Abstract
Murine (Hepa1c1c7) hepatoma cells are a suitable in vitro system for investigating the regulation of chemoprotective enzymes by selenazolidines, novel L-selenocysteine prodrugs developed as potential chemopreventive agents. They are less sensitive to the cytotoxic effects of both selenite and the less toxic selenazolidines than rat hepatoma (H4IIE) cells. All four selenazolidine 4-carboxylic acid (SCA) derivatives examined elevated thioredoxin reductase (Txnrd1), alpha-class glutathione transferases (Gsta), and UDP-glucuronosyltransferase (Ugt)1a6 mRNAs. NAD(P)H-quinone oxidoreductase (Nqo1) was induced by the three 2-alkyl derivatives (2-cyclohexylSCA, 2-butylSCA, and 2-methylSCA) but not SCA itself. Transcripts of mu- and pi-class glutathione transferases were induced only by 2-cyclohexylSCA and 2-butylSCA. Only Gsta and Txnrd1 transcripts were elevated by L-selenomethionine, L-selenocystine, or Se-methyl-L-selenocysteine. Txnrd1, Gsta, Nqo1, and Gstp responses to selenazolidines were all abolished by actinomycin D while Ugt1a6 responses were not. Induction responses to the selenazolidines were also eliminated (most) or reduced (Txnrd1 by 2-methylSCA) by cycloheximide, with the exception of Ugt1a6. The Ugt1a6 mRNA levels in the presence of SCAs and cycloheximide were similar to those with cycloheximide alone, and were almost double those of vehicle-treated cells. Thus, Hepa1c1c7 cells appear to provide a viable platform for the study of protective enzyme regulation by selenocompounds, and with the exception of Ugt1a6, the mRNA elevations from selenazolidines are transcriptionally dependent.
Keywords: Selenium, selenazolidines, selenocysteine prodrugs, thioredoxin reductase, NAD(P)H-quinone oxidoreductase, glutathione transferase, UDP-glucuronosyltransferase, cycloheximide, actinomycin D, Hepa1c1c7, H4IIE
INTRODUCTION
Among the desirable characteristics of cancer chemopreventive agents is an ability to increase the activities of chemoprotective enzymes to protect against chemical insults and/or an ability to decrease bioactivation activities. Changes in these parameters have been investigated in vivo for several of the synthetic organoselenium compounds developed with chemopreventive intent. Effects on inactivation and chemoprotective enzymes have received the greatest attention. Reported investigations include studies with phenylenebis(methylene)selenocyanate isomers (Reddy et al., 1992, Tanaka et al., 1997, Sohn et al., 1999, Prokopczyk et al., 2000), benzylselenocyanate, and its glutathione conjugate (Kawamori et al., 1998), and diphenylmethylselenocyanate (Ghosh et al., 2005, Das and Bhattacharya, 2005). Effects on bioactivation enzyme activities (cytochrome P450s) were investigated for phenylenebis(methylene)selenocyanate isomers (Sohn et al., 1999, Prokopczyk et al., 2000) following demonstration of their inhibitory activity in vitro (Shimada et al., 1997).
To establish and screen for enzyme altering properties, and the mechanism(s) by which they arise, we had need of an in vitro model that is responsive, and responsive at concentrations of the selenium-containing compounds where cell viability is not compromised. This cell viability consideration is of importance for chemopreventive agents where a pro-apoptotic effect on tumor cells is also another desirable property.
Reported in vitro studies of chemoprotective enzyme modification by seleno-compounds provide little in the way of precedence for the selection of a suitable cell model. Several available cell lines have been utilized but most studies have concerned themselves with only a single, or a very narrow range of enzymes and often only a single or very limited number of selenium compounds. The most extensively studied compound has been inorganic selenite which has been investigated for effects on two protective enzymes (glutathione peroxidase, thioredoxin reductase) in human HepG2 (Helmy et al., 2000, Morgan et al., 2002, et al., 2003a, Zhang et al., 2003) and in mouse Hepa1c1c7 (Hintze et al., 2003a) hepatoma cell lines, in a Chinese hamster lung fibroblast (V79) cell line (Short et al., 2003), and a mouse fibrosarcoma WEHI 164 cell line (Reszka et al., 2005). In addition to the hepatoma cells, selenite effects in human cell lines have been investigated in colon cancer (HT-29; Berggren et al., 1997), breast cancer (MCF-7) and lung cancer (A549) cells and Jurkat and HL-60 leukemia cells (Gallegos et al., 1997). Of the organoselenium-compounds, selenium-containing amino acids and some derivatives have been examined for thioredoxin reductase changes in HT-29 human colon cancer cells (Berggren et al., 1997), for glutathione peroxidase changes in V79 Chinese hamster lung fibroblasts (Short et al., 2003), and for glutathione transferase changes in rat H35 Reuber rat hepatoma cells (t'Hoen et al., 2002). 1,4 Phenylenebis(methylene)selenocyanate has been examined for glutathione transferase changes in mouse hepatoma (Hepa1c1c7) cells (Lee et al., 1999), human lung adenocarcinoma cells (El -Bayoumay et al., 2006) and alongside unsubstituted, 2-oxo- and 2-methyl-selenazolidine 4-carboxylic acids, for glutathione peroxidase changes in the V79 Chinese hamster lung fibroblasts (Short et al., 2003). Methylseleninic acid has been examined for its effects on enzyme expression in a human prostate cancer cell line (LNCaP) and increases in several Phase 2 enzymes, most notably NQO1 were identified (Zhao et al., 2004).
With the paucity of information available for precedence, this present study seeks to show that the easily propagated mouse Hepa1c1c7 hepatoma cell line offers a suitable in vitro platform for the study of the regulation of a wide range of chemoprotective enzymes by organic selenium-containing compounds. In providing evidence of increased levels of mRNAs of many chemoprotective enzymes by several members of a series of test compounds, and at concentrations far removed from their IC50 value, it also provides an opportunity to investigate the mechanism or mechanisms by which these elevations occur.
MATERIALS AND METHODS
Cell culture and treatments
Mouse Hepa1c1c7 cells (CRL-2026) and rat H4IIE cells (CRL-1548) were obtained from ATCC (Manassas, VA) and maintained in antibiotic/antimycotic-free Minimum Essential Medium Eagle (Sigma, St Louis, MO), supplemented with 10% fetal bovine serum (HyClone, Logan, UT) in 75-cm2 tissue culture flasks at 37°C under a 5% CO2 humidified environment. The H4IIE cells were cultured using the same medium but with 10% bovine calf serum in addition, according to the suppliers recommendation. When confluent, L-selenazolidines [2-methyl-, 2 butyl-, 2-cyclohexyl-, and 2-unsubstituted selenazolidine-4(S)-carboxylic acid (SCA)], synthesized as described elsewhere (Xie et al., 2001; El-Sayed et al., 2006), L-selenomethionine, L-selenocystine, or Se-methyl-L-selenocysteine (Acros Organics, Morris Plains, NJ) were added at a final concentration of 100 μ M in DMSO vehicle (0.1%, final concentration) and incubated for 18 (mRNA studies) or 24 (viability studies) hours. The structures of these compounds are shown in Figure 1. In the cytotoxicity/viability determinations, the cells were grown in 6-well plates and a Cell Counting Kit-8 (Dojindo Molecular Technologies Inc, Gaithersburg, MD), which utilizes the detection of the dehydrogenase-dependent reduction of a highly water soluble tetrazolium salt, WST-8 [(2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium] in the presence of an intracellular to extracellular electron mediator (1-methoxy-5-methylphenazinium methylsulfate) to a yellow formazan with an absorption maximum at 460 nm, as a monitor of living cells. In the mRNA studies, the cells were washed with buffer, homogenized in TRIzol (Invitrogen; Carlsbad, CA) and frozen at −80°C for later RNA isolation. In the mechanism studies, cells were treated with either actinomycin-D or cycloheximide (Sigma, St Louis, MO) at a final concentration of 50 nM in dimethyl sulfoxide (DMSO) (0.1%) for one hour prior to the addition of selenazolidines (100 μ M, 18 hours).
Figure 1.
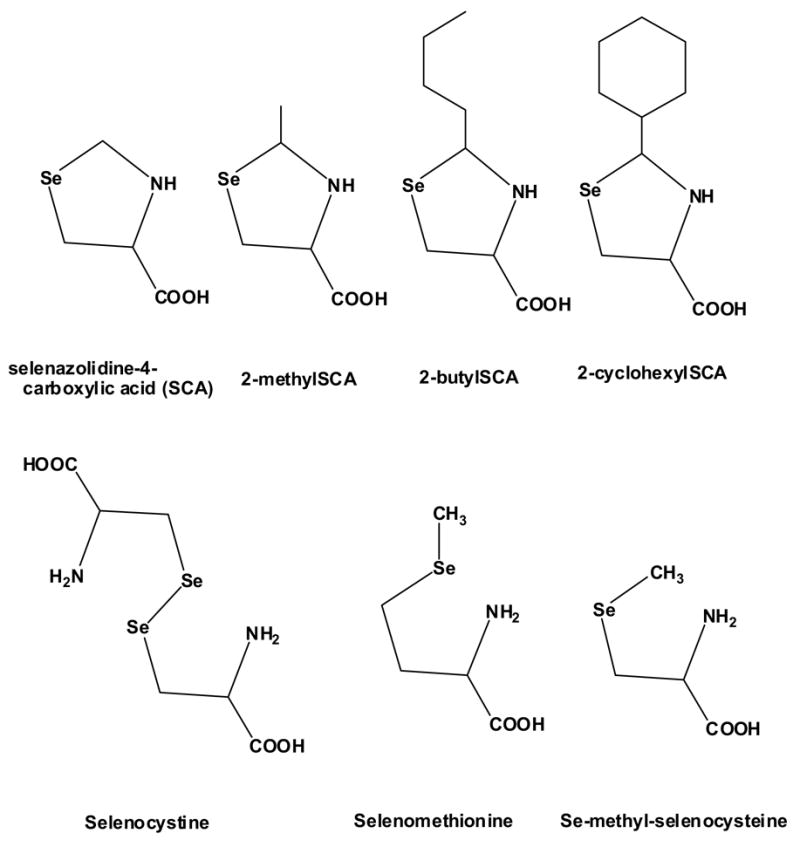
The chemical structure of organoselenium compounds investigated in Hepa1c1c7 cells in vitro.
mRNA quantification
mRNA levels were determined by Northern blotting with 20 μ g of total RNA isolated by TRIzol extraction. Gel electrophoresis, nucleic acid transfer to membranes, and 32P probe labeling were all performed as described previously (Le et al., 1996). The cDNA probe fragments for the mu- and alpha-class Gsts shared >75% homology with five out of six family members and three out of four family members, respectively, and were therefore optimal for capturing changes within the classes. The pi-class Gst probe shared 98% homology between the two members of this family. The sequences and homologies of all probes are described more fully elsewhere (El-Sayed et al., 2006). Hybridized blots were washed under high stringency conditions, and the density of the autoradiogram bands was quantified also as described previously (El-Sayed et al., 2006). All mRNA bands were normalized to the same-sample cyclophilin mRNA band and changes expressed as fold change from cells exposed only to DMSO vehicle, which was not significantly different from cells where no vehicle was added to the medium.
Statistical analysis of data
Results are expressed as the mean ± SEM of 3 biological replicates. Statistical analyses were performed using ANOVA, followed by Fisher’s protected least significant difference multiple range test. Differences were considered significant at P values of < 0.05.
RESULTS
Two hepatoma cell lines, mouse Hepa1c1c7 and rat H4IIE were initially utilized for toxicity evaluation of the selenazolidines. The rat H4IIE cell line was more sensitive to the toxic effects of selenium compounds than the mouse cell line (Figure 1). In both cell lines, the selenazolidines were less toxic than selenite which reduced viability by 50% at ~40 μ M and 100 μ M in 24h in H4IIE and Hepa1c1c7 cells, respectively. In Hepa1c1c7 cells, all the selenazolidines resulted in an enhanced cell viability at low concentrations, maximally between 25 and 50 μ M, whereas in H4IIE cells, enhanced viability was only seen with 2-cyclohexylSCA. For all selenium compounds, cell viability decreased with increasing concentration. No organoselenium compound up to 200 μ M decreased cell viability past 50% in the H4IIE cells. In the Hepa1c1c7 cells, although viability decreased below the highest enhanced level at 25–50 μ M, no organoselenium compound (up to 200 μ M) reduced cell viability below that seen in the absence of any added selenium. Using a much extended range of concentrations (not shown), IC50 values in Hepa1c1c7 cells were determined to be 550 μ M for 2-methylSCA, 610 μ M for 2-butylSCA and 2-cyclohexylSCA and >1 mM for SCA
When Hepa1c1c7 cells were exposed to the selenazolidines (100 μ M,18 hours), the mRNAs of thioredoxin reductase (Txnrd1), alpha-class glutathione transferases (Gsta), and UDP-glucuronosyltransferase (Ugt)1a6 were elevated by all selenazolidines (Figure 2; top panel). NAD(P)H-quinone oxidoreductase (Nqo1) was induced by only the three 2-alkyl selenazolidines; 2-butylSCA, 2-cyclohexylSCA, and 2-methylSCA (Figure 3; top panel). Transcripts of mu- and pi-class glutathione transferases were each induced by two selenazolidines, 2-cyclohexylSCA and 2-butylSCA. By comparison, the only change seen after exposure to the selenoamino acids; L-selenomethionine, L-selenocystine, or Se-methyl-L-selenocysteine, was an elevation of Gsta by all three and Txnrd1 by L-selenocystine and Se-methyl-L-selenocysteine (Table 1). These compounds were not investigated further.
Figure 2.
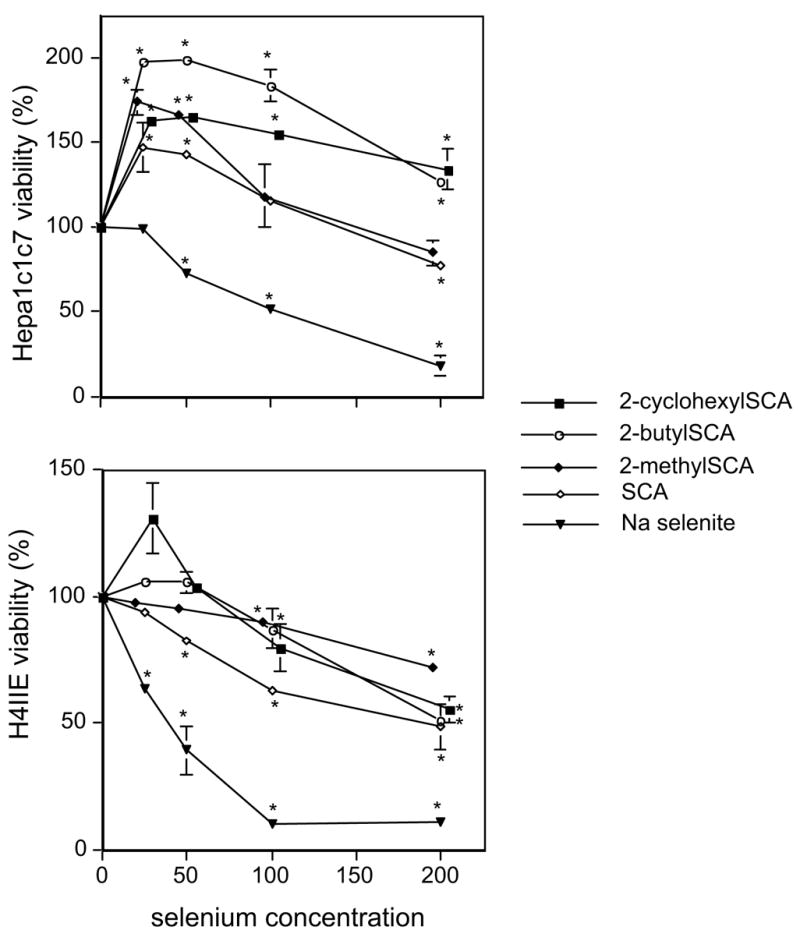
Effect of 24-hour exposure to selenazolidines and selenite on the viability of mouse (Hepa1c1c7) and rat (H4IIE) hepatoma cells.
Data are the mean of three replicates and error bars are ± SEM. 2-CyclohexylSCA and 2-methylSCA concentration values have been slightly displaced laterally for clarity.
* statistically different ( p< 0.05) from no-selenium, vehicle (DMSO) control.
Figure 3.
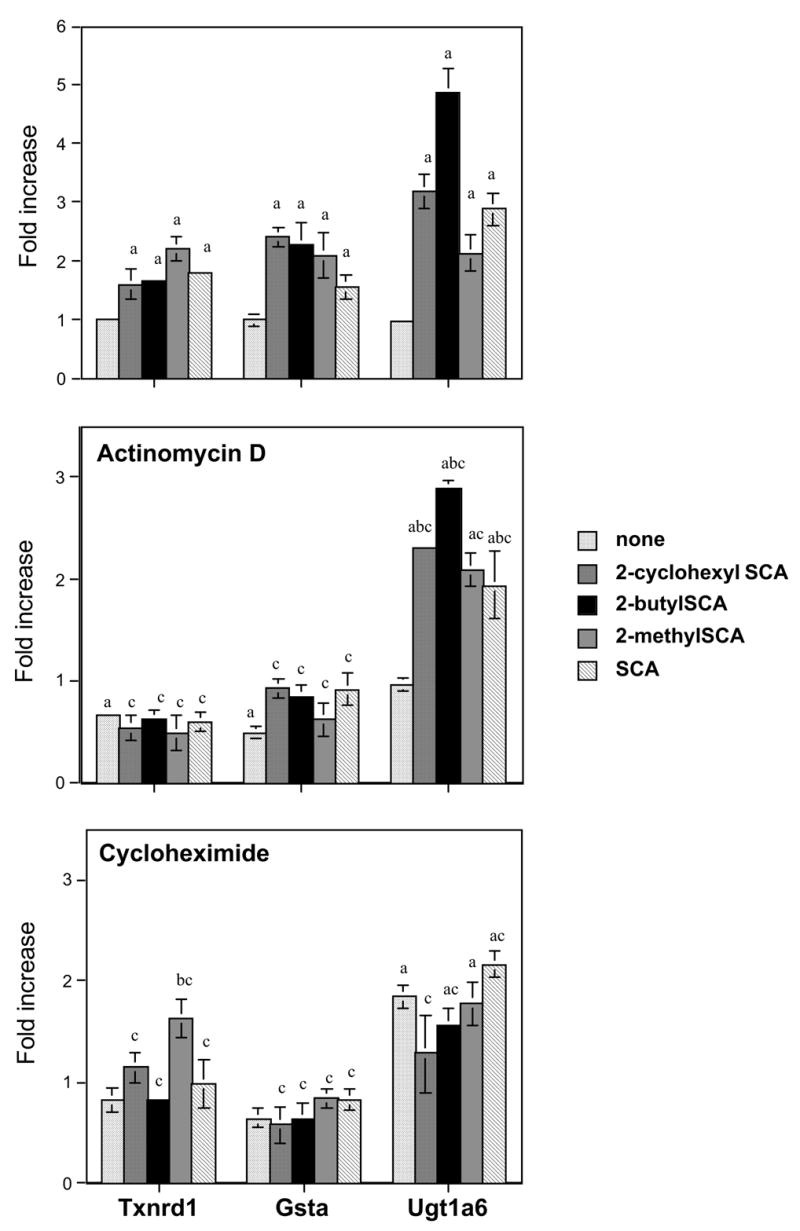
Effect of 18-hour exposure of Hepa1c1c7 cells to selenazolidines on the gene transcript levels of thioredoxin reductase (Txnrd1), alpha-class glutathione transferases (Gsta) and UDP-glucuronosyltransferase1a6 (Ugt1a6).
Data are the mean of three replicates and error bars are ± SEM. Fold increase is relative to DMSO (vehicle) controls, shown as "none" in the top panel. The center panel indicates mRNA levels in the presence of 50 nM actinomycin D and the lower panel the mRNA levels in the presence of 50 nM cycloheximide. Letters above the columns indicate significant (p<0.05) differences: 'a' from DMSO controls, 'b' from same-panel (−)-selenium (+)-inhibitor, and 'c' from values in the absence of any inhibitor (i.e., top panel).
Table 1.
Effect of selenoamino acids on chemoprotective enzymes mRNAs in Hepa1c1c7 cells.
| mRNA | L-selenocystine | L-selenomethionine | Se-methyl-L-selenocysteine |
|---|---|---|---|
| (fold change from DMSO [vehicle] controls) | |||
| Txnrd1 | 1.86 ± .14** | 0.89 ± .18 | 2.39 ± .43** |
| Gsta1/2/3* | 1.78 ± .12** | 1.87 ± .06** | 1.62 ± .28** |
| Ugt1a6 | 1.12 ± .21 | 0.53 ± .13 | 1.53 ± .36 |
| Nqo1 | 1.53 ± .26 | 0.71 ± .18 | 1.37 ± .24 |
| Gstm1/2/3/5/6* | 0.82 ± .21 | 0.80 ± .16 | 0.98 ± .40 |
| Gstp1/2* (2.4kb) | 0.62 ± .08 | 0.51 ± .17 | 0.91 ± .26 |
| Gstp1/2* (1.2kb) | 1.19 ± .26 | 1.24 ± .09 | 1.92 ± .86 |
members with >75% homology to Northern blot probes utilized. Data are the mean of three replicates ± SEM
statistically significant elevations from vehicle (DMSO) controls
Mechanistic studies were performed using the transcription inhibitor actinomycin-D and the translation inhibitor cycloheximide. Both agents carry some toxicity burden at concentrations required to exert their inhibitory effect. In preliminary range-finding experiments, it was established that 50 nM was an acceptable concentration. The 19-hour viability of the cells at this concentration, compared to cells in the absence of inhibitor, was 90% for cycloheximide and 81% for actinomycin-D. For the enzymes under study, actinomycin-D at 50 nM caused statistically significant reductions in all gene transcripts with the exception of Ugt1a6 (Figures 2 and 3; center panels). For those showing reductions, the decreases ranged from 34% (Txnrd1) to 64 and 74% for the two Gstp transcripts. Cycloheximide decreased the level of Gstm and the 1.2kb Gstp transcripts (Figure 3; lower panel), and increased the transcript level of Ugt1a6 (Figure 2; lower panel). Other apparent decreases by cycloheximide were not statistically significant.
In the presence of actinomycin-D, the responses (elevations) of Txnrd1 and Gsta mRNAs to all selenazolidines, were eliminated (Figure 2; center panel) while the elevations in Ugt1a6 mRNA, although reduced in magnitude for all except with 2-methylSCA treatment, remained present. The elevations in Gstm and Gstp transcripts by 2-cyclohexylSCA and 2-butylSCA were also eliminated in the presence of actinomycin D (Figure 3; center panel), as were the elevations of Nqo1 by 2-cyclohexylSCA, 2-butylSCA, and 2-methylSCA.
In the presence of cycloheximide and selenazolidines, all the transcript elevations seen with the selenazolidines alone were either eliminated for most or significantly reduced (2-methylSCA induction of Txnrd1), with the exception of Ugt1a6 (Figures 2 and 3; lower panels). For Ugt1a6, the elevations by all selenazolidines except 2-methylSCA were significantly reduced from changes seen in the absence of cycloheximide, although for all except 2-cyclohexylSCA, were still significantly elevated from cells not exposed to any selenazolidine or inhibitor. However, because cycloheximide alone elicited an almost doubling of the transcript levels of Ugt1a6, the transcript levels with selenazolidines in addition were not significantly elevated above that.
DISCUSSION
In both rodent hepatoma cell lines investigated, all the selenazolidines were less toxic than selenite. This differential concurs with studies in Chinese hamster lung fibroblasts (V79) where IC50 values for selenite, 2-methylSCA, and SCA were 17, 79, and 317 μ M, respectively (Short et al., 2003). With IC values of 550 μ M and >1mM for 2-methylSCA and SCA respectively in Hepa 1c1c7cells, hepatoma cells appear more resistant to selenocompound toxicity than fibroblasts. For all of the selenazolidines, the murine hepatoma IC50 values are much in excess of the 100 μ M of selenite, supporting the general observations that organoselenium compounds are less toxic than inorganic selenium compounds. The low toxicity of the 2-alkyl selenazolidines contrasts with 1,4 phenylenebis(methylene)selenocyanate, another chemopreventive agent, which in the V79 cell study was of similar toxicity to selenite. In addition to toxicity, selenium appears to have growth- or viability-promoting properties at low concentrations. This phenomenon, seen here with hepatoma cells, has also been seen with bovine endothelial cells (Wei et al., 1986), bovine granulosa cells (Basini and Tamanini, 2000), and gastric adenocarcinoma cells (Verma et al., 2004).
All four of the selenazolidines investigated and also L-selenocystine and Se-methyl-L-selenocysteine increased Txnrd1 transcript levels. Inorganic selenium-dependent elevations of thioredoxin reductase mRNA had previously been observed in human hepatoma (HepG2; Morgan et al., 2002) but not in mouse fibrosarcoma (WEHI 164; Reszka et al., 2005) cell lines. Organoselenium compound effects on thioredoxin reductase in cell lines at the transcript level have not been reported. Both organoselenium and selenite studies have investigated enzyme activity changes, however. Increased thioredoxin reductase activity following exposure to selenomethionine, Se-methylselenocysteine and selenite was elicited in HT-29 human colon cancer cells (Berggren et al., 1997) and the selenite increases in thioredoxin reductase activity (and mRNA levels) were also observable in MCF-7 breast cancer and A549 lung cancer cells but not in Jurkat and HL-60 leukemia cells (Gallegos et al., 1997). Selenite-dependent increases in thioredoxin reductase activity were subsequently demonstrated to occur in both human HepG2 and mouse Hepa1c1c7 hepatoma cell lines (Hintze et al., 2003a, Hintze et al., 2003b, Zhang et al., 2003) and in the human endothelial-like immortalized cell line (EAhy926; Lewin et al., 2002). From our Hepa1c1c7 studies with actinomycin D, transcriptional upregulation is a mechanism that could contribute to the increases in activity seen for this selenoprotein, and from the influence of cycloheximide, requires ongoing protein synthesis for the transcriptional upregulation to occur.
Increased mRNA levels of alpha-class and pi-class glutathione transferases following exposure to Se-allylselenocysteine were observed in rat H35 Reuber rat hepatoma cells (t'Hoen et al., 2002). Our observations in Hepa1c1c7 cells add Se-methylselenocysteine, selenocystine, selenomethionine, and four selenazolidines to the list of inducers of Gsta in hepatoma cells in vitro and two selenazolidines to the list of Gstp inducers. The current study demonstrates that Gstm is also inducible by organoselenium compounds, in this case, by the same compounds able to induce Gstp. All the Gst mRNA elevations appear dependent on transcriptional upregulation, and by a mechanism that requires ongoing protein synthesis. We have not investigated whether increased levels of Gst mRNAs result in elevations in enzyme activity but Lee et al., (1999), investigating 1,4-phenylenebis(methylene)selenocyanate effects in mouse Hepa1c1c7 cells were unable to observe any increase in glutathione transferase (CDNB) activity.
Selenium compound effects on NQO and UGTs in in vitro systems appear to have been the subject of one reported investigation to date (Zhao et al., 2004). In vivo, in rat, hepatic NQO activity was increased by 1,4-phenylenebis(methylene)selenocyanate (Tanaka et al., 1997), and hepatic UGT activity was increased by 1,2-, 1,3-, and 1,4-phenylenebis(methylene)selenocyanate (Sohn et al., 1999). UGT activity was not elevated by 1,4-phenylenebis(methylene)selenocyanate in mouse, however (Prokopczyk et al., 2000). Our present studies in hepatoma cells demonstrate that mouse Nqo l is responsive to some selenocompounds (three 2-alkyl selenazolidines) but not others (SCA, L-selenocystine, L-selenomethionine, Se-methyl-L-selenocysteine). Zhao et al., ( 2004) reported NQO1 mRNA and activity induction by methylseleninic acid in a human prostate cancer cell line. The induction by the 2-alkyl selenazolidines is transcription-dependent and also requires ongoing protein synthesis. The Ugt1a6 transcript level was elevated by all four selenazolidines, but not any of the selenoamino acids. The mechanism behind this mRNA elevation is unclear. Some elevation may arise from increased transcription since for all selenazolidines except 2-methylSCA, the magnitude of the increase was significantly diminished in the presence of actinomycin D. That some elevation was still evident would suggest that additional mechanisms are operating, however, possibly mRNA stabilization. Such a mechanism has been implicated in maintaining hepatic glutathione peroxidase mRNA levels in a selenium-sufficient as compared to a selenium-deficient diet in rats (Christensen and Burgener, 1992). The elevation seen in the Ugt1a6 transcript with cycloheximide present provides evidence of additional unusual features in the regulation of this gene; in this case, the possibility of the presence of an inhibitory transcription factor.
Explanations as to why the SCAs induce in the same manner as selenocystine, but with other responses in addition, can only be speculated upon. It could relate to the different chemical nature of the compound added to the cells (Figure 1), or to the biochemical redox consequences of generating selenocysteine, the common point of convergence. The formation of selenocysteine from the spontaneous hydrolysis of 2-alkylSCAs and proline dehydrogenase action on SCA carries no redox burden while generation from selenocystine requires consumption of cellular reducing equivalents, (glutathione) which in some manner might impede enzyme induction. Against this explanation is the equivalency of the pattern of induction of selenocystine and Se-methylselenocysteine (Table 1) despite that only the former depletes glutathione: the latter compound is likely processed in the hepatoma cell in much the same manner as in kidney cells by cysteine conjugate β -lyase/glutamine transaminase K (Commandeur, et al., 2000). The β -elimination reaction, and hence the absence of any selenocysteine formation from Se-methylselenocysteine adds additional support to the idea that the inductions elicited by all the selenium compounds are in response to the parent compound to which the cells are initially exposed. Alternatively it could be a response to a common late-stage metabolite, such as methylselenol, that is capable of being formed to varying extents and at various rates from all the compounds investigated.
Rodent tumor models continue to be mainstay for the evaluation of chemopreventive compounds. These test systems are time-consuming and expensive so a means of pre-selecting only the most promising compounds for evaluation is desirable. Chemopreventive compound effects on the regulation of selected enzymes are often used as a key indication of potential efficacy, and in vitro cell-based systems can be an ideal system in which to evaluate these effects. The present study demonstrates that in the study of chemoprotective enzymes, and their regulation by organoselenium compounds, the Hepa1c1c7 cell line appears to be a responsive system that is able to differentiate between compounds and at concentrations where the cell line is relatively insensitive to any overt cytotoxic effects.
Figure 4.
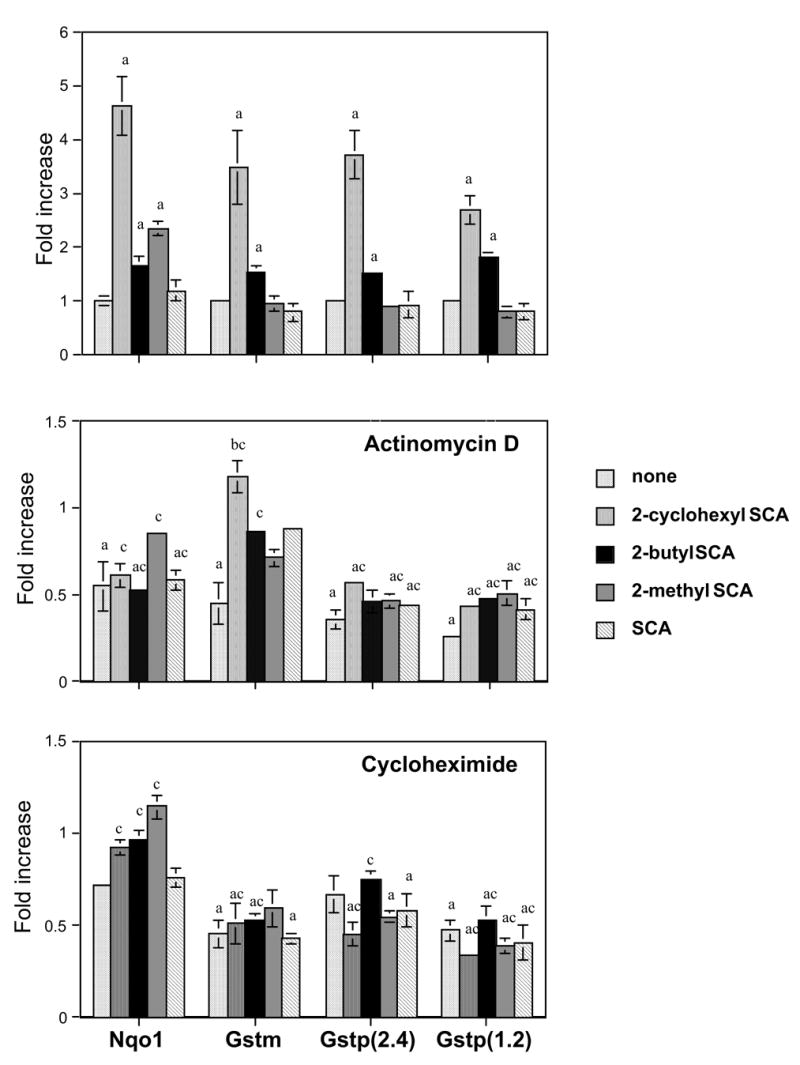
Effect of 18-hour exposure of Hepa1c1c7 cells to selenazolidines on the gene transcript levels of NAD(P)H quinone oxidoreductase (Nqo1), mu-class glutathione transferases (Gstm) and 2.4 kb and 1.2kb transcripts of pi-class glutathione transferases (Gstp).
Data are the mean of three replicates and error bars are ± SEM. Fold increase is relative to DMSO (vehicle) controls, shown as "none" in the top panel. The center panel indicates mRNA levels in the presence of 50 nM actinomycin D and the lower panel the mRNA levels in the presence of 50 nM cycloheximide. Letters above the columns indicate significant (p<0.05) differences: 'a' from DMSO controls, 'b' from same-panel (−)-selenium (+)-inhibitor, and 'c' from values in the absence of any inhibitor (i.e., top panel).
Acknowledgments
This project was supported by a USPHS Grant GM 058913.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basini G, Tamanini C. Selenium stimulates estradiol production in bovine granulosa cells: possible involvement of nitric oxide. Domest Anim Endocrinol. 2000;18:1–17. doi: 10.1016/s0739-7240(99)00059-4. [DOI] [PubMed] [Google Scholar]
- Berggren M, Gallegos A, Gasdaska J, Powis G. Cellular thioredoxin reductase activity is regulated by selenium. Anticancer Res. 1997;17:3377–3380. [PubMed] [Google Scholar]
- Christensen MJ, Burgener KW. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J Nutr. 1992;122:1620–1626. doi: 10.1093/jn/122.8.1620. [DOI] [PubMed] [Google Scholar]
- Commandeur JN, Andreadou I, Rooseboom M, Out M, de Leur LJ, Groot E, Vermeulen NP. Bioactivation of selenocysteine Se-conjugates by a highly purified rat renal cysteine conjugate beta-lyase/glutamine transaminase K. J Pharmacol Exp Ther. 2000;294:753–761. [PubMed] [Google Scholar]
- Das RK, Bhattacharya S. Anti-tumour promoting activity of diphenylmethyl selenocyanate against two-stage mouse skin carcinogenesis. Asian Pac J Cancer. 2005;6:181–188. [PubMed] [Google Scholar]
- El-Bayoumy K, Das A, Narayanan B, Narayanan N, Fiala ES, Desai D, Rao CV, Amin S, Sinha R. Molecular targets of the chemopreventive agent 1,4-phenylenebis(methylene) selenocyanate in human non small cell lung cancer. Carcinogenesis. 2006;27:1369–1376. doi: 10.1093/carcin/bgi328. [DOI] [PubMed] [Google Scholar]
- El-Sayed WM, Aboul-Fadl T, Lamb J, Roberts JC, Franklin MR. Acute effects of novel selenazolidines on murine chemoprotective enzymes. Chem Biol Interact. 2006 doi: 10.1016/j.cbi.2006.05.002. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Gallegos A, Berggren M, Gaskaska JR, Powis G. Mechanisms of the regulation of thioredoxin reductase activity in cancer cells by the chemopreventive agent selenium. Cancer Res. 1997;57:4965–4970. [PubMed] [Google Scholar]
- Ghosh S, Das RK, Sengupta A, Bhattacharya S. Inhibition of azoxymethane-induced aberrant crypt foci in rat by diphenylmethyl selenocyanate through downregulation of COX-2 and modulation of glutathione transferase and lipid peroxidation. Biol Trace Elem Res. 2005;105:171–185. doi: 10.1385/BTER:105:1-3:171. [DOI] [PubMed] [Google Scholar]
- Helmy MH, Ismail SS, Fayed H, El-Bassiouni EA. Effect of selenium supplementation on the activities of glutathione metabolizing enzymes in human hepatoma Hep G2 cell line. Toxicology. 2000;144:57–61. doi: 10.1016/s0300-483x(99)00190-0. [DOI] [PubMed] [Google Scholar]
- Hintze KJ, Keck AS, Finley JW, Jeffery EH. Induction of hepatic thioredoxin reductase activity by sulforaphane, both in Hepa1c1c7 cells and in male Fisher 344 rats. J Nutr Biochem. 2003a;14:173–179. doi: 10.1016/s0955-2863(02)00282-6. [DOI] [PubMed] [Google Scholar]
- Hintze KJ, Wald KA, Zeng H, Jeffery EH, Finley JW. Thioredoxin reductase in human hepatoma cells is transcriptionally regulated by sulforaphane and other electrophiles via an antioxidant response element. J Nutr. 2003b;133:2721–2727. doi: 10.1093/jn/133.9.2721. [DOI] [PubMed] [Google Scholar]
- Kawamori T, El-Bayoumy K, Ji BY, Rodriguez JG, Rao CV, Reddy BS. Evaluation of benzyl selenocyanate glutathione conjugate for potential chemopreventive properties in colon carcinogenesis. Int J Oncol. 1998;13:29–34. doi: 10.3892/ijo.13.1.29. [DOI] [PubMed] [Google Scholar]
- Le HT, Lamb JG, Franklin MR. Drug metabolizing enzyme induction by benzoquinolines, acridine, and quinacrine; tricyclic aromatic molecules containing a single heterocyclic nitrogen. J Biochem Toxicol. 1996;11:297–303. doi: 10.1002/(SICI)1522-7146(1996)11:6<297::AID-JBT5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Lee SK, Song L, Mata-Greenwood E, Kelloff GJ, Steele VE, Pezzuto JM. Modulation of in vitro biomarkers of the carcinogenic process by chemopreventive agents. Anticancer Res. 1999;19:35–44. [PubMed] [Google Scholar]
- Lewin MH, Arthur JR, Riemersma RA, Nicol F, Walker SW, Millar EM, Howie AF, Beckett GJ. Selenium supplementation acting through the induction of thioredoxin reductase and glutathione peroxidase protects the human endothelial cell line EAhy926 from damage by lipid hydroperoxides. Biochim Biophys Acta. 2002;1593:85–92. doi: 10.1016/s0167-4889(02)00333-6. [DOI] [PubMed] [Google Scholar]
- Morgan KT, Ni H, Brown HR, Yoon L, Qualls CW, Jr, Crosby LM, Reynolds R, Gaskill B, Anderson SP, Kepler TB, Brainard T, Liv N, Easton M, Merrill C, Creech D, Sprenger D, Conner G, Johnson PR, Fox T, Sartor M, Richard E, Kuruvilla S, Casey W, Benavides G. Application of cDNA microarray technology to in vitro toxicology and the selection of genes for a real-time RT-PCR-based screen for oxidative stress in Hep-G2 cells. Toxicol Pathol. 2002;30:435–451. doi: 10.1080/01926230290105613. [DOI] [PubMed] [Google Scholar]
- Prokopczyk B, Rosa JG, Desai D, Amin D, Sohn OS, Fiala ES, El-Bayoumy K. Chemoprevention of lung tumorigenesis induced by a mixture of benzo(a)pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone by the organoselenium compound 1,4-phenylenebis(methylene)selenocyanate. Cancer Lett. 2000;161:35–46. doi: 10.1016/s0304-3835(00)00590-5. [DOI] [PubMed] [Google Scholar]
- Reddy BS, Rivenson A, Kulkarni N, Upadhyaya P, El-Bayoumy K. Chemoprevention of colon carcinogenesis by the synthetic organoselenium compound 1,4-phenylenebis(methylene) selenocyanate. Cancer Res. 1992;52:5635–5640. [PubMed] [Google Scholar]
- Reszka E, Gromadzinska J, Stanczyk M, Wasowicz W. Effect of selenium on expression of selenoproteins in mouse fibrosarcoma cells. Biol Trace Elem Res. 2005;104:165–172. doi: 10.1385/BTER:104:2:165. [DOI] [PubMed] [Google Scholar]
- Shimada T, El-Bayoumy K, Upadhyaya P, Sutter TR, Guengerich FP, Yamazaki H. Inhibition of human cytochrome P450-catalyzed oxidations of xenobiotics and procarcinogens by synthetic organoselenium compounds. Cancer Res. 1997;57:4757–4764. [PubMed] [Google Scholar]
- Short MD, Xie Y, Li L, Cassidy PB, Roberts JC. Characteristics of selenazolidine prodrugs of selenocysteine: toxicity and glutathione peroxidase induction in V79 cells. J Med Chem. 2003;46:3308–3313. doi: 10.1021/jm020496q. [DOI] [PubMed] [Google Scholar]
- Sohn OS, Fiala ES, Upadhyaya P, Chae YH, El-Bayoumy K. Comparative effects of phenylenebis(methylene)selenocyanate isomers on xenobiotic metabolizing enzymes in organs of female CD rats. Carcinogenesis. 1999;20:615–621. doi: 10.1093/carcin/20.4.615. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Makita H, Kawabata K, Mori H, El-Bayoumy K. 1,4-Phenylenebis(methylene) selenocyanate exerts exceptional chemopreventive activity in rat tongue carcinogenesis. Cancer Res. 1997;57:3644–3648. [PubMed] [Google Scholar]
- t’Hoen PA, Rooseboom M, Bijsterbosch MK, van Berkel TJ, Vermeulen NP, Commandeur JN. Induction of glutathione S-transferase mRNA levels by chemopreventive selenocysteine Se-conjugates. Biochem Pharmacol. 2002;63:1843–1849. doi: 10.1016/s0006-2952(02)00987-5. [DOI] [PubMed] [Google Scholar]
- Verma A, Atten MJ, Attar BM, Holian O. Selenomethionine stimulates MAPK (ERK) phosphorylation, protein oxidation, and DNA synthesis in gastric cancer cells. Nutr Cancer. 2004;49:184–190. doi: 10.1207/s15327914nc4902_10. [DOI] [PubMed] [Google Scholar]
- Wei X, Wright GC, Jr, Sokoloff L. The effect of sodium selenite on chondrocytes in monolayer culture. Arthritis Rheum. 1986;29:660–664. doi: 10.1002/art.1780290511. [DOI] [PubMed] [Google Scholar]
- Xie Y, Short MD, Cassidy PB, Roberts JC. Selenazolidines as novel organoselenium delivery agents. Bioorg Med Chem Lett. 2001;11:2911–2915. doi: 10.1016/s0960-894x(01)00590-x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Svehlikova V, Bao Y, Howie AF, Beckett GJ, Williamson G. Synergy between sulforaphane and selenium in the induction of thioredoxin reductase 1 requires both transcriptional and translational modulation. Carcinogenesis. 2003;24:497–503. doi: 10.1093/carcin/24.3.497. [DOI] [PubMed] [Google Scholar]
- Zhao H, Whitfield ML, Xu T, Botstein D, Brooks JD. Diverse effects of methylselenininc acid on the transcriptional program of human prostate cancer cells. Mol Biol Cell. 2004;15:506–519. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]


