± 9 s. This interference indicates that Aer2-166 competes with the PAS domain from native Aer for the upstream signaling component, or a down-stream component, such as the C-terminal domain of Aer or an oligomeric Aer complex. It is likely that Aer2-166 binds to an upstream component since overproduction of Aer2-166 did not inhibit chemotaxis to cobalt or aspartate. These responses are processed by the Tar chemoreceptor, which has a C-terminal domain that is homologous to that of Aer (Le Moual and Koshland, 1996).
Selection of PAS residues for mutagenesis
An alignment of more than 300 sequences from the PAS domain superfamily identified conserved and variable residues in the PAS fold (Figure 3; Taylor and Zhulin, 1999). Molecular modeling (Pellequer et al., 1998) and the three dimensional coordinates from three PAS domains that have been crystallized (Borgstahl et al., 1995; Gong et al., 1998; Morais Cabral et al., 1998; Pellequer et al., 1999) revealed how the conserved residues stabilize the structure. On the other hand, PAS is a multifunctional superfamily, and sequence variability detected in comparative analysis is likely to contribute to the functional diversity (Taylor and Zhulin, 1999). Therefore, in selecting residues to mutate, we considered both conserved and variable residues that were likely to contribute to the PAS fold and the Aer functional specificity, respectively. The A–E subdomain (Figure 4) constitutes a PAS core that may contain a site for protein-protein interactions and signaling, the G–I subdomain constitutes the β-scaffold that maintains the structural integrity of the PAS domain, and the F helical connector forms a link between the PAS core and the β-scaffold (Pellequer et al.,1998). The greatest variability in length and sequence within PAS domains is in the EF and FG loops (see Figure 1 in Taylor and Zhulin, 1999; Gong et al., 1998; Pellequer et al., 1998; Morais Cabral et al., 1998). The attachment sites for known cofactors in PAS domains are in the EF loop, and/or F helix (Borgstahl et al., 1995; Gong et al., 1998). The variable regions in PAS domains may determine the unique ligand-binding properties of individual PAS domains (Pellequer et al., 1998).
Fig. 3.
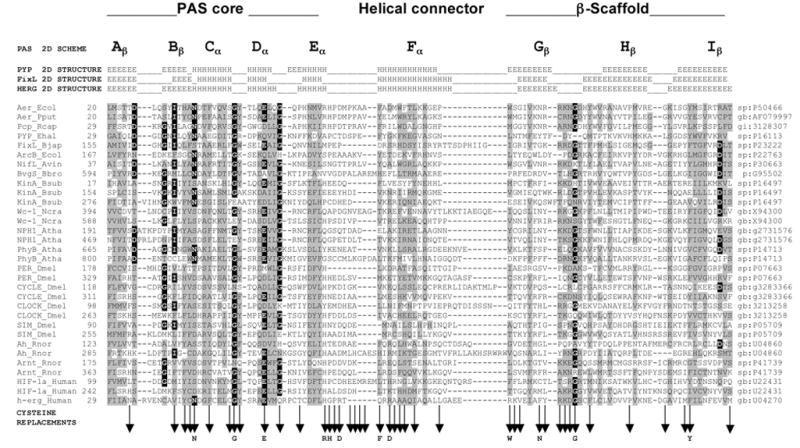
Multiple alignment of selected PAS domains, showing the secondary structures of PYP, FixL and HERG, and sub-domains of the crystallographic structure of PYP (adapted from Taylor and Zhulin, 1999). The variable N-terminal cap is not included in the alignment. Residues in reverse contrast are identical in >50% of the sequences of >300 PAS domains. Shaded residues are similar in >75% of the sequences. Vertical arrows beneath the alignment indicate cysteine replacements. A residue shown below a vertical arrow indicates that the cysteine replacement mutation at this site produced a defective aerotaxis phenotype. A complete alignment of PAS domain sequences is maintained at www.llu.edu/medicine/micro/PAS
Fig. 4.
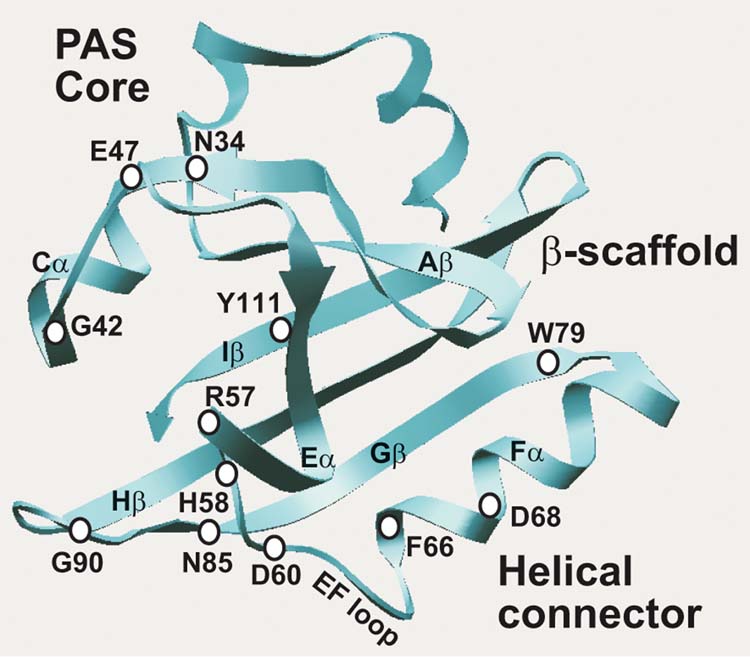
Scheme of the Aer PAS domain illustrating the major structural features and predicted position of critical residues for signal transduction.
Using cysteine-replacement mutagenesis we serially substituted 42 selected residues in the PAS domain of the His6x-Aer protein encoded in the pAVR2 plasmid (Figure 3). We chose to replace the residues with cysteine rather than alanine because solvent accessibility studies with the cysteine mutations can be used to elucidate protein structure. None of the three cysteines found in the native Aer protein are in, or adjacent to, the PAS domain. Two cysteines (Cys193 and Cys203) are in the putative membrane domain and one (Cys253) is in the C-terminal domain.
PAS domain residues important for signal transduction
The Aer and Tsr proteins are independent transducers for aerotaxis in E. coli (Rebbapragada et al., 1997). To evaluate the impact of a mutation on the Aer-mediated pathway we expressed the mutant protein in BT3312 (aer tsr) cells which have a null phenotype for aerotaxis. The pAVR2 plasmid expressing a His-tagged Aer under the control of the trc promoter and the pGH1 plasmid expressing untagged native Aer were similarly effective in complementing BT3312 (data not shown). We mutated the His6x-tagged aer gene to facilitate purification of the mutated proteins for follow-up studies. Expression of the mutated Aer proteins was confirmed by Western immunoblot analysis (data not shown).
Three biological screens were used to evaluate the phenotype of Aer mutants. Ring formation in succinate semi-soft agar is inhibited in cells that have a null phenotype for Aer (Bibikov et al., 1997). The mutated Aer proteins supported a range of responses on succinate plates (Figure 5A). Mutants that were defective, or partially defective, in migration on succinate plates were tested for aerotaxis in a capillary assay (Zhulin et al., 1996). Bacteria that are positive for aerotaxis form a band at the preferred oxygen concentration near the capillary opening. The intensity of the aerotaxis band was evaluated semi-quantitatively and scored as 0, +, ++ or +++ (Figure 5B). Mutants that scored less than +++ were evaluated quantitatively using a temporal oxygen gradient assay (Rebbapragada et al., 1997), in which the duration of the response to a decrease and increase in oxygen concentration was observed under a microscope. E. coli has a three-dimensional random walk motility pattern, alternating between smooth swimming and tumbling in isotropic medium. In response to a decrease (21% to 0) in oxygen concentration, bacteria that are wild-type for aerotaxis tumbled continuously for 20 to 30 seconds then resumed random swimming. In response to an oxygen increase (0 to 21%), wild-type cells swam smoothly for about 150 seconds. Of the 42 mutations in the Aer PAS domain that were made by cysteine replacement mutagenesis (Figure 3), only 12 mutations (N34C, G42C, E47C, R57C, H58C, D60C, F66C, D68C, W79C, N85C, G90C, Y111C) produced an aerotaxis phenotype that was different from the phenotype of cells that are wild-type for aerotaxis and chemotaxis (Figure 4). Of the 8 residues in the Aer EF loop, only 2 (H58, D60) were essential for aerotaxis; the P59C, P61C, K63C, A64C and A65C mutants retained aerotaxis. However, the length of the EF loop appeared to be critical. Aerotaxis was abolished when Ala65 was deleted, or an additional alanine was inserted after Ala65.
Fig. 5.
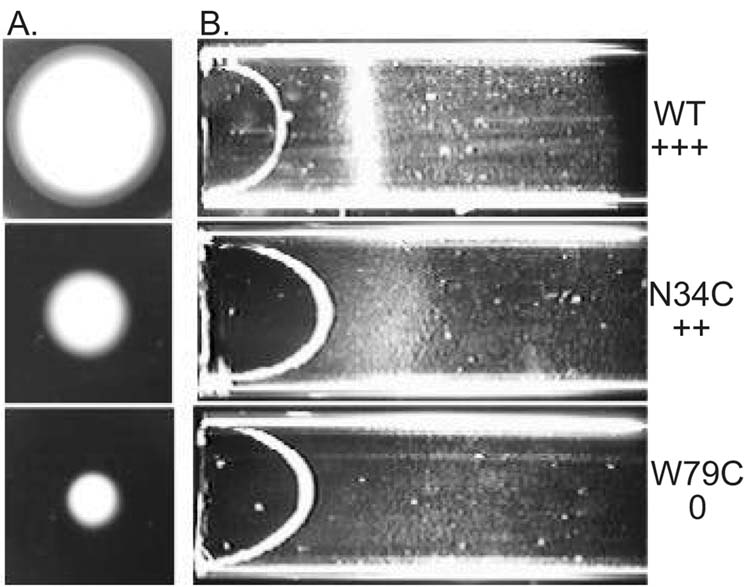
Comparison of aerotaxis responses in succinate semi-soft agar (A), and a capillary (B). A. Cells (5 μl) in mid-logarithmic phase were inoculated in the center of a succinate semi-soft agar plate (H1 medium, 30 mM succinate, 0.28% agar, 100 μg/ml ampicillin) and incubated at 30º C for 24 hours. Cells with a wild type aerotaxis phenotype formed a distinct convex ring (top) that is absent in mutants that have a null phenotype for aerotaxis (bottom). Other mutants were partially defective in aerotaxis (center). B. Cells (O.D.600 = 0.45) in Luria-Bertani medium were loaded into an optically flat capillary. Band formation is shown for each of the strains used in (A).
The responses of E. coli aer mutants that had diminished or no responses to oxygen in temporal gradients, are shown in Table I. Each of the Aer mutants listed in Table I had a normal tumbling frequency and a random-walk swimming pattern prior to a change in oxygen concentration. This indicated that the mutation disrupted the aerotaxis signaling pathway but did not cause a permanent signaling bias (signal on or signal off). A small residual response to an oxygen increase was observed in G42C and E47C strains, whereas other mutants had a null phenotype for aerotaxis. Residues Gly42, Glu47, and Gly90 are conserved in the PAS core and GH loop. Residues Asp68 and Trp79 are conserved in the F helix, and G strand, respectively. Residues Arg57, His58 and Asp60 are variable residues in and near the putative EF loop (Figure 4).
Table I.
Behavior of Aer mutants with decreased aerotactic responses and normal pre-stimulus signaling.a
| Aer | Response time (s) | |
|---|---|---|
| O2 ↑ | O2 ↓ | |
| wild type | 158 ± 16 | 24 ± 7 |
| G42C | 10 ± 3 | NR |
| E47C | 12 ± 3 | 9 ± 1 |
| R57C | NR | NR |
| H58C | NR | NR |
| D60C | NR | NR |
| D68C | NR | NR |
| W79C | NR | NR |
| G90C | NR | NR |
The duration of the responses to an oxygen increase and decrease were determined using the temporal oxygen gradient assay.
NR, no response.
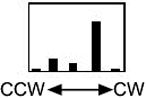
Three mutations (N34C, F66C, N85C) that diminished the aerotaxis responses also increased the tumbling frequency in unstimulated cells, indicating a constant signal-on bias. This was confirmed by measuring the rotation pattern of bacteria tethered to a glass slide by anti-flagella antibody (Table II). The flagellar motors rotate the bodies of tethered wild-type cells predominantly in the counterclockwise (CCW) direction, corresponding to linear swimming in free-swimming cells (Block et al., 1982). A histogram (Table II) of the rotation pattern was typical of wild type for BT3352 (BT3312 pAVR2) cells expressing the Aer protein with the native sequence (Ames and Parkinson, 1994), but bacteria expressing Aer with the N34C, F66C or N85C mutation had a CW bias. This implies that the mutated Aer is in a signal-on (CW) conformation, even in the absence of an external signal. The mutants are also partially defective in processing the aerotaxis signal. Asn34 and Phe66 are conserved residues in the Cα and Fα helices, respectively, and Asn85 is a variable residue in the Gβ strand (Figure 4).
Table II.
Behavior of Aer mutants with altered pre-stimulus swimming bias.a
| Response time (s) | Rotation
|
||
|---|---|---|---|
| Aer | O2↑ | O2↓ | patternb |
| wild type | 158 ± 16 | 24 ± 7 |

|
| N34C | 24 ± 5 | 12 ± 3 |

|
| F66C | 52 ± 11 | 11 ± 4 |

|
| N85C | 32 ± 8 | 10 ± 4 |
|
Response times were determined as described in Table I.
Rotation of unstimulated tethered cells was observed for 15s. The histogram represents the number of cells that rotated 100% CCW, >50% CCW, randomly, >50% CW, 100% CW.
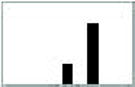
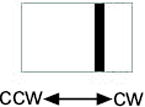
The AerY111C mutant had inverted aerotaxis responses in the temporal gradient assay (Table III). In response to an oxygen increase, cells expressing the native Aer sequence gave a positive (smooth-swimming) response whereas cells expressing AerY111C displayed a negative (tumbling) response. Conversely, in response to an oxygen decrease, bacteria expressing wild-type Aer gave a negative response and bacteria expressing AerY111C gave a positive response. In an isotropic environment the rotation pattern for the Y111C mutant was similar to the normal response shown in the first histogram in Table II. The histograms in Table III show the flagellar rotation patterns for AerY111C and the control immediately after an oxygen increase and an oxygen decrease. Compared to the prestimulus rotation pattern, an oxygen increase causes a CW bias in Y111C and a CCW bias in the control. Residue Tyr111 is centrally located in the I strand of the PAS domain β-scaffold (Figure 4) and appears to be a critical residue in signaling by the FAD-PAS domain (see below).
Table III.
Inversion of the aerotactic responses in AerY111C mutant.a
| Response time (s) | Rotation pattern | |||
|---|---|---|---|---|
| Aer | O2↑ | O2↓ | O2↑ | O2↓ |
| wild type | 158 ± 16 (smooth) | 24 ± 7 (tumbling) |

|
|
| Y111C | 25 ± 2 (tumbling) | 41 ± 4 (smooth) |
|
|
Responses and rotation patterns were measured as described in Table II, with the exception that rotation was observed immediately after an oxygen increase or decrease.
Other amino acid substitutions were made at Tyr111 of Aer to investigate the mechanism of signal inversion in the AerY111C mutant (Figure 6). Substituting Phe for Tyr111 resulted in little loss of activity, but 75% of the smooth swimming response was lost after Arg, His, Ala or Ser substitution. Ala and Ser, residues are closest in size to Cys, but only the AerY111C mutant had inverted responses.
Fig. 6.
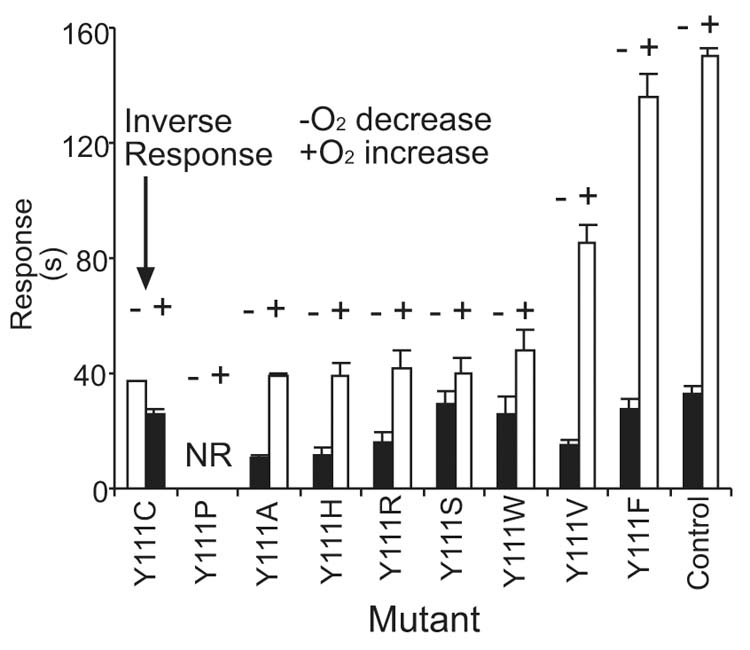
Behavioral responses of Aer mutants with substitutions for Tyr111. BT3312 cells were transformed with a plasmid that encoded a mutated aer gene and the duration of the aerotaxis responses were determined. Control, BT3352 (BT3312 pAVR2);
 , smooth swimming;
, smooth swimming;
 , tumbling; −, oxygen decrease; +, oxygen increase; NR, no response.
, tumbling; −, oxygen decrease; +, oxygen increase; NR, no response.
PAS domain residues involved in FAD binding
Several of the mutant Aer proteins were purified and the FAD levels were determined. However, the lability of FAD in the purified proteins led us to question whether FAD-binding was diminished as a result of the mutation or a loss of FAD during purification. To avoid this uncertainty, we measured the increase in total membrane FAD content when Aer was overproduced. The basal level of FAD was 0.15 nmoles/mg membrane protein (approximately 2,500 FAD molecules/cell) in strain BT3352. Concentrations reached 1.2 nmoles FAD/mg membrane protein when aer expression was induced by 0.6 mM IPTG. There was no significant change in FMN, riboflavin or non-covalently bound heme in membrane fractions. The FAD concentration in the cytosolic fraction did not increase significantly, indicating that FAD is not significantly leached from the membranes into the cytoplasm. The increase in FAD was approximately equimolar with the increase in Aer monomer in the membranes as estimated by Western blot analysis (data not shown). These findings indicated that FAD synthesis is closely regulated and confirmed the validity of the method of Bibikov et al., (1997). We used this method to estimate FAD bound to mutant Aer proteins. FAD measurements were normalized by reporting the FAD ratio in cells that were induced and uninduced for Aer.
We determined the membrane FAD content in aer tsr cells transformed with plasmids derived from pAVR2 that expressed one of 12 mutated Aer proteins (Figure 7). Only four mutants, Aer R57C, H58C, D60C and D68C, did not have a statistically significant increase in the FAD content. These mutants also had a null aerotaxis phenotype (Table I). All four mutations reside in, or near, the EF loop of the PAS domain. Either FAD binds to the mutated residues or the mutation induces a conformational change that prevents FAD binding.
Fig. 7.
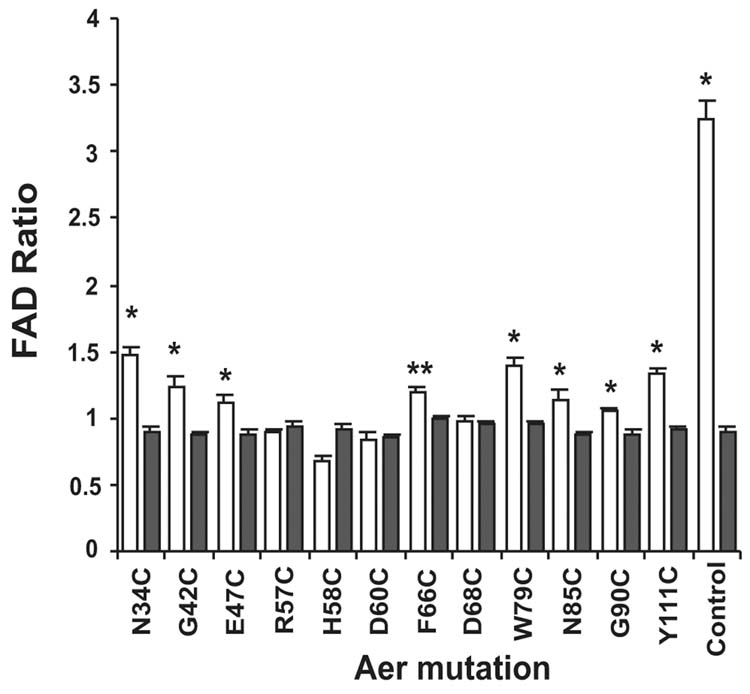
FAD binding by Aer mutants. Histograms represent the increase in membrane FAD when mutant Aer proteins are overexpressed. BT3312 (aer tsr) cells, transformed with a plasmid that encoded a mutant Aer protein, were grown and divided into two flasks. Aer overexpression was induced in one flask by 0.6 mM IPTG. The ratio of total membrane FAD in the induced and uninduced cells was determined (Experimental procedures).
 , Aer alleles;
, Aer alleles;
 , negative control (BT3312 pProEX). Positive control, BT3352 (aer+). Each transformant was analyzed in duplicate on three separate days. The results were analyzed by one-way analysis of variance that compared the relative increase in FAD in each mutant strain to the FAD ratio in the negative control. *, significant increase in FAD (p < 0.01); **, significant increase in FAD (p< 0.05).
, negative control (BT3312 pProEX). Positive control, BT3352 (aer+). Each transformant was analyzed in duplicate on three separate days. The results were analyzed by one-way analysis of variance that compared the relative increase in FAD in each mutant strain to the FAD ratio in the negative control. *, significant increase in FAD (p < 0.01); **, significant increase in FAD (p< 0.05).
The properties of the Arg57, His58 and Asp60 residues that are important in FAD binding (Figure 7) were investigated by additional site-directed mutagenesis. Replacement of Arg57 with Lys did not result in a loss of aerotaxis and, by inference, FAD binding (Table IV). This suggests that the positive charge on residue 57 may be important. Partial activity of Aer was retained when Asp60 was replaced by Glu, Gln and Asn. The carbonyl oxygen of Asp60 may be a critical atom. Aer was inactivated when His58 was replaced by Lys, which also has a positive charge, and was partially active when Tyr was substituted for His58, suggesting that the charge of His58 may not be important for FAD binding.
Table IV.
Effects on behavior of substitutions at Arg57, His58 and Asp60.a
| Aer | Response time (s) | |
|---|---|---|
| O2↑ | O2↓ | |
| wild type | 158 ± 16 | 24 ± 7 |
| R57C | NR | NR |
| R57K | 120 ± 5 | 30 ± 4 |
| H58C | NR | NR |
| H58K | NR | NR |
| H58Y | 88 ± 9 | 13 ± 2 |
| D60C | NR | NR |
| D60E | 125±4 | 28±5 |
| D60N | 117±6 | 32±4 |
Measurement of responses is described in Table I.
Several mutants with a normal aerotaxis phenotype were tested for FAD binding. However, none of the mutants increased the membrane FAD content to the levels observed for wild-type Aer (data not shown). One explanation for this finding is that mutants that signal normally at low expression levels could have subtle structural differences that become noticeable only after amplifying the construct. The differences might include impaired assembly into the membrane, altered association with components that stabilize FAD binding or increased susceptibility to proteolysis.
Discussion
The results of serial cysteine substitution at 42 residues in the Aer PAS domain demonstrate that cofactor binding and the “active site” for signaling are centered around the EF loop and PAS core region (Figure 4). This is similar to the PYP protein (Pellequer et al., 1998), and differs from the FixL PAS domain, in which cofactor interactions and signaling are centered around the FG loop and F helix (Gong et al., 1998).
Spatial relationship of the critical residues
Twelve residues (Asn34, Gly42, Glu47, Arg57, His58, Asp60, Phe66, Asp68, Trp79, Asn85, Gly90, Tyr111) that are involved in signal transduction in the Aer PAS domain were identified. To visualize the spatial relationship of the mutations, we modeled the three-dimensional structure of the Aer PAS domain on the crystal structure of PYP. Using the multiple alignment of 300 PAS domains (Taylor and Zhulin, 1999; www.llu.edu/medicine/micro/PAS), the coordinates of PYP (Protein Data Bank ID:2PYP) as a template, and the ProMod II program (Guex and Peitcsch, 1997) we generated a structural model for the Aer PAS domain (Figure 8). The modeled Aer PAS domain has a five-stranded antiparallel β-barrel structure that is similar to PAS domains in PYP, FixL and HERG (Pellequer et al., 1999). The overall fold resembles a left-handed glove in which the fingers enclose a pocket. Projecting into the pocket are most of the Aer residues that are involved in FAD binding and signal transduction (Figure 8). The fingers of the glove are formed primarily from the β-barrel, the palm from α-helical loops and the thumb from β-sheets, as described previously for FixL (Gong et al., 1999)
Fig. 8.
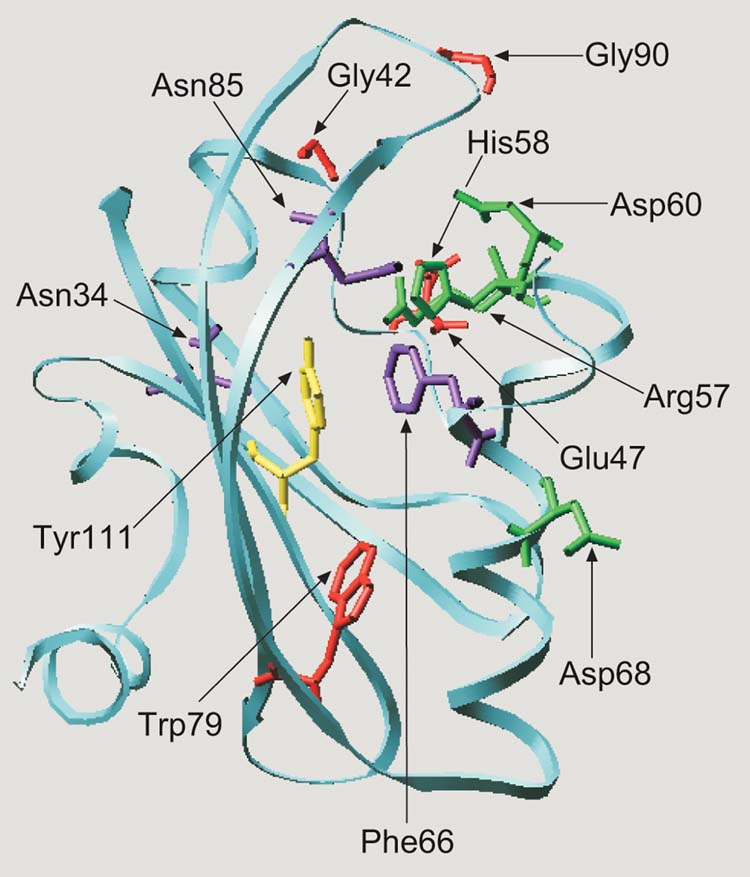
Hypothetical structural model for the E.coli Aer protein visualizing the predicted position of residues involved in aerotaxis. The Aer PAS domain (residues 1 to 119) was aligned with the PYP sequence (residues 1 to 125). The alignment was adjusted to minimize energy based on a sudo-Sippl field and submitted to the ProMod II program using the SWISS-PDB viewer. The figure was generated using the MOLSCRIPT program (Kraulis, 1991). Cysteine replacement of the residues shown as stick models produced a null aerotaxis phenotype (red), a loss of FAD binding and a null phenotype (green), inverted responses (yellow) and a CW signaling bias (purple).
Mutated residues in Aer that are highly conserved within the PAS superfamily (Figure 3 and Taylor and Zhulin, 1999) appear to be critical in forming the PAS fold. For example, in other PAS domains the Asn34 equivalent stabilizes the BC turn structure by forming hydrogen bonds to three backbone nitrogen atoms (Pellequer et al., 1998, 1999). Gly42 ends the C helix; Gly90 forms the turn in the GH loop.
Putative FAD binding site and “active site”
Four of the Aer PAS domain residues (Arg57, His58, Asp60 and Asp68) that are involved in FAD binding (Figures 7, 8) and signaling (Table I) are in or near the EF loop, which forms one boundary of the Aer major pocket. Residues involved in FAD binding (green), signaling (purple) and Tyr111 (yellow) are clustered. Bibikov et al. (2000) found that Arg104 (Hβ) is also a determinant for FAD binding. Cysteine replacement of Tyr111 resulted in inverted aerotaxis responses (Table III). We propose that FAD is bound in the pocket and that residues in contact with the isoalloxazine ring of FAD transduce redox changes into conformational changes in the PAS domain. The latter transmit the aerotaxis signal to the C-terminal domain of Aer. The interaction of Tyr111 with FAD may modulate the mid point potential of FAD (see below). The Phe66 and Asn85 residues (Figure 8) are presumably part of the signal transduction pathway because the F66C and N85C mutants were locked in CW signaling mode (Table II).
The FAD binding residues, Arg 57, His 58, and Asp60 are variable residues (Figure 3). However, histidine and aspartate residues in positions corresponding to His58 and Asp60 are conserved in a subset of 40 PAS proteins, including Aer homologs from Pseudomonas putida (N. N. Nichols and C. S. Harwood, personal communication) and other proteobacteria (I. B. Zhulin, unpublished observation), the WC-2 protein of Neurospora crassa, and PER, ARNT and CLOCK proteins from mammals (www.llu.edu/medicine/micro/PAS). The PAS domain of the Aer protein is a prototype for redox-sensing PAS domains, and possibly for this large and diverse subfamily of PAS domains, in which the . . . HXD. . . motif is present at the C-terminal end of the EF loop.
We were unsuccessful in demonstrating FAD binding to the Aer2-166 PAS domain fragment. An Aer R235C mutation in the linker region of the C-terminal domain also abolished FAD binding (Q. Ma and B.L.Taylor, unpublished observation). It is possible that association of the PAS domain with the C-terminal region of Aer or a partner protein is necessary to stabilize the PAS domain structure. This is supported by the finding by Bibikov et al. 2000 that both the linker region and the region between the PAS domain and the membrane domain are required for FAD binding to Aer.
Role of FAD in sensing
The signal for aerotaxis originates when oxygen binds to the terminal oxidase of the electron transport system (Taylor and Zhulin, 1998). We postulate that the Aer PAS domain interacts with one of the electron transport components. The reduction potential of the FAD cofactor in Aer is expected to be within a range that would permit electron exchange with ubiquinone/menaquinone via the electron transport system. Maximal rates of respiration (saturating electron donor and optimal electron acceptor) favor formation of the semi-quinone form of ubiquinone (menaquinone). If the FAD cofactor of Aer is in equilibrium with the quinones of the respiratory system, the semi-quinone form (FADH·) would be predominant at maximal respiration. These steady state conditions are associated with CCW rotation of the flagella and swimming by the bacteria, suggesting that FADH· excites a CCW signal in the protein. Aerobic E. coli that are starved for an electron donor have a fully oxidized electron transport system, and show a smooth-swimming response to the introduction of an electron donor (Zhulin et al., 1997a). In the absence of oxygen and alternative electron acceptors, cells have a fully reduced electron transport system and show a smooth-swimming response to the addition of oxygen (Table II). These results can be explained if the fully oxidized quinone (FAD) or fully reduced quinol (FADH2) form, respectively, send a CW signal in Aer.
The range of inverse responses that result from mutations in bacterial chemotaxis and aerotaxis has been reviewed recently and possible mechanisms proposed (Taylor and Johnson, 1997; Jung and Spudich, 1998). The inverted aerotactic response caused by the Y111C mutation can be explained by our model for aerotaxis, if it is assumed that the mutation shifted the redox potential of the FAD cofactor so that it is fully oxidized (the quinone state) during maximal rates of electron transport. A reductive shift in the electron transport chain that would normally shift FADH· to the quinol state and cause a CW signal, will now shift FAD quinone to the semi-quinone state and cause a CCW signal, i.e. an inverted response.
We propose that the signal propagation to and from the Aer PAS domain is by protein-protein interactions. The present studies suggest three possible PAS domain sites, for isologous or heterologous protein-protein interaction. One site surrounds Glu47, which corresponds to PYP residue Gln56 in a cluster of charged residues that form a putative protein-protein interaction site in PYP. Other sites are the hydrophobic side chains of residues Val56 and Met57, and residues Ile82 and Val83 at the surface of Aer. These putative hydrophobic interaction sites are different from the site of the hydrophobic patch in the HERG PAS domain that has been implicated in binding to the transmembrane region of the channel (Morais Cabral et al., 1998; Chen et al., 1999).
Experimental procedures
Strains and construction of plasmids
E. coli RP437 (aer+ tsr+, J.S. Parkinson) and its derivatives RP5882 (tsr, Callahan et al., 1987) and BT3312 (aer tsr) were grown at 30°C in Luria-Bertani medium containing thiamine (1 μM) and supplemented with ampicillin (100 μg/ml) when needed. The aer tsr double mutant BT3312 was generated by P1 transduction of thr+Δtsr-7021 from RP5882 into UU1117 (Bibikov et al., 1997) recipient cells. The wild type aer gene was amplified by PCR from the pGH1 plasmid (Rebbapragada et al., 1997) with the following primers: sense, 5’CCAGGCGCCTCTTCTCATCCGTATGTC 3’; antisense, 5’ACGCGTCGACTTAATGCAGTACCGTCACCGC 3’. The N-terminal primer lacks the methionine start codon and includes an EheI restriction site; the C-terminal primer includes a SalI site after a stop codon (bold). The pAVR2 plasmid, which expresses His6x-Aer under the control of the trc promoter and lacIq, was constructed by subcloning the 1.5 kb EheI-SalI PCR fragment containing the aer gene into the EheI and SalI sites of the pProEXHTa expression vector (GIBCO) which encodes a N-terminal His6x tag. The pAVR1 plasmid was obtained using PCR to introduce a stop codon (bold) and XbaI restriction site after Aer residue 166 in pAVR2 (Primers: sense 5’ CGCTGGCGGGCGCGTTAGTCTAGAGTGATGACCCTGATG 3’ antisense 5’ CATCAGGGTCATCACTCTAGACTAACGCGCCCGCCAGCG 3’), and excising a 1 kb XbaI-XbaI fragment corresponding to Aer residues 167–506. All constructs were confirmed by automated DNA sequencing.
The Aesr chimera (pAVR4) encodes a fusion of Aer residues 2–260 and Tsr residues 271–550 (Figure 1). Primers for a PCR fragment encoding Tsr residues 271–551 contained AatII (sense) and XbaI (antisense) restriction sites: sense 5’ CGTACCGTCGGTGACGTCCGCAACGGGGCC3’, antisense 5’CCCCTCTAGATTAAAATGTTTCCCAGTTCTCCTC3’. A silent mutation encoding an AatII restriction site at Aer codon 260 was incorporated into pAVR2. Primers: sense 5’CTAAATAACGACGTCTCAAGCCAG3’, antisense 5’CTGGCTTGAGACGTCGTTAATTAG3’. An AatII-XbaI fragment encoding Aer261-506 was excised from pAVR2 and the AatII-XbaI PCR fragment encoding Tsr271-551 ligated in its place.
Mutagenesis and complementation
Cysteine was serially substituted at 42 residues in Aer, which was encoded in the pAVR2 plasmid (Figure 3). The mutations were generated by Pfu polymerase PCR using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, California). Additional point mutations, insertions and deletions were also generated with this method. The primer sequences used are available at www.llu.edu/medicine/micro/faculty/primers.htm.
Complementation in trans by mutated Aer proteins was evaluated by transforming E. coli BT3312 (aer tsr), with a plasmid that expressed the mutated Aer protein. When pAVR2 ( aer+) was uninduced, the aerotaxis response in BT3352 (BT3312 pAVR2) cells to an 0 to 21% increase in oxygen concentration was longer (158 ± 16 s) than the response of RP5882 (aer+ tsr) cells (90 ± 1) indicating that the Aer levels in BT3352 were about 50% higher than in RP5882 (Rebbapragada et al., 1997). The phenotypes of mutated plasmid aer genes were evaluated in BT3312 cells without IPTG induction.
Purification of the Aer protein
Cultures (1 l) of E. coli BT3352 were grown to OD600 = 0.6. Aer expression was induced (0.6 mM IPTG) for 3 h. Cells were disrupted in a French press. Following centrifugation at 15,000 x g for 20 minutes, membranes were sedimented (485,000 x g, 20 min) and frozen at −80°C for future use. Membranes were resupended in buffer containing 1% Triton X-100 and the Aer protein was isolated by nickel NTA affinity chromatography (Qiagen, Chatsworth, CA) following the manufacturer’s protocol. Protein concentration was determined by the Bradford assay (Bio-Rad, Richmond, CA). Protein samples were stored at −20°C prior to analysis.
Anti-Aer antibody and Western blot analysis
Rabbit sera raised against the Aer2-166 fragment were prepared by Zymed Technologies (San Francisco, CA). Aer2-166 was isolated from a culture of E. coli ALS294 (rpoH, kindly supplied by R. Schwanner, University of Georgia) transformed with pAVR1 (Aer2-166). For Western blot analysis we followed the procedures of the reagent manufacturers using goat anti-rabbit IgG H+L horseradish peroxidase conjugated antibody (Bio-Rad) as a secondary antibody. Bands were visualized by Super Signal Chemiluminescent Substrate Detection reagents (Pierce Chemical, Rockford, IL).
Isolation of membranes and FAD analysis
His6x-Aer and its mutant derivatives were induced in E. coli aer tsr strains. Cultures (600 ml) were grown to mid-logarithmic phase and divided, IPTG (1 mM) was added to one flask and growth at 30ºC continued for 3 h. Harvested cells were resuspended in 40 mM Tris-HCl pH 8.0 containing complete protease inhibitor cocktail (EDTA-free, Boehringer Mannheim), and disrupted with five cycles of freeze thaw and sonification. Membranes were prepared as before, and membrane proteins resolved by SDS-PAGE and Western blot analysis.
The purified His6x-tagged Aer protein was diluted 1:1 in 10 M urea and centrifuged through a 10 kDa Centricon filter (Millipore). The filtrate was analyzed on a Jupiter C18 5μm reverse phase HPLC column (Phenomenex, Torrance, CA) using an ammonium acetate/methanol (pH 5.9) gradient. Peaks and spectra were monitored with a diode array spectrophotometer (Shimadzu). Peak purity was confirmed by overlaying the spectra of the upslope, apex and downslope. The retention times and peak areas of the samples were compared to a FAD standard.
Alternatively the increase in the total membrane FAD content was determined. Membranes were extracted five times with chloroform. The aqueous phase was filtered through a 0.2 μm nylon-66 microfilterfuge tube (Rainin, Woburn, MA) prior to HPLC analysis.
Behavioral Assays
Bacteria were screened for aerotaxis on semi-soft succinate swarm plates containing 100 μg/ml ampicillin (Bibikov et al., 1997). For the capillary assay, cells grown to OD600 = 0.4–0.45 were loaded into an optically flat capillary, and the aerotactic band formation was observed and video-recorded using a dark-field video microscope (Zhulin et al., 1996). To quantify the aerotactic response to gradients of oxygen and nitrogen, a temporal assay for aerotaxis was timed by inspection in a double blind study (Rebbapragada et al., 1997).
Tethered cell behavioral assays followed the procedure of Block et al., (1982) using anti-flagella IgG donated by J.S. Parkinson (University of Utah). The rotation of 50 individual cell bodies was observed for 15 second each and scored as published (Ames and Parkinson, 1994).
Acknowledgments
We thank John S. Parkinson for providing bacterial stains and anti-flagella antibody, Ryan Schwanner for donating the ALS294 strain, Brenton Reading, Marie Gilles-Gonzalez, and J. S. Parkinson for helpful discussions and Caroline Harwood, Qinhong Ma and Sarah Herrmann for communicating results before publication. We are also grateful to Jack Scarbrough and Galina Repik for excellent technical support. This work was supported by grants from the National Institute of General Medical Sciences (GM29481) and Loma Linda University to B.L.T.
References
- Ames P, Parkinson JS. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov SI, Barnes LA, Gitin Y, Parkinson JS. Domain organization and FAD-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. 2000 doi: 10.1073/pnas.100118697. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Segall JE, Berg HC. Impulse responses in bacterial chemotaxis. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- Borgstahl GE, Williams DR, Getzoff ED. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- Callahan AM, Frazier BL, Parkinson JS. Chemotaxis in Escherichia coli: construction and properties of lambda tsr transducing phage. J Bacteriol. 1987;169:1246–1253. doi: 10.1128/jb.169.3.1246-1253.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zou A, Splawski I, Keating MT, Sanguinetti MC. Long QT syndrome-associated mutations in the per-arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem. 1999;274:10113–10118. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- Jung KH, Spudich JL. Suppressor mutation analysis of the sensory rhodopsin-I transducer complex: insights into the color-sensing mechanism. J Bacteriol. 1998;180:2033–2042. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Kraulis P. A program to produce both detailed and schematic plots of proteins. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Le Moual H, Koshland DEJ. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED. Photoactive yellow protein: a structural prototype for the three- dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellequer JL, Brudler R, Getzoff ED. Biological sensors: More than one way to sense oxygen. Curr Biol. 1999;9:R416–R418. doi: 10.1016/s0960-9822(99)80257-7. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. PAS: a multifunctional domain family comes to light. Curr Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Perspectives on oxygen sensing. Cell. 1999;98:281–284. doi: 10.1016/s0092-8674(00)81957-1. [DOI] [PubMed] [Google Scholar]
- Soderback E, Reyes-Ramirez F, Eydmann T, Austin S, Hill S, Dixon R. The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Johnson MS. Rewiring a receptor: negative output from positive input. FEBS Lett. 1998;425:377–381. doi: 10.1016/s0014-5793(98)00253-1. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. In search of higher energy: metabolism-dependent behaviour in bacteria. Mol Microbiol. 1998;28:683–690. doi: 10.1046/j.1365-2958.1998.00835.x. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB, Johnson MS. Aerotaxis and other energy-sensing behavior in bacteria. Annu Rev Microbiol. 1999;53:103–128. doi: 10.1146/annurev.micro.53.1.103. [DOI] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulin IB, Bespalov VA, Johnson MS, Taylor BL. Oxygen taxis and proton motive force in Azospirillum brasilense. J Bacteriol. 1996;178:5199–5204. doi: 10.1128/jb.178.17.5199-5204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulin IB, Rowsell EH, Johnson MS, Taylor BL. Glycerol elicits energy taxis of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1997;179:3196–3201. doi: 10.1128/jb.179.10.3196-3201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulin IB, Taylor BL, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]


